Abstract
Arkadia was originally identified as a protein that enhances signalling activity of Nodal and induces mammalian nodes during early embryogenesis; however, the mechanisms by which Arkadia affects transforming growth factor-β (TGF-β) superfamily signalling have not been determined. Here we show that Arkadia is widely expressed in mammalian tissues, and that it enhances both TGF-β and bone morphogenetic protein (BMP) signalling. Arkadia physically interacts with inhibitory Smad, Smad7, and induces its poly-ubiquitination and degradation. In contrast to Smurf1, which interacts with TGF-β receptor complexes through Smad7 and degrades them, Arkadia fails to associate with TGF-β receptors. In contrast to Smad7, expression of Arkadia is down-regulated by TGF-β. Silencing of the Arkadia gene resulted in repression of transcriptional activities induced by TGF-β and BMP, and accumulation of the Smad7 protein. Arkadia may thus play an important role as an amplifier of TGF-β superfamily signalling under both physiological and pathological conditions.
Keywords: BMP/signal transduction/Smad/TGF-β/ubiquitination
Introduction
Ubiquitin-dependent protein degradation plays critical roles in a wide variety of biological processes, including signal transduction, cell cycle progression and transcriptional regulation (Hershko and Ciechanover, 1998). Ubiquitination of proteins is induced by three distinct enzymes: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases. Proteins poly-ubiquitinated by these enzymes are subsequently degraded by 26S proteasomes. In the ubiquitin-proteasome pathways, E3 ubiquitin ligases play key roles in the recognition of target proteins and degradation by 26S proteasomes. Of E3 ligases in mammals, RING finger type and HECT type have been well characterized (Laney and Hochstrasser, 1999).
Transforming growth factor-β (TGF-β) is a member of a large family of structurally related cytokines, including activins, Nodal, and bone morphogenetic proteins (BMPs). TGF-β superfamily cytokines bind to type II and type I serine/threonine kinase receptors, and transmit signals through Smad proteins (Heldin et al., 1997; Derynck and Zhang, 2003; Shi and Massague, 2003). Receptor-regulated Smads (R-Smads) are directly activated by type I receptor kinases, form heteromeric complexes with common-partner Smads (Co-Smads) and regulate transcription of target genes in the nucleus. TGF-β, activin and Nodal bind to their corresponding receptors and activate Smads 2 and 3, whereas BMPs and anti-Müllerian hormone phosphorylate Smads 1, 5 and 8 (Miyazawa et al., 2002). In contrast, inhibitory Smads (I-Smads) repress TGF-β superfamily signalling through multiple mechanisms, including interaction with activated type I receptors and prevention of R-Smad phosphorylation (Hanyu et al., 2001; Murakami et al., 2003).
Ubiquitin-proteasome pathways play critical roles in regulation of TGF-β superfamily signalling. R-Smads, including Smad2 and Smad3, are degraded by ubiquitin-proteasome pathways in a ligand-dependent manner, resulting in termination of TGF-β signalling (Lo and Massagué, 1999). Smad3 activated by TGF-β is degraded by ROC1–SCF complexes in the nucleus (Fukuchi et al., 2001). Jab1 has recently been shown to interact directly with Co-Smad, Smad4, and to induce its ubiquitin-dependent degradation (Wan et al., 2002).
Smurf1 and Smurf2 are HECT type E3 ligases and contain WW domains, which bind to PY motifs in target proteins (Zhu et al., 1999; Lin et al., 2000; Zhang et al., 2001). Smurfs interact with certain R-Smads, including Smad1 and Smad5, and induce their degradation in a ligand-independent manner; Smurfs have therefore been suggested to limit the intracellular pools of R-Smads in order to regulate the magnitude of TGF-β superfamily signalling. In addition, Smurfs bind to I-Smads, Smad6 and Smad7. They stimulate export of I-Smads from the nucleus to the cytoplasm, recruit I-Smads to TGF-β and BMP receptors, and induce degradation of these receptors (Kavsak et al., 2000; Ebisawa et al., 2001; Murakami et al., 2003). Thus, Smurfs support inhibitory activities of I-Smads in TGF-β superfamily signalling pathways.
Arkadia was originally isolated through gene-trap insertion mutagenesis in mice, and was shown to be an intracellular protein that is essential for induction of mammalian nodes (Episkopou et al., 2001). Arkadia is similar to no known proteins; however, since it contains a RING finger domain in its C-terminal region, Arkadia has been suggested to act as an E3 ubiquitin ligase. Genetic analyses by mating heterozygous Arkadia and Nodal mice revealed that Arkadia and Nodal are involved in the same signalling pathways, and that Arkadia acts through Nodal in the induction of nodes. Using Xenopus assays, Niederlander et al. (2001) demonstrated that Arkadia modulates signalling activities of Nodal, leading to the induction of Spemann’s organizer in Xenopus embryos.
Here, we examined the mechanisms of action of Arkadia in the regulation of TGF-β superfamily signalling. Arkadia enhanced TGF-β as well as BMP signalling. It physically interacted with Smad7, and induced degradation of Smad7, but not that of TGF-β receptors. Thus, although both Smurfs and Arkadia physically interact with Smad7, they have opposite functions in regulation of TGF-β superfamily signalling. Episkopou et al. (2001) reported that Arkadia is broadly expressed in mouse embryos. In addition to embryonic tissues, we show that Arkadia is expressed ubiquitously in adult tissues, and that its expression is repressed by TGF-β. Thus, Arkadia may fine-tune signalling activities of the TGF-β superfamily cytokines under various physiological and pathological conditions.
Results
Arkadia enhances TGF-β superfamily signalling
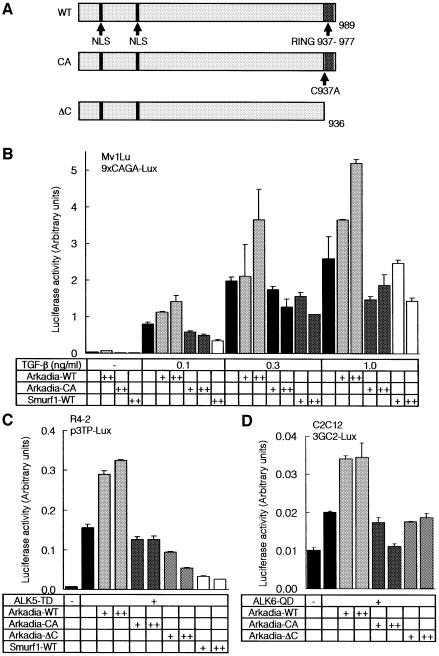
We prepared a mouse Arkadia construct with a mutation at the C-terminal RING finger domain (Arkadia-CA), in which Cys937 was replaced with alanine, resulting in a lack of E3 ubiquitin ligase activity. We also generated a mutant with truncation of the C-terminal 53 amino acid residues, including the RING finger domain (Arkadia-ΔC) (Figure 1A).
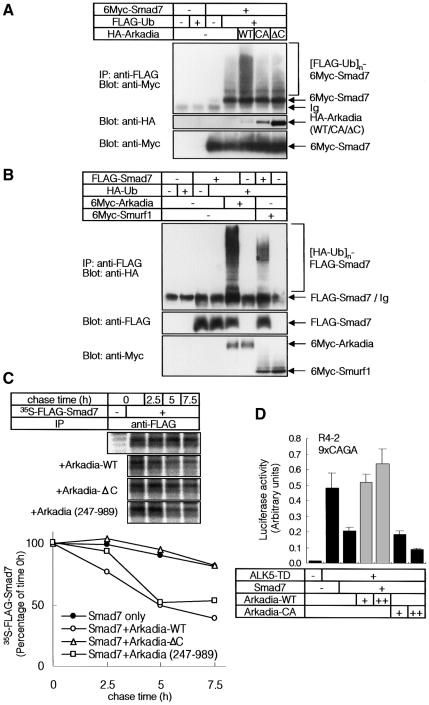
Fig. 1. Arkadia enhances transcriptional activities of TGF-β superfamily signalling. (A) Schematic representation of the structures of wild type (WT) Arkadia, and Arkadia-CA and ΔC mutants. (B and C) Effects of wild-type Arkadia and Arkadia mutants on the transcriptional activity of TGF-β (B) and constitutively active TβR-I (ALK5-TD) (C) were examined using 9xCAGA-Lux (B) and p3TP-lux assays (C). Wild-type (B) or R mutant (R4-2) (C) Mv1Lu cells were co-transfected with the luciferase constructs and various combinations of indicated cDNAs. + and ++ represent 0.1 and 0.5 µg of DNAs transfected in Mv1Lu cells (B), respectively, and 0.2 and 0.5 µg of DNAs transfected in R mutant cells (C), respectively. (D) Effects of wild-type Arkadia and Arkadia mutants on the transcriptional activity of constitutively active BMP type I receptor (ALK6-QD) were examined using 3GC2-Lux assays. C2C12 cells were co-transfected with the luciferase construct and various combinations of indicated cDNAs. + and ++ represent 0.1 and 0.5 µg of DNAs, respectively, transfected in C2C12 cells.
Analyses using TGF-β-responsive promoter-reporter constructs revealed that wild-type Arkadia enhanced the transcriptional activity induced by TGF-β, or by a constitutively active TGF-β type I receptor (ALK5-TD), in a dose-dependent fashion (Figure 1B and C). In contrast, neither Arkadia-CA nor Arkadia-ΔC enhanced the transcription induced by TGF-β or ALK5-TD, and they slightly inhibited the transcriptional activities of TGF-β and ALK5-TD. These findings suggest that the C-terminal RING finger domain is important for the enhancement of TGF-β signalling by Arkadia. In contrast, Smurf1 suppressed the transcription induced by TGF-β or ALK5-TD (Figure 1B and C), in agreement with previous reports (Ebisawa et al., 2001; Murakami et al., 2003). When a BMP-responsive promoter-reporter construct, 3GC2-Lux (Ishida et al., 2000), was used, Arkadia enhanced the transcriptional activity of a constitutively active BMP type I receptor, ALK6-QD, whereas the Arkadia mutants failed to do so (Figure 1D), indicating that Arkadia regulates the signalling activities of BMP as well as those of TGF-β.
Arkadia physically interacts with I-Smads
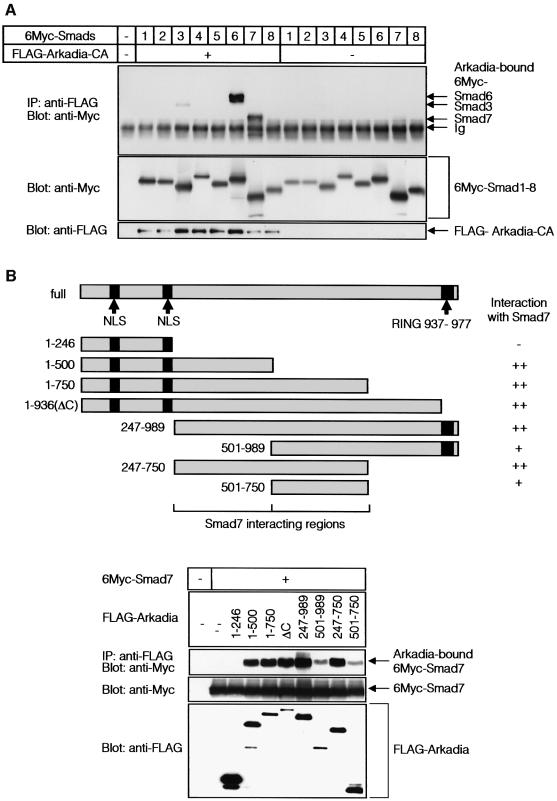
In order to elucidate the mechanisms of enhancement of TGF-β superfamily signalling by Arkadia, we examined whether Arkadia physically interacts with Smads. Arkadia-CA was used in this experiment, since wild-type Arkadia may induce rapid degradation of Smads when they bind to each other. When COS7 cells were transfected with FLAG-Arkadia-CA and 6Myc-tagged Smads, and their interaction was determined by immunoprecipitation of Arkadia followed by immunoblotting of Smads, Arkadia-CA interacted strongly with I-Smads, Smad6 and Smad7 (Figure 2A). Arkadia-CA also interacted with Smad3 weakly, but not with other Smads.
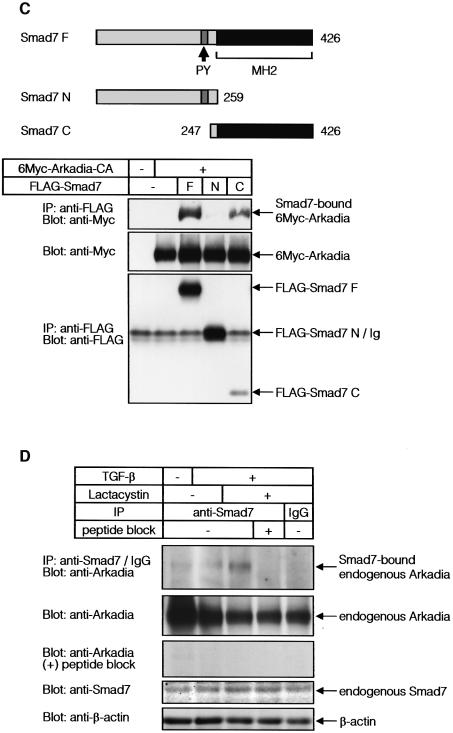
Fig. 2. Arkadia binds to I-Smads. (A) Interaction between Smads and Arkadia. COS7 cells transfected with or without FLAG-Arkadia and 6Myc-tagged Smads were subjected to FLAG-immunoprecipitation (IP) followed by Myc-immunoblotting (Blot). The top panel shows the interaction and the lower two panels show the expression of each protein as indicated. (B and C) Regions responsible for the interaction between Smads and Arkadia. Structures of deletion mutants of Arkadia (B) and Smad7 (C) are shown in the upper figure parts. Transfected COS7 cells were subjected to FLAG- immunoprecipitation followed by Myc-immunoblotting. Cell were treated with 2.5 µM lactacystin in (B). (D) Interaction between endogenous Smad7 and Arkadia was examined in HaCaT cells treated or not with TGF-β (1 ng/ml) for 8 h, and 10 µM lactacystin was added during the last 3 h, where indicated. Cells were subjected to immunoprecipitation using Smad7 antibody or control IgG, followed by immunoblotting using anti-Arkadia antibody. The Smad7 peptide used for immunization of a goat was used in the fourth lane from the left. The top panel shows the interaction and the lower three panels the expression of each protein, as indicated.
We then studied the interaction between Arkadia and Smad7 in more detail. Arkadia appeared to interact with Smad7 through multiple regions; deletion mutants of Arkadia containing amino acids 247–500 and those containing amino acids 501–750, interacted with Smad7 (Figure 2B), although the interaction of the latter was weaker than that of the former. Of the regions in Smad7, the C-terminal MH2 domain, but not the N-terminal region, interacted with Arkadia (Figure 2C).
Interaction of endogenous Arkadia and Smad7 proteins was examined in HaCaT keratinocytes, which express Arkadia (see Figures 5A and 6A). These cells were treated with TGF-β to induce expression of Smad7. Smad7 immunoprecipitation followed by Arkadia immunoblotting revealed that endogenous Arkadia was only weakly co-immunoprecipitated with Smad7, probably because Smad7 bound to Arkadia was rapidly degraded. When the cells were treated with a proteasomal inhibitor, lactacystin, endogenous Arkadia was found to be efficiently co-immunoprecipitated with Smad7 (Figure 2D).
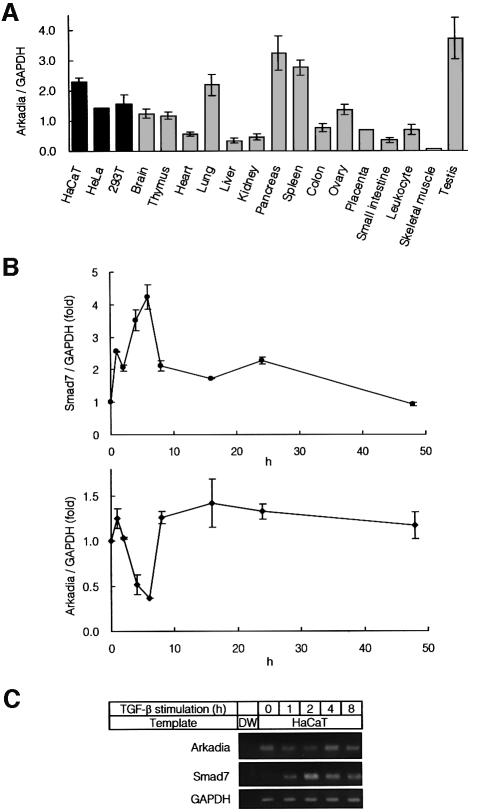
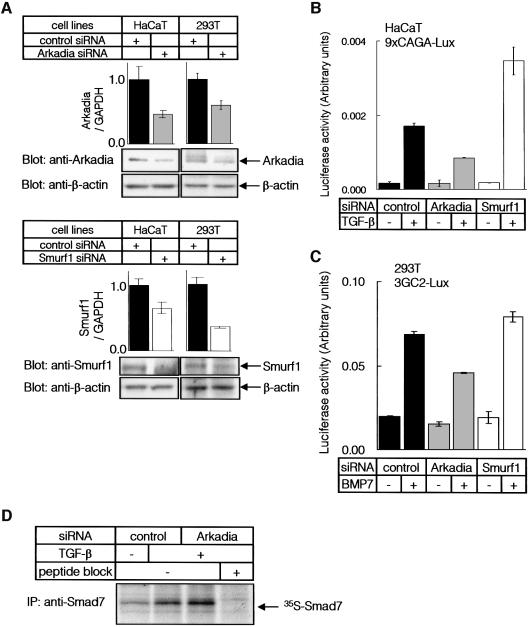
Fig. 5. Expression of Arkadia in various tissues. (A) Quantitative real-time PCR analyses using human cell lines and human adult tissues. Expression level of Arkadia in each tissue was normalized by that of GAPDH. (B) Quantitative real-time PCR of HaCaT cells treated with 1 ng/ml of TGF-β. Expression of Smad7 and Arkadia was examined. (C) RT–PCR analysis of expression of Arkadia and Smad7. HaCaT cells were treated as described in (B), and RNA samples were analysed by RT–PCR. Distilled water (DW) was used as a control.
Fig. 6. Knock-down of Arkadia and Smurf1 in mammalian cells. (A) HaCaT and 293T cells were transfected with control, Arkadia or Smurf1 siRNA, and expression levels of Arkadia or Smurf1 RNAs were determined by quantitative real-time PCR analysis (top panels). Expression levels of Arkadia or Smurf1 proteins were determined by immunoblot analysis (lower panels). Expression of β-actin was determined as a loading control. (B and C) HaCaT (B) or 293T cells (C) were transfected with siRNAs and promoter–reporter constructs as indicated. Cells were treated with TGF-β (0.3 ng/ml) (B) or BMP7 (500 ng/ml) (C) for 24 h, and luciferase activities were measured as in Figure 1B. (D) Accumulation of Smad7 protein by knock-down of the Arkadia gene. HaCaT cells were transfected or not with Arkadia siRNA, and treated with 1 ng/ml TGF-β for 7.5 h. Endogenous Smad7 protein was immunoprecipitated after metabolic labelling of the cells. Cell lysates from equal numbers of cells were applied to each lane, and analysed by SDS–PAGE followed by autoradiography. Experiments were repeated with essentially the same results.
Arkadia induces poly-ubiquitination of Smad7
Since the C-terminal RING finger domain is critical for the enhancement of TGF-β superfamily signalling by Arkadia, we examined whether Arkadia induces poly-ubiquitination and degradation of Smads. We found that Arkadia strongly induced poly-ubiquitination of Smad7, but only weakly induced that of Smad2 and Smad3 in transfected cells (data not shown). Ubiquitination of Smad7 was observed when wild-type Arkadia, but not the Arkadia mutants, were used (Figure 3A), suggesting critical roles of the C-terminal RING finger domain of Arkadia in Smad7 ubiquitination. Although Smurf1 induced ubiquitination of Smad7, as previously reported (Ebisawa et al., 2001), Arkadia was much more potent than Smurf1 in doing so (Figure 3B).
Fig. 3. Arkadia induces ubiquitin-dependent degradation of Smad7. (A) Arkadia induces ubiquitination of Smad7. 293T cells were transfected with the indicated plasmids, and treated with 2.5 µM of lactacystin for 24 h before cell lysis. Lysates from cells were subjected to anti-FLAG immunoprecipitation followed by anti-Myc immunoblotting. Poly-ubiquitination species of Smad7 ([FLAG-Ub]n-6Myc-Smad7) are indicated in the top panel. (B) Arkadia was more potent than Smurf1 in ubiquitination of Smad7. 293T cells were treated as in (A). Lysates from cells were subjected to anti-FLAG immunoprecipitation followed by anti-HA immunoblotting. (C) Arkadia induced rapid turnover of Smad7. COS7 cells were transfected with FLAG-Smad7 and Arkadia (WT, 247–989 or ΔC). [35S]methionine- and cysteine-labelled cell lysates were immunoprecipitated by FLAG antibody. Immune complexes were subjected to SDS–PAGE and examined using a Fuji BAS 2500 Bio-Imaging Analyzer (Fuji Photo Film). The autoradiographic signals were quantified and the values plotted relative to 0-h values. (D) Effects of Arkadia on the transcriptional activity of ALK5-TD in the presence of Smad7. R mutant Mv1Lu cells were co-transfected with the 9xCAGA-Lux and various combinations of ALK5-TD, Smad7, Arkadia-WT and Arkadia-CA cDNAs. + and ++ represent 0.2 and 0.4 µg of DNAs, respectively, transfected into R mutant Mv1Lu cells.
We next examined whether Arkadia induces degradation of Smad7. As shown in Figure 3C, turnover of Smad7 was enhanced in the presence of Arkadia, but not in the presence of Arkadia-ΔC. Moreover, we found that in the presence of Smad7, wild-type Arkadia, but not Arkadia-CA, strongly enhanced transcription induced by ALK5-TD (Figure 3D). These findings suggest that Arkadia induces proteasome-dependent degradation of Smad7, which may lead to amplification of TGF-β superfamily signalling. Interestingly, Arkadia (247–989), which lacks the N-terminal nuclear localization signals (NLS) (see Figure 2B), also efficiently induced degradation of Smad7, similar to the wild-type Arkadia (Figure 3C). Consistent with this finding, Arkadia (247–989) enhanced transcription induced by ALK5-TD in the 9xCAGA-Lux assay (data not shown).
Arkadia–Smad7 complex is not recruited to the TGF-β receptor complexes
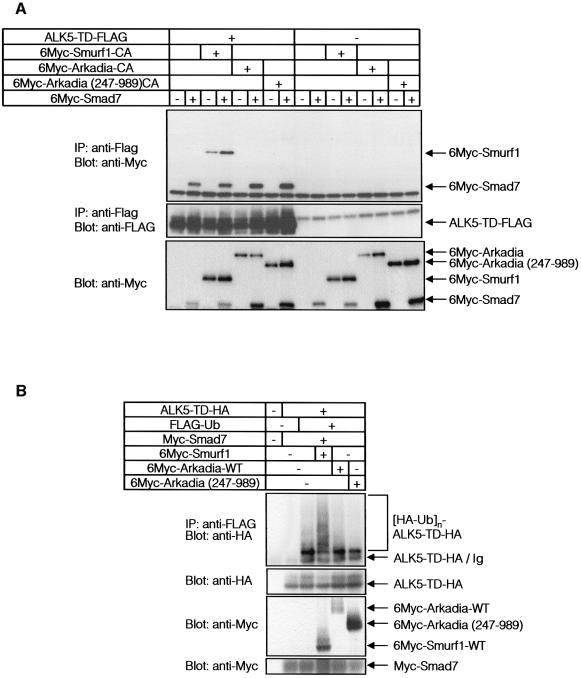
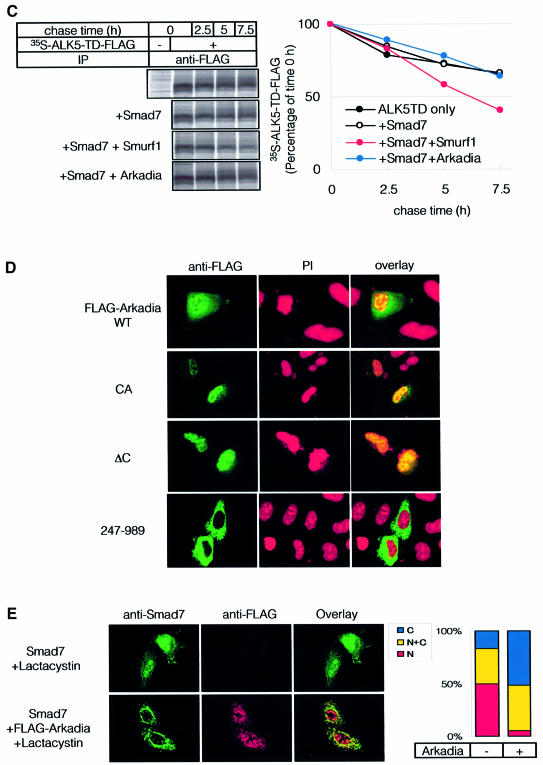
Smurf1 is recruited to the TGF-β receptor complexes through Smad7, and induces ubiquitination and degradation of ALK5 (Ebisawa et al., 2001). We examined whether Arkadia associates with TGF-β receptor complexes through Smad7. COS7 cells were transfected with Smad7, ALK5-TD, and Arkadia or Smurf1, and the cell lysates were subjected to immunoprecipitation of ALK5-TD, followed by immunoblotting of Smad7, Arkadia and Smurf1. As shown in Figure 4A, Smad7 was efficiently co-immunoprecipitated with ALK5-TD. Smurf1 was also co-immunoprecipitated with ALK5-TD, and this was enhanced in the presence of Smad7. In contrast, Arkadia was co-immunoprecipitated with ALK5-TD in neither the presence nor the absence of Smad7. Arkadia (247–989), which lacks the N-terminal NLSs and is located in the cytoplasm (see Figure 4D), failed to interact with ALK-5TD (Figure 4A). In agreement with this finding, neither wild-type Arkadia nor Arkadia (247–989) induced poly-ubiquitination of ALK5-TD, whereas Smurf1 induced it (Figure 4B). Moreover, Smad7–Smurf1 induced the degradation of ALK5-TD, as previously reported (Ebisawa et al., 2001), whereas Smad7–Arkadia or Smad7 alone did not significantly affect the turnover of ALK5-TD protein (Figure 4C). These results indicate that both Smurf1 and Arkadia interact with Smad7; however, Smurf1 interacts with TGF-β receptors through Smad7 and induces their degradation, whereas Arkadia neither interacts with TGF-β receptors nor induces their degradation. These results are consistent with the findings that Smurf1 represses the transcriptional activity of TGF-β through degradation of TGF-β receptors, while Arkadia has an effect opposite to Smurf1, since it degrades Smad7 but not TGF-β receptors (see Figure 1B and C).
Fig. 4. Smurf1, but not Arkadia, recruits Smad7 to ALK5. (A) Association of Arkadia or Smurf1 with ALK5-TD in the presence of Smad7. COS7 cells were transfected with the indicated plasmids. Lysates from cells were subjected to FLAG-immunoprecipitation (IP) followed by Myc-immunoblotting (Blot). (B) Ubiquitination of TGF-β type I receptor (ALK5-TD) is induced by Smurf1 but not by Arkadia. 293T cells were transfected with the indicated plasmids, and treated as in Figure 3A. Lysates from cells were subjected to anti-FLAG immunoprecipitation followed by anti-HA immunoblotting. (C) Addition of Smad7 and Arkadia did not induce rapid turnover of ALK5-TD. COS7 cells were transfected with ALK5-TD-FLAG in the presence or absence of Smad7, Smurf1 and Arkadia. Metabolic labelling and pulse-chase analysis were performed as in Figure 3C. (D) Subcellular localization of Arkadia. HeLa cells were transfected with FLAG-Arkadia alone [wild type, CA and ΔC mutants, and Arkadia (247–989)]. Cells were fixed and stained as described in Materials and methods. Anti-FLAG staining for Arkadia (green) and nuclear staining by PI (red) were conducted. (E) Subcellular localization of Arkadia and Smad7. Cells were transfected with Smad7 with or without FLAG-Arkadia (wild type) in the presence of 10 µM lactacystin. Anti-Smad7 staining (green) and anti-FLAG staining for Arkadia (red) were conducted (left panel). The distribution of Smad7 in cells transfected or not with FLAG-Arkadia was scored as nuclear (N), nuclear and cytoplasmic (N+C), or cytoplasmic (C), and presented graphically (right panel). Experiments were repeated with essentially the same results.
Arkadia contains NLSs in the N-terminal region and was shown to be located in the nucleus in Xenopus embryos (Niederlander et al., 2001). Consistent with this, wild-type Arkadia and the Arkadia mutants (Arkadia-CA and ΔC) were located in the nucleus in mammalian cells (Figure 4D). In contrast, the Arkadia mutant (247–989) lacking NLSs (see Figure 2B) was detected in the cytoplasm, indicating that NLSs are important for nuclear localization of Arkadia. However, since Arkadia (247–989) induced degradation of Smad7 and enhanced the transcription induced by TGF-β signals (Figure 3C and data not shown), nuclear localization of Arkadia may not be essential for degradation of Smad7.
We next examined the localization of Smad7 in the absence and presence of Arkadia. As previously reported (Ebisawa et al., 2001), Smad7 was observed in the nuclei of HeLa cells, which were not affected by a proteasome inhibitor, lactacystin. When Arkadia was co-transfected, Smad7 was detected in the cytoplasm in ∼50% of the cells (Figure 4E). In most other cells, Smad7 was observed in the cytoplasm and nucleus. Arkadia was co-localized with Smad7 in the cytoplasm, although it also remained in the nucleus. These findings suggest that Arkadia induces cytoplasmic localization of Smad7, and that degradation of Smad7 may occur in the cytoplasm as well as in the nucleus.
Arkadia is widely expressed in adult tissues
Expression of Arkadia in mammalian tissues was examined by quantitative real-time PCR. As shown in Figure 5A, Arkadia is expressed in all three human cell lines tested. In addition, analyses of mRNAs obtained from various human tissues showed that Arkadia is widely expressed in human tissues, including testis, spleen, pancreas and lung.
Smad7 has been shown to be induced by TGF-β; it thus regulates TGF-β signalling through a negative feedback loop (Miyazono, 2000). When HaCaT cells were treated with TGF-β, and expression of Smad7 and Arkadia was analysed by quantitative real-time PCR, Smad7 was transiently induced (Figure 5B). In contrast, expression of Arkadia was transiently repressed upon stimulation by TGF-β. Similar results were obtained by RT–PCR analysis (Figure 5C). Thus, expression of Smad7 and Arkadia is reciprocally regulated, and expression of Arkadia may be suppressed under conditions in which Smad7 activity is required.
Arkadia decreases the intracellular pool of Smad7 protein
In order to study the functions of endogenous Arkadia in mammalian cells, we knocked-down the expression of endogenous Arkadia protein by using small interfering RNAs (siRNAs). Arkadia and Smurf1 siRNAs decreased their respective mRNA levels and proteins in both HaCaT cells and 293T cells (Figure 6A). Silencing of the Arkadia gene resulted in repression of the transcriptional activities induced by TGF-β as well as by BMP (Figure 6B and C), which is in clear contrast to silencing of the Smurf1 gene. Thus, endogenous Arkadia in mammalian cells may play a role in the amplification of transcriptional activities induced by TGF-β superfamily proteins.
We next examined the effects of Arkadia on expression of Smad7 protein. Expression of the Arkadia gene was knocked-down by Arkadia siRNA, and Smad7 protein was detected by immunoprecipitation of metabolically labelled HaCaT cells. Smad7 was induced by TGF-β, and silencing of the Arkadia gene resulted in accumulation of endogenous Smad7 protein (Figure 6D). These findings suggest that Arkadia degrades Smad7 under physiological conditions.
Discussion
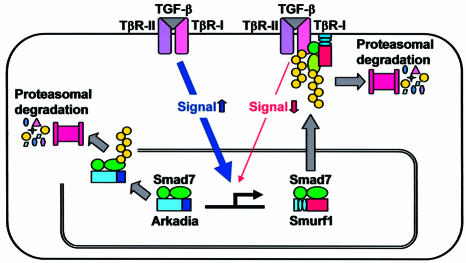
Arkadia was isolated as an intracellular protein that enhances signalling by Nodal (Episkopou et al., 2001). However, the mechanisms of its effects on Nodal signalling have yet to be determined. Arkadia has NLSs in its N-terminal region, and a RING finger domain at its C-terminal end. The present study revealed that Arkadia functions as an E3 ubiquitin ligase on Smad7. Moreover, since Arkadia is not recruited to TGF-β receptors, it degrades only Smad7, resulting in enhancement of TGF-β superfamily signalling. In contrast, Smurf1 efficiently targets Smad7 to TGF-β receptors, leading to degradation of the receptors (Figure 7). Smurf1 has recently been found to contain a nuclear export signal (Tajima et al., 2003), and to target the Smurf1–Smad7 complex to the plasma membrane through its N-terminal C2 domain (Suzuki et al., 2002). In contrast, Arkadia is located in the nucleus through its NLSs, and an Arkadia mutant lacking the NLSs was observed in the cytoplasm. However, the Arkadia mutant without NLSs failed to interact with, and induce degradation of, TGF-β receptors, suggesting that targeting of Smad7–Smurf1 complex to the plasma membrane may be critical for degradation of the receptors.
Fig. 7. Mechanisms of action of Smurf1 and Arkadia through I-Smads. Smurf1 induces degradation of TGF-β type I receptor (TβR-I/ALK5) through interaction with Smad7, leading to inhibition of TGF-β signalling. In contrast, Arkadia degrades Smad7, but not TβR-I/ALK5, resulting in amplification of TGF-β signalling.
Smurf1 binds to the PY motif of Smad7 through its WW domains. In contrast, multiple regions in the middle portion of Arkadia appeared to be responsible for interaction with Smad7. Arkadia contains no known motifs, except for the N-terminal NLSs and the C-terminal RING finger domain. It binds to the C-terminal MH2 domains of I-Smads, which are structurally similar to the corresponding regions of other Smads. Thus, it will be important to determine how Arkadia specifically interacts with I-Smads. In mammals, Smad6 preferentially inhibits BMP signalling, while Smad7 inhibits both TGF-β/activin and BMP signalling (Hata et al., 1998; Itoh et al., 1998; Hanyu et al., 2001). In the present study, we have shown that Arkadia enhances both TGF-β and BMP signalling. Although Arkadia binds to both Smad6 and Smad7, amplification of Nodal and TGF-β signalling by Arkadia may therefore be mediated by ubiquitin-dependent degradation of Smad7, but not by that of Smad6.
Signalling by TGF-β superfamily cytokines is regulated by various mechanisms (Miyazono, 2000). Extracellular antagonists, including noggin, chordin and follistatin, neutralize the effects of BMPs and activin. In addition, I-Smads and transcriptional co-repressors, including c-Ski and SnoN, downregulate the signalling activities of Smads inside cells. These antagonists may play critical roles in limiting the spatio-temporal range of TGF-β superfamily signalling. Intriguingly, several molecules have been shown to enhance TGF-β superfamily signalling through regulating the activity of these antagonists. A metalloprotease, Tolloid, cleaves chordin and enhances BMP signalling activities (Marques et al., 1997; Piccolo et al., 1997; Srinivasan et al., 2002). Anaphase-promoting complex (APC) interacts with Smad3 and induces degradation of SnoN bound to Smad3, resulting in enhancement of TGF-β signalling (Stroschein et al., 2001; Wan et al., 2001). Smurf2 was also shown to degrade SnoN when it binds to Smad2, leading to enhancement of TGF-β signalling under certain conditions (Bonni et al., 2001). In contrast to these molecules, the function of Arkadia is unique, since it interacts with I-Smads and amplifies both TGF-β/activin and BMP signalling pathways by degradation of I-Smads.
Many proteins have been shown to interact with Smads (Miyazawa et al., 2002); however, only a few molecules bind specifically to I-Smads. STRAP and YAP65 bind to Smad7 and enhance its inhibition of TGF-β signalling (Datta and Moses, 2000; Ferrigno et al., 2002). Recently, Tob proteins have been shown to repress BMP signalling through interaction with I-Smads (Yoshida et al., 2003). In contrast, associated molecule with the SH3 domain of STAM (AMSH) has been shown to interact with Smad6 and suppress its inhibition of BMP signalling by preventing the interaction of Smad6 with BMP type I receptors and R-Smads (Itoh et al., 2001). AMSH is a cytoplasmic protein; although its physiological roles have yet to be determined, these findings, together with our results, suggest that activities of I-Smads are regulated through multiple mechanisms.
Expression of I-Smads is regulated by various mechanisms. TGF-β and BMP induce the expression of Smad7; they thus terminate their own signals by negative feedback loops. In addition, certain growth factors, laminar shear stress, interferon-γ, NF-κB signalling and ultraviolet irradiation have been shown to induce the expression of Smad7 (Miyazono et al., 2003). Smurfs facilitate the inhibitory activities of I-Smads; although the mechanisms regulating Smurf1 expression have not yet been reported, Smurf2 has recently been shown to be upregulated in advanced esophageal squamous carcinoma (Fukuchi et al., 2002). In contrast, amplification of TGF-β superfamily signalling may be crucial under certain conditions for achieving sufficient magnitude of signalling by TGF-β superfamily cytokines. Although Arkadia was originally identified as a protein that induces a node and node-derived mesendoderm during early embryogenesis, it appears to serve as one such amplifier of TGF-β superfamily signalling in various tissues. Determination of the mechanisms of action of Arkadia and regulation of its expression may thus be important in understanding how TGF-β superfamily signalling is regulated under various physiological and pathological conditions.
Materials and methods
cDNA construction
Mouse Arkadia cDNA (DDBJ/EMBL/GenBank accession no. AF330197) was cloned using a PCR-based approach with a mouse control cDNA (Clontech) as template. Construction of Arkadia mutants was also performed using a PCR-based approach. The original constructs of constitutively active forms of ALK5 and ALK6 (ALK5-TD and ALK6-QD), Smad1 to -8, Smurf1, Smurf1-CA, Smad7 mutants and ubiquitin (Ub) cDNAs were described previously (Imamura et al., 1997; Kawabata et al., 1998; Ebisawa et al., 2001; Hanyu et al., 2001).
Cell culture and transfection
COS7, HeLa, wild-type and R mutant of Mv1Lu, 293T, HaCaT and C2C12 cells were cultured as described previously (Ishida et al., 2000; Tajima et al., 2003). Transfection of DNA was performed using FuGENE6 (Roche Applied Science) unless otherwise indicated.
Luciferase assay
Cells were transiently transfected with various combinations of promoter–reporter constructs, expression plasmids and pcDNA3. Luciferase activity was measured using the Dual-Luciferase Reporter System (Promega) as described previously (Tajima et al., 2003).
Immunoprecipitation and immunoblotting
COS7 cells or 293T cells were transiently transfected using FuGENE6, and incubated for 24 h before analysis. To determine endogenous interaction between Arkadia and Smad7, HaCaT cells treated with TGF-β (1 ng/ml) for 8 h were used. Cells were lysed with Nonidet P-40 (NP-40) lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40). Immunoprecipitation and immunoblotting were performed as described previously (Ebisawa et al., 2001). For inhibition of proteasomal degradation, cells were incubated with 2.5 µM of lactacystin (Kyowa Medex) for 24 h unless otherwise indicated.
Preparation of antibodies
Goat anti-Smad7 (N19; sc-7004) and goat anti-Smurf1 (F20; sc-14906) antibodies were purchased from Santa Cruz Biotechnology. For detection of endogenous Smad7 in metabolically labelled HaCaT cells, anti-Smad7 antiserum was prepared by immunizing a rabbit with a chemically synthesized peptide (CKAVRGAKGHHHPH) conjugated to keyhole lympet hemocyanin (Calbiochem) using succinimidyl 4-(p-maleimidophenyl)butyrate (Pierce Chemical Co.). Anti-Arkadia antiserum was prepared by immunizing a rabbit with mouse Arkadia (amino acid residues 1–203), expressed as a fusion protein with glutathione S-transferase.
Metabolic labelling and pulse-chase analysis
COS7 cells transfected with DNAs were labelled for 10 min at 37°C with 42.9 µCi/ml [35S]methionine and cysteine (Amersham) in methionine- and cysteine-free Dulbecco’s modified Eagle’s medium (DMEM), and chased in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM methionine and 0.5 mM cysteine for the time periods indicated, as described previously (Fukuchi et al., 2001). For detection of endogenous Smad7 protein, HaCaT cells were transfected or not with Arkadia siRNA. The cells were then treated or not with 1 ng/ml TGF-β, and labelled for 7.5 h at 37°C with 0.2 mCi/ml [35S]methionine and cysteine. Cells were then lysed and subjected to immunoprecipitation followed by SDS–PAGE. The gels were fixed, dried, and examined using a Fuji BAS 2500 Bio-Imaging Analyzer (Fuji Photo Film) or by autoradiography.
Immunofluorescence labelling
Immunohistochemical staining of FLAG-Arkadia and Smad7 was performed as described previously (Fukuchi et al., 2001). Cell nuclei were stained by 4,6-diamidino-2-phenylindole (PI). Intracellular localization was determined by confocal laser scanning microscopy.
Quantitative real-time PCR analysis and RT–PCR
Total RNAs were extracted using TRIZOL (Invitrogen). First-strand cDNAs were synthesized using the ThermoScript RT-PCR System (Invitrogen) with oligo(dT)20 primers. Quantitative real-time RT–PCR analysis was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The primer sequences used were as follows: human Arkadia: forward 5′-CCACATAGGATGCACCCAAAC, reverse 5′-AATTCCCAGTTCCCAGGCA; human Smurf1: forward 5′-GTCCAGAAGCTGAAAGTCCTCAGA, reverse 5′-CACGGAATTTCACCATCAGCC; human Smad7: forward 5′-CCTTAGCCGACTCTGCGAACTA, reverse 5′-CCAGATAATTCGTTCCCCCTGT; and human GAPDH: forward 5′-GAAGGTGAAGGTCGGAGTC, reverse 5′-GAAGATGGTGATGGGATTTC. RT–PCR analysis was performed as described by Nishihara et al. (2002) with minor modifications. The primer sequences used were as follows: human Arkadia: forward 5′-CATCCTCACTTGGCCCATTATCAC, reverse 5′-GTCCCTTCTTCCCCATCTTGTTTG; and human Smad7: forward 5′-CCTTAGCCGACTCTGCGAACTA, reverse 5′-TGCATAAACTCGTGGTCATTGG. The primers for human GAPDH were the same as those used for quantitative real-time RT–PCR.
RNA interference and oligonucleotides
siRNAs were introduced into cells (Kisielow et al., 2002) using the Lipofectamine2000 reagent (Invitrogen) according to the manufacturer’s instructions with 120 pmol of siRNA and 4 µl of transfection reagent per well of a 12-well tissue culture plate for 293T, or 200 pmol of siRNA and 10 µl of transfection reagent per well of a six-well plate for HaCaT. For detection of endogenous Smad7 protein, HaCaT cells in a 10-cm culture dish were transfected with 720 pmol of siRNA and 36 µl of transfection reagent. Expression vectors were co-transfected with siRNAs. The following 21-mer oligonucleotide pairs were used for RNA interfererence: Arkadia siRNA from nucleotides 518–538 (DDBJ/EMBL/GenBank accession number AF330197), 5′-AAGUGGGGAAUGAAUUCUCUC-3′, 5′-GAGAGAAUUCAUUCCCCACUU-3′; and Smurf1 siRNA from nucleotides 411–431 (DDBJ/EMBL/GenBank accession number XM_166483), 5′-AAGAACCUUGCAAAGAAAGAC-3′, 5′-GUCUUUCUUUGCAAGGUUCUU-3′. siRNA from Euglena gracilis chloroplast DNA between s16S and 16S (DDBJ/EMBL/GenBank accession number X05005), 5′-AAGCGCGCAAAGUAGGAUUCG-3′, 5′-CGAAUCCUACUUUGCGCGCUU-3′, was used as a control. Annealing was performed as described previously (Elbashir et al., 2001).
Acknowledgments
Acknowledgements
This research was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by grants from Nippon Boehringer Ingerheim (VRIA), the Takeda Science Foundation and the Japanese Research Foundation for Clinical Pharmacology.
References
- Bonni S., Wang,H.R., Causing,C.G., Kavsak,P., Stroschein,S.L., Luo,K. and Wrana,J.L. (2001) TGF-β induces assembly of a Smad2–Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol., 3, 587–595. [DOI] [PubMed] [Google Scholar]
- Datta P.K. and Moses,H.L. (2000) STRAP and Smad7 synergize in the inhibition of transforming growth factor β signaling. Mol. Cell. Biol., 20, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R. and Zhang,Y.E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature, 425, 577–584. [DOI] [PubMed] [Google Scholar]
- Ebisawa T., Fukuchi,M., Murakami,G., Chiba,T., Tanaka,K., Imamura,T. and Miyazono,K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem., 276, 12477–12480. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Episkopou V., Arkell,R., Timmons,P.M., Walsh,J.J., Andrew,R.L. and Swan,D. (2001) Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature, 410, 825–830. [DOI] [PubMed] [Google Scholar]
- Ferrigno O., Lallemand,F., Verrecchia,F., L’Hoste,S., Camonis,J., Atfi,A. and Mauviel,A. (2002) Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-β/Smad signaling. Oncogene, 21, 4879–4884. [DOI] [PubMed] [Google Scholar]
- Fukuchi M., Imamura,T., Chiba,T., Ebisawa,T., Kawabata,M., Tanaka,K. and Miyazono,K. (2001) Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol. Biol. Cell, 12, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M. et al. (2002) High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res., 62, 7162–7165. [PubMed] [Google Scholar]
- Hanyu A., Ishidou,Y., Ebisawa,T., Shimanuki,T., Imamura,T. and Miyazono,K. (2001) The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J. Cell Biol., 155, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Lagna,G., Massagué,J. and Hemmati-Brivanlou,A. (1998) Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev., 12, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C.H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Imamura T., Takase,M., Nishihara,A., Oeda,E., Hanai,J., Kawabata,M. and Miyazono,K. (1997) Smad6 inhibits signalling by the TGF-β superfamily. Nature, 389, 622–626. [DOI] [PubMed] [Google Scholar]
- Ishida W., Hamamoto,K., Kusanagi,K., Yagi,K., Kawabata,M., Takehara,K., Sampath,T.K., Kato,M. and Miyazono,K. (2000) Smad6 is a Smad1/5-induced Smad inhibitor: Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem., 275, 6075–6079. [DOI] [PubMed] [Google Scholar]
- Itoh S., Landstrom,M., Hermansson,A., Itoh,F., Heldin,C.H., Heldin,N.E. and ten Dijke,P. (1998) Transforming growth factor β1 induces nuclear export of inhibitory Smad7. J. Biol. Chem., 273, 29195–29201. [DOI] [PubMed] [Google Scholar]
- Itoh F., Asao,H., Sugamura,K., Heldin,C.H., ten Dijke,P. and Itoh,S. (2001) Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J., 20, 4132–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata M., Inoue,H., Hanyu,A., Imamura,T. and Miyazono,K. (1998) Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J., 17, 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak P., Rasmussen,R.K., Causing,C.G., Bonni,S., Zhu,H., Thomsen,G.H. and Wrana,J.L. (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF-β receptor for degradation. Mol. Cell, 6, 1365–1375. [DOI] [PubMed] [Google Scholar]
- Kisielow M., Kleiner,S., Nagasawa,M., Faisal,A. and Nagamine,Y. (2002) Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem. J., 363, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney J.D. and Hochstrasser,M. (1999) Substrate targeting in the ubiquitin system. Cell, 97, 427–430. [DOI] [PubMed] [Google Scholar]
- Lin X., Liang,M. and Feng,X.H. (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J. Biol. Chem., 275, 36818–36822. [DOI] [PubMed] [Google Scholar]
- Lo R.S. and Massagué,J. (1999) Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nat. Cell Biol., 1, 472–478. [DOI] [PubMed] [Google Scholar]
- Marques G., Musacchio,M., Shimell,M.J., Wunnenberg-Stapleton,K., Cho,K.W. and O’Connor,M.B. (1997) Production of a DPP activity gradient in the early Drosophila embryo through the opposing actions of the SOG and TLD proteins. Cell, 91, 417–426. [DOI] [PubMed] [Google Scholar]
- Miyazono K. (2000) Positive and negative regulation of TGF-β signaling. J. Cell Sci., 113, 1101–1109. [DOI] [PubMed] [Google Scholar]
- Miyazawa K., Shinozaki,M., Hara,T., Furuya,T. and Miyazono,K. (2002) Two major Smad pathways in TGF-β superfamily signaling. Genes Cells, 7, 1191–1204. [DOI] [PubMed] [Google Scholar]
- Miyazono K., Suzuki,H. and Imamura,T. (2003) Regulation of TGF-β signaling and its roles in progression of tumors. Cancer Sci., 94, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami G., Watabe,T., Takaoka,K., Miyazono,K. and Imamura,T. (2003) Inhibition of BMP signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell, 14, 2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederlander C., Walsh,J.J., Episkopou,V. and Jones,C.M. (2001) Arkadia enhances nodal-related signalling to induce mesendoderm. Nature, 410, 830–834. [DOI] [PubMed] [Google Scholar]
- Nishihara N., Watabe,T., Imamura,T. and Miyazono,K. (2002) Functional heterogeneity of bone morphogenetic protein receptor-II mutants found in patients with primary pulmonary hypertension. Mol. Biol. Cell, 13, 3055–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Agius,E., Lu,B., Goodman,S., Dale,L. and De Robertis,E.M. (1997) Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell, 91, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. and Massague,J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell, 113, 685–700. [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Rashka,K.E. and Bier,E. (2002) Creation of a Sog morphogen gradient in the Drosophila embryo. Dev. Cell, 2, 91–101. [DOI] [PubMed] [Google Scholar]
- Stroschein S.L., Bonni,S., Wrana,J.L. and Luo,K. (2001) Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev., 15, 2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C., Murakami,G., Fukuchi,M., Shimanuki,T., Shikauchi,Y., Imamura,T. and Miyazono,K. (2002) Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem., 277, 39919–39925. [DOI] [PubMed] [Google Scholar]
- Tajima Y., Goto,K., Yoshida,M., Shinomiya,K., Sekimoto,T., Yoneda,Y., Miyazono,K. and Imamura,T. (2003) Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-β signaling by Smad7. J. Biol. Chem., 278, 10716–10721. [DOI] [PubMed] [Google Scholar]
- Wan Y., Liu,X. and Kirschner,M.W. (2001) The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol. Cell, 8, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Wan M., Cao,X., Wu,Y., Bai,S., Wu,L., Shi,X., Wang,N. and Cao,X. (2002) Jab1 antagonizes TGF-β signaling by inducing Smad4 degradation. EMBO Rep., 3, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y. et al. (2003) Tob proteins enhance inhibitory Smad-receptor interactions to repress BMP signaling. Mech. Dev., 120, 629–637. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chang,C., Gehling,D.J., Hemmati-Brivanlou,A. and Derynck,R. (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl Acad. Sci. USA, 98, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Kavsak,P., Abdollah,S., Wrana,J.L. and Thomsen,G.H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature, 400, 687–693. [DOI] [PubMed] [Google Scholar]