Abstract
CSR1 (cellular stress response 1), a newly characterized tumor-suppressor gene, undergoes hypermethylation in over 30% of prostate cancers. Re-expression of CSR1 inhibits cell growth and induces cell death, but the mechanism by which CSR1 suppresses tumor growth is not clear. In this study, we screened a prostate cDNA library using a yeast two-hybrid system and found that the cleavage and polyadenylation-specific factor 3 (CPSF3), an essential component for converting heteronuclear RNA to mRNA, binds with high affinity to the CSR1 C terminus. Further analyses determined that the binding motifs for CPSF3 are located between amino acids 440 and 543. The interaction between CSR1 and CPSF3 induced CPSF3 translocation from the nucleus to the cytoplasm, resulting in inhibition of polyadenylation both in vitro and in vivo. Downregulation of CPSF3 using small interfering RNA induced cell death in a manner similar to CSR1 expression. A CSR1 mutant unable to bind to CPSF3 did not alter CPSF3 subcellular distribution, did not inhibit its polyadenylation activity and did not induce cell death. In summary, CSR1 appears to induce cell death through a novel mechanism by hijacking a critical RNA processing enzyme.
Keywords: CSR1, CPSF3, cell death, RNA polyadenylation
Introduction
Prostate cancer is a leading cause of cancer-related deaths for men in the United States (Jemal et al., 2007). Although the majority of prostate cancers are not lethal, with only 15–20% of prostate cancers progressing to metastatic and life-threatening disease, almost 30 000 men die of this disease each year (Isaacs, 1997; Jemal et al., 2005, 2008). The high incidence of this disease among an aging population makes treatment, and especially prevention, a priority. However, our understanding of the molecular mechanisms underlying prostate cancer remains quite incomplete.
In previous studies, we analysed the expression of 37 000 human genes and identified CSR1, a cellular stress-response protein, as a protein significantly downregulated in prostate cancer (Luo et al., 2002; Yu et al., 2004, 2006). CSR1 is located at 8p21–22, a locus that is frequently deleted in several human malignancies, including prostate cancer, head and neck squamous cell carcinoma and lung cancer (Ohata et al., 1993; Wistuba et al., 1999; Kurimoto et al., 2001; Coon et al., 2004; Gallucci et al., 2006). One study suggested that CSR1 protects cells from mutational damage of oxidative-free radicals by increasing their metabolism (Han et al., 1998). Recently, we identified CSR1 as a putative tumor suppressor for prostate cancer (Yu et al., 2006). Further analyses demonstrated that CSR1 is frequently methylated in prostate cancer. CSR1 methylation, and, to a lesser extent, inactivation of CSR1 expression, are associated with a high rate of prostate cancer metastasis, regardless of tumor differentiation. Restoring CSR1 expression, in prostate cancer cell lines where CSR1 is methylated, suppresses tumor growth and metastasis both in vitro and in an animal model. The underlying mechanism of CSR1 tumor-suppressor activity, however, is not understood.
In normal cells, CSR1, which is located primarily in the cytoplasm near the cell membrane, has three putative functional domains: a membrane-spanning region, an α-helix coiled-coil domain and a collagen-like domain. In this study, we found that the C terminus of CSR1 binds in vitro and in vivo with cleavage and polyadenylation-specific factor 3 (CPSF3), an enzyme necessary for polyadenylation of RNA (Ryan et al., 2004; Dominski et al., 2005a), which is localized to the nucleus to process heteronuclear RNA into mRNA. For an interaction between CPSF3 and CSR1 to occur, one of these proteins has to redistribute to its non-physiological compartment, that is, either CSR1 is relocated to the nucleus or CPSF3 travels to the cytoplasm. Herein, we examine the localization of both these proteins, as well as the polyadenlyation of RNA in the cell, and the impact of CSR1 expression on cell death. CSR1 may protect genetic material through multiple pathways, by accelerating the metabolism of mutagens, while inducing metabolic cell death by limiting the availability of functional mRNA. This study offers a new insight into CSR1-mediated tumor-suppressor activity.
Results
We used a yeast two-hybrid screening system to identify proteins that interact with CSR1, and therefore may serve as downstream signaling molecules. The CSR1 C terminus (156 amino acids) was ligated into the pGBKT7 vector to create the pBD-CSR1c plasmid, expressing a fusion protein containing the DNA-binding domain of GAL4 and 156 amino acids of the CSR1 C terminus. This construct was used to screen a human prostate library using the Matchmaker GAL4 two-hybrid system 3, as described in the Materials and methods section. After three rounds of metabolic screening, 21 positive colonies were identified. After restriction enzyme digestions, several redundant clones were eliminated; 12 unique clones were identified and sequenced. One of these clones contained a cDNA encoding CPSF3.
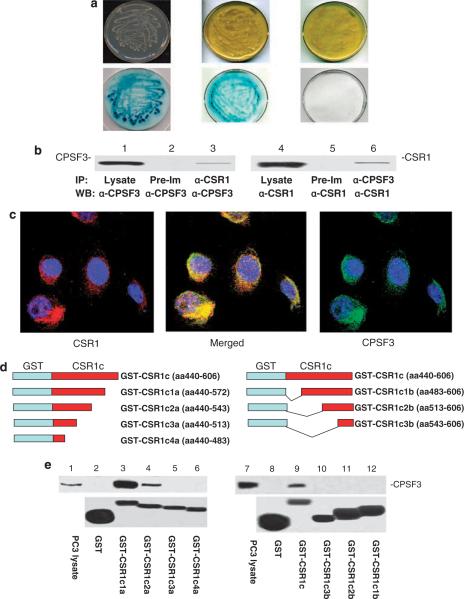
To validate the yeast two-hybrid screening results, pAD-CPSF3 and pBD-CSR1c were co-transfected into yeast AH109 cells, grown in high-stringency medium (SD-Ade/-His/-Leu/-Trp), and tested for β-galactosidase activity. As shown in Figure 1a, yeast cells harboring pBD-CSR1c and pAD-CPSF3 grew in high-stringency medium agar plates and showed positive galactosidase activity. The negative control, cells co-transfected with pBD-Lamin C and pAD-T antigen, failed to grow in the high-stringency selective medium. These cells grew normally in low-stringency medium selective only for plasmids (SD-Leu/-Trp), and were negative for galactosidase activity (Figure 1a). This finding suggests that CPSF3-binding activity is mediated by a region located in the C terminus of CSR1. To verify the interaction, an in vivo CSR1–CPSF3-binding analysis was performed using the protein extracts from PC3 cells transfected with pCMV-CSR1. As shown in Figure 1b, co-immunoprecipitation of CSR1 and CPSF3 was readily apparent; detection of CSR1/CPSF3 complex was found in either CSR1 or CPSF3 immunoprecipitates. Double immunostaining of CSR1 and CPSF3 (Figure 1c) revealed that CSR1 and CPSF3 were colocalized in the cytoplasm or at the plasma membrane of cells expressing CSR1. To investigate whether CSR1 and CPSF3 bind in a cell-free system, the C terminus of CSR1 was ligated into pGEX-5X-3 to create a vector expressing GST (glutathione S-transferase)-CSR1c (156 amino acids from CSR1)-fusion protein. An in vitro-binding assay confirmed that CPSF3 binds to the CSR1 C terminus (Figure 1d). A series of deletion mutants with partial deletions of the N or C terminus of CSR1c were created to determine the motifs necessary for CPSF3 binding. Analysis of the binding data (Figure 1d) from these mutants demonstrates that a stretch of 103 amino acids (440–543) in the C terminus of CSR1 is essential for its interaction with CPSF3.
Figure 1.
CSR1 (cellular stress response 1) interacts with cleavage and polyadenylation-specific factor 3 (CPSF3). (a) Growth of yeast AH109 strain harboring pBD-CSR1c and pAD-CPSF3 on high-stringency agar plates (upper left) and β-galactosidase activity (lower left), pBD-p53 and pAD-T antigen on high-stringency agar plates (upper center) and β-galactosidase activity (lower center), pBD-lamin C and pAD-T antigen on low-stringency agar plates (upper right) and β-galactosidase activity (lower right). (b) Co-immunoprecipitation of CSR1 and CPSF3. Proteins from PC3 cells transfected with pCMV-CSR1 were immunoprecipitated with antibodies against CSR1 (α-CSR1), CPSF3 (α-CPSF3) and pre-immune serum (Pre-im), and immunoblotted with the indicated antibodies. (c) Colocalization of CSR1 and CPSF3. PC3 cells transfected with pCMV-CSR1 were immunostained simultaneously with antibodies specific for CSR1 (rhodamine) and CPSF3 (fluorescein). (d) Constructs of glutathione S-transferase (GST) fusion proteins. GST-CSR1c and its deletion mutants CSR1C1a–4a and 1b–3b were constructed as indicated. (e) GST and GST-CSR fusion proteins were purified through a glutathione column as described in the Materials and methods section. Binding assays were performed with protein extracts from PC3 cells. The bound CPSF3 was separated by electrophoresis on a 10% SDS–polyacrylamide gel electrophoresis and immunoblotted with antibodies specific for CPSF3. The upper panel: Immunoblot with anti-CPSF3 antibodies;the lower panel: Coumassie staining of GST-CSR1 fusion proteins in a polyacrylamide gel.
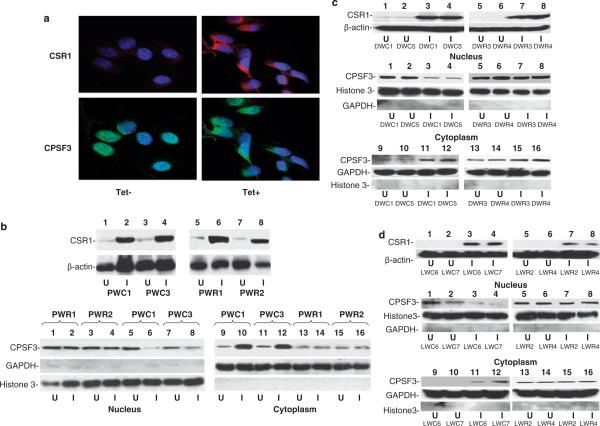
To investigate whether the interaction between CSR1 and CPSF3 alters the subcellular localization of either protein, pCDNA4-CSR1/pCDNA6 was transfected into PC3, DU145 and LNCaP cells to obtain stable transformed cell lines inducible for CSR1 expression. Two colonies from each transfection (PWC1 and PWC3 for PC3 transfection, DWC1 and DWC5 for DU145, and LWC6 and LWC7 for LNCaP) were selected for further analysis. CSR1 expression was induced in the PWC1 cell line, a PC3 cell subclone transfected with the pCDNA4-CSR1/pCDNA6 plasmid. Subcellular fractionation revealed that CPSF3 was predominantly a nuclear protein when there is minimal CSR1 expression (Figures 2a and b). However, nuclear CPSF3 is reduced dramatically with concomitant increase of cytosolic one when CSR1 expression is induced. Similar translocation of CPSF3 to the cytoplasm was found in LNCaP and DU145 cells that express CSR1 (Figures 2c and d). To examine the change in CPSF3 distribution, immunofluorescent staining, using antibodies specific for CPSF3, was performed on cell lines transfected with pCDNA4-CSR1/pCDNA6; the results demonstrate that CSR1 expression induces redistribution of CPSF3 from its primary location, the nucleus, to the cytoplasm (Figure 2a). The CPSF3 staining, after CSR1 induction, is not diffuse but patchy, suggesting that CPSF3 is bound in a complex. To investigate whether redistribution of CPSF3 is dependent on its interaction with CSR1, we generated a mutant CSR1 that does not bind CPSF3 due to deletion of part of the CPSF3 interaction motifs (amino acids 483–513). Expression of this mutant in all three cell lines (PWR1 and PWR2-PC3, DWR3 and DWR4-DU145 and LWR2 and LWR4-LNCaP) failed to induce redistribution of CPSF3 to the cytoplasm (Figures 2b–d). Thus, CSR1/CPSF3 interaction is essential for redistributing CPSF3, and might be an important factor in CSR1 tumor suppression.
Figure 2.
Redistribution of cleavage and polyadenylation-specific factor 3 (CPSF3) upon induction of cellular stress response 1 (CSR1) expression. (a) Immunofluorescence staining of PWC1 cells. PWC1 cells (PC3 harboring pCDNA4-CSR1/pCDNA6) treated with or without tetracycline were stained with antibodies specific for CSR1 or CPSF3. The images were obtained after incubation with rhodamine-conjugated anti-rabbit (CSR1) and fluorescein-conjugated anti-goat secondary antibodies. (b) Immunoblot analysis of CPSF3 distribution. The upper panel: CSR1 expression in PWC1, PWC3, PWR1 and PWR2 cells. The lower panel: nuclear and cytoplasmic proteins from PC3 harboring wild-type (PWC1 and PWC3) or mutant (PWR1 and PWR2) were separated by electrophoresis on an 8% SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted with antibodies specific for CPSF3, GAPDH or histone 3. (c) Immunoblot analysis of CPSF3 distribution in DU145 cells. The upper panel: CSR1 expression in DWC1 (DU145 harboring pCDNA4-CSR1/pCDNA6, lanes 1 and 3), DWC5 (lanes 2 and 4) or mutant cells (DWR3 (lanes 5 and 7) and DWR4 (lanes 6 and 8)). The lower panel: nuclear and cytoplasmic proteins from DU145 cells expressing wild-type CSR1 (DWC1 (lanes 1, 3, 9 and 11) and DWC5 (lanes 2, 4, 10 and 12)) or mutant CSR1 (DWR3 (lanes 5, 7, 13 and 15) and DWR4 (lanes 6, 8, 14 and 16)) were separated by electrophoresis on an 8% SDS–PAGE and immunoblotted with antibodies specific for CPSF3, GAPDH or histone 3. (d) Immunoblot analysis of CPSF3 distribution in LNCaP cells. The upper panel: CSR1 expression in LWC6 (LNCaP harboring pCDNA4-CSR1/pCDNA6, lanes 1 and 3) and LWC7 (lanes 2 and 4) or mutants (LWR2 (lanes 5 and 7) and LWR4 (lanes 6 and 8)) cells. The lower panel: nuclear and cytoplasmic proteins from LNCaP harboring wild-type (LWC6 (lanes 1, 3, 9 and 11) and LWC7 (lanes 2, 4, 10 and 12)) or mutant (LWR2 (lanes 5, 7, 13 and 15) and LWR4 (lanes 6, 8, 14 and 16)) CSR1 were separated by electrophoresis on an 8% SDS–PAGE and immunoblotted with antibodies specific for CPSF3, GAPDH or histone 3. U: uninduced;I: induced with tetracycline.
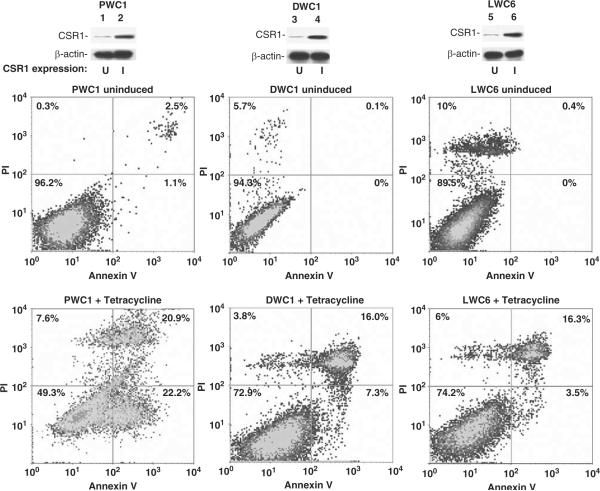
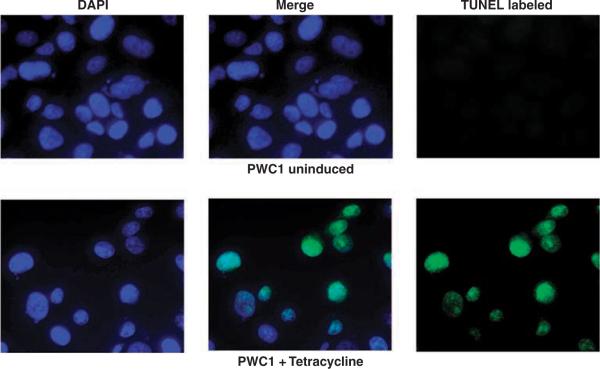
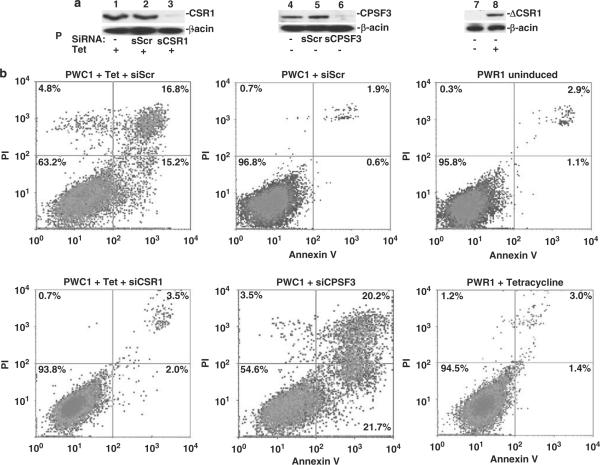
Expression of CSR1 is induced by stresses such as UV irradiation or exposure to toxic chemicals. By analogy to p53, a tumor-suppressor gene that is also a stress-activated protein, the possible effects of CSR1 expression on cell death were investigated. Four of the tetracycline-inducible pCDNA4-CSR1 clones (of PC3 transformants) were selected for cell death analysis by annexin V binding to externalized phosphatidylserine residues. Induction of CSR1 expression in PC-3 cells increased cell death 5- to 10-fold, with both necrosis and apoptosis occurring (Figure 3 and Table 1). Similar findings were obtained for LNCaP and DU145 cells (Figure 3). The fragmentation of genomic DNA in CSR1-induced cells was detected by the TUNEL (terminal dUTP nick-end labeling) assay, confirming that the cells were undergoing apoptotic changes (Figure 4). Tetracycline-induced cell death was largely reversed by a small interfering RNA (siRNA) specific for CSR1 (Figure 5 and Table 1), further validating the role of CSR1 in inducing cell death. Interestingly, a siRNA specific for CPSF3 induced cell death to a similar degree to CSR1 induction (Figure 5 and Table 1). To investigate whether disrupting the CSR1/CPSF3 interaction will abrogate the cell death induced by CSR1, cell lines harboring the CSR1 mutant that does not interact with CPSF3 were treated with tetracycline (Figure 5 and Table 1). All cell lines harboring the mutant (PWR1, PWR2, DWR3, DWR4, LWR2 and LWR4) did not die when exposed to tetracycline, implying that reducing functional CPSF3 in the nucleus through redistribution is a critical mechanism of CSR1-induced cell death.
Figure 3.
Fluorescence-activated cell sorting (FACS) analysis of cell death induced by cellular stress response 1 (CSR1) expression. PWC1 (PC3 cells expressing wild-type CSR1) or DWC1 (DU145 cells expressing wild-type CSR1), or LWC6 cells (LNCaP cells expressing wild-type CSR1) were treated or not with tetracycline. These cells were stained with Alexa Fluor 488-labeled annexin V and propidium iodide (PI). The cells were sorted and quantified in a LSR-II flow cytometer as described in the Materials and methods section. The upper panel: immunoblot of CSR1 expression;the middle and lower panels: density plots of FACS analyses of samples from the upper panel.
Table 1.
Cell death induced by CSR1
| CSR1 induction | PI staining alone |
Annexin V alone |
Annexin V+PI |
|||
|---|---|---|---|---|---|---|
| −CSR1 (%) | +CSR1 (%) | −CSR1 (%) | +CSR1 (%) | −CSR1 (%) | +CSR1 (%) | |
| PWC1 | 0.3 | 7.6 | 1.1 | 22.2 | 2.5 | 20.9 |
| PWC3 | 0.5 | 7.5 | 1.3 | 19.8 | 1.3 | 18.7 |
| PDC1 | 1.0 | 8.5 | 0.9 | 20.8 | 1.0 | 19.9 |
| PDC2 | 1.2 | 7.9 | 0.7 | 21.6 | 0.5 | 18.5 |
| DWC1 | 5.7 | 3.8 | 0 | 7.3 | 0.1 | 16.0 |
| DWC5 | 6.9 | 4.4 | 1.3 | 8.3 | 0.6 | 15.7 |
| LWC6 | 10 | 6 | 0 | 3.6 | 0.4 | 16.3 |
| LWC7 | 9.8 | 6.8 | 0.3 | 3.5 | 0.9 | 15.9 |
| PWC1+scramble siRNA | 0.7 | 4.8 | 0.6 | 15.2 | 1.9 | 16.8 |
| PWC1+CSR1 siRNA | 0.5 | 0.7 | 3.0 | 2.0 | 3.7 | 3.5 |
| PWC1+CPSF3 siRNA | 3.5 | 3.4 | 21.7 | 20.1 | 20.2 | 19.2 |
| PDC1+scramble siRNA | 1.2 | 8.2 | 0.5 | 20.1 | 1.4 | 19.3 |
| PDC1+CSR1 siRNA | 1.3 | 2.2 | 1.0 | 2.9 | 1.5 | 3.1 |
| PDC1+CPSF3 siRNA | 9.5 | 9.9 | 16.1 | 16.6 | 16.0 | 16.8 |
| PWR1 | 2.8 | 3.1 | 0.8 | 0.9 | 1.0 | 1.3 |
| PWR2 | 2.5 | 2.8 | 0.7 | 0.5 | 1.1 | 0.8 |
| DWR3 | 8.9 | 7.9 | 2.9 | 1.1 | 1.4 | 1.1 |
| DWR4 | 7.3 | 10.6 | 2.3 | 1.7 | 2.4 | 4.4 |
| LWR2 | 10.7 | 11.3 | 0.8 | 0.9 | 0.7 | 1.4 |
| LWR4 | 12.8 | 10.5 | 1.1 | 1.4 | 0.6 | 0.9 |
Abbreviations: CPSF3, cleavage and polyadenylation-specific factor 3; CSR1, cellular stress response 1; PI, propidium iodide; siRNA, small interfering RNA.
PWC1, PWC3, PDC1 and PDC2 are four different isolates of PC3 cells harboring pCDNA4-CSR1/pCDNA6 constructs.
Figure 4.
Terminal dUTP nick-end labeling (TUNEL) analysis of cell death induced by cellular stress response 1 (CSR1) expression. PWC1 cells were induced with (lower panel) or without (upper panel) tetracycline. The fragmented genomic DNA was labeled with fluorescein dUTP and viewed with a fluorescence microscope as described in the Materials and methods section.
Figure 5.
Interaction with cleavage and polyadenylation-specific factor 3 (CPSF3) is required for cellular stress response 1 (CSR1)-mediated cell death. (a) Immunoblot analysis of CSR1, CPSF3 and mutant CSR1 expression (ΔCSR1) expression. PWC1 cells (lanes 1 through 6) were transfected with scramble small interfering RNA (siRNA) (sScr, lanes 2 and 5) or siRNA specific for CSR1 (sCSR1, lane 3) or siRNA specific for CPSF3 (sCPSF3, lane 6), and treated with tetracycline (lanes 1–3) or not (lanes 4–6), or PWR1 (PC3 harboring mutant CSR1) cells were treated with tetracycline (lane 8) or not (lane 7). (b) Fluorescence-activated cell sorting analysis of cell death in cells used in (a). These cells were stained with Alexa Fluoro 488-labeled annexin V and propidium iodide (PI). The cells were sorted and quantified in a LSR-II flow cytometer as described in the Materials and methods section.
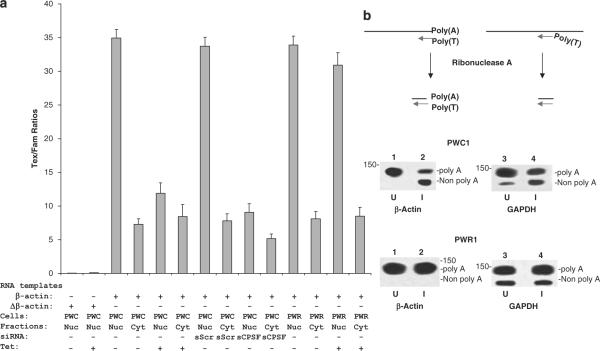
RNA cleavage and polyadenylation, necessary for the production of mRNA, is a complex procedure involving multiple proteins. CPSF3 is one of the critical factors involving in pre-mRNA processing. To investigate whether redistribution of CPSF3 from the nucleus to the cytoplasm has an adverse effect on RNA processing, in vitro polyadenylation assays were performed using protein purified from the nucleus or the cytoplasm of CSR1-induced cells. A 737 base β-actin RNA containing the polyadenylation acceptor site labeled with Famuridine was incubated with the nuclear or cytoplasmic protein extracts to examine the polyadenylation activity. The results demonstrate that induction of CSR1 expression decreased nuclear polyadenylation activity by threefold (Figure 6), whereas activity from the cytoplasmic fraction was similar whether or not CSR1 expression was induced. In contrast, when the Δβ-actin RNA, which lacks the polyadenylation acceptor site (AAUAAA), was used as a template, no polyadenylation activity was detected, confirming the specificity of the assay. In comparison with wild-type CSR1, polyadenylation assays using the nuclear extract from PWR1 cells, which are transfected with the mutant CSR1 that does not interact with CPSF3, showed no significant decrease in polyadenylation activity upon tetracycline induction, supporting the evidence that CSR1/CPSF3 interaction is required to inhibit the nuclear polyadenylation activity.
Figure 6.
Decreased nuclear polyadenylation activity following induction of cellular stress response 1 (CSR1) expression. (a) In vitro polyadenylation of nuclear proteins and cytoplasmic proteins from PWC1 (PWC) and PWR1 (PWR) cells. A β-actin RNA or its AAUAAA deleted mutant (Δβ-actin) labeled with Fam-uridine were polyadenylated with nuclear (nuc) or cytoplasmic (cyt) proteins from PWC1 or PWR1 cells in the presence of Texas Red-conjugated ATP. Incorporation of Texas Red-conjugated ATP was normalized by comparing it to Fam-labeled RNA. Each condition was analysed in quadruplicate. (b) RNase protection analysis of polyadenylation status of β-actin and GAPDH. Total RNA was extracted from PWC1 or PWR1 cells with or without induction of CSR1 expression. The RNA was hybridized with biotin-labeled probes corresponding to the 3′ ends of β-actin and GAPDH, which contain 24 thymidine bases at the 5′ end, and treated with RNase A. The protected RNA was separated and visualized by electrophoresis in 4.75% polyacrylamide gel as described in the Materials and methods section. The upper panel: diagram of RNase protection assays;the lower panel: RNase protection blots.
To investigate whether in vivo polyadenylation activity was decreased in cells that expressed CSR1, the polyadenylation of two housekeeping genes, β-actin and glyceraldehydes 3-phosphate dehydrogenase (GAPDH), was examined in PWC1 cells using an RNAse protection assay. Biotin-labeled probes of RNA containing a 24 base polythymidine at their 5′ end were generated for both β-actin and GAPDH. These probes were hybridized to total RNA extracted from PWC1 cells treated with or without tetracycline to quantify polyadenylation of these two genes. As demonstrated in Figure 6b, there were substantial increases in the fraction of both β-actin and GAPDH that was not polyadenylated, suggesting a general decrease in mRNA in CSR1-expressing cells. In contrast, no such effect was seen in cells induced to express mutant CSR1.
Discussion
To our knowledge, this is the first study demonstrating that a protein induces cell death through limiting the processing of RNA. Several lines of evidences support the hypothesis that inhibition of CPSF3 activity is the mechanism of CSR1-mediated cell death. First, interaction of CSR1 and CPSF3 induced redistribution of CPSF3 from the nucleus to the cytoplasm. The displaced CPSF3 could not access heteronuclear RNA to convert it into messenger RNA, as demonstrated by the lower polyadenylation activity of nuclei isolated from cells with induced CSR1 expression, and higher levels of unpolyadenylated RNA for housekeeping genes. Second, knocking down CPSF3 using siRNA generated a similar pattern of cell death in PC3 cells to that induced by CSR1 induction. Third, expression of a CSR1 mutant that lacks CPSF3-binding activity does not result in translocation of CPSF3, and also could not induce cell death. These conclusions are further strengthened by the recent finding that downregulation of a certain CPSF3 homolog induced cell growth arrest in HeLa cells (Dominski et al., 2005b). The significance of the CSR1/CPSF3 interaction is twofold: first, it raises the possibility that other RNA processing enzymes are involved in cell death or cell growth regulation; second, it suggests that redistributing these enzymes is a prospective therapeutic approach to kill cancer cells.
The physiological role of CSR1 is not known. In contrast to p53, a tumor-suppressor gene that responds to cell stress such as radiation and chemical assault, there is substantial expression of CSR1 in mature prostate gland acinar cells. This observation suggests that CSR1 probably has a role beyond a cell stress-response gene. Most likely, CSR1 limits the overgrowth of acinar cells by inducing cell death. Even though CSR1 has only a short stretch of amino acids considered as a plausible extracellular domain, and it is not likely that it interacts with a ligand directly, it is quite possible that CSR1 acts as signal transducer for a membrane receptor. If so, the membrane receptor with which CSR1 interacts is likely a receptor known to be involved in cell growth and cell death regulation. The identification of the putative CSR1 coupling receptor and its ligand are critical to gain insight into the CSR1-mediated signaling mechanism.
Materials and methods
Plasmid construction
For construction of the pBD-CSR1c fusion proteins, a mutagenic primer set (TGTGGCCTATAATCATATGAA TGTCACCATCCTACGAGGTGCC and TTCATTTCAAG CAAAGTCGACGCCTGGATCTGCTCTGCGCCCCTC) was designed to create two restriction sites, Nde1 and Sal1 (New England Biolabs, MA, USA), in the C terminus 156 amino acids of CSR1, so that the PCR product could be ligated into a the pGBK7T vector (Clontech, Mountain View, CA, USA). A PCR reaction was performed on cDNA template of the donor prostate (Clontech) using the following conditions: 94 °C for 1 min followed by 35 cycles of 94 °C for 30 s, 68 °C for 3 min and a final 3 min extension step at 68 °C. The PCR product was cleaved with Nde1 and Sal1, gel purified and ligated into a similarly cleaved pGBKT7 vector. The fusion protein contained 156 amino acids from the CSR1 C terminus. The construct was transformed into One Shot competent cells (Invitrogen, Carlsbad, CA, USA). Plasmid DNA was extracted from selected transformed cells and digested with Nde1 and Sal1 to detect the presence of the insert. The coding frame was confirmed by automated sequencing.
For construction of pGST-CSR1c, a GST fusion protein, a mutagenic primer set (AGGAATTCAGTGGACACACAG CATGGAG/ATGCGGCCGCAGCCCTCCTCAGTAGAAG CT) was designed to create an EcoR1 and a Not1 cleavage site within the CSR1 coding region that encodes a 156 amino-acid region of the C terminus of CSR1. PCR was performed using these primers under the following conditions: 94 °C for 1 min followed by 35 cycles of 94 °C for 30 s, 68 °C for 3 min and a final 10 min extension step at 68 °C. The PCR product was subsequently gel purified and ligated into a pCR2.1 TA cloning vector (Invitrogen). The plasmid DNA was transformed into Escherichia coli. The plasmid DNA from the selected transformants was cleaved with EcoR1 and Not1, and ligated into a similarly cleaved pGEX-5x-3 vector in frame. A series of deletions including 5′ or 3′ deletions of pGST-CSR1c were performed using the primer sets listed in Table 2 (also, see Figure 1d for amino-acid sequences in the constructs). The procedures for generating these mutants were similar to those described for pGST-CSR1c. The pGST-CSR1c and its mutants were transformed into E. coli BL21 cells for recombinant protein production.
Table 2.
Primer sequences for constructing pGST-CSR1C deletion mutants
| Name | Sequences |
|---|---|
| GST-CSR1C1a | aggaattcagtggacacacagcatggag/tcagtcagtcacgatgcggccgctggtgaccctgtcttccctg |
| GST-CSR1C2a | aggaattcagtggacacacagcatggag/tcagtcagtcacgatgcggccgctatgtccccttttgggcct |
| GST-CSR1C3a | aggaattcagtggacacacagcatggag/tcagtcagtcacgatgcggccgcaacagggcccctttct |
| GST-CSR1C4a | aggaattcagtggacacacagcatggag/tcagtcagtcacgatgcggccgctttcgggcctctgccgc |
| GST-CSR1C1b | tgggatccccaggaattccgggcccccagggccagaa/atgcggccgcagccctcctcagtagaagct |
| GST-CSR1C2b | tgggatccccaggaattccggccctcgagggttccca/atgcggccgcagccctcctcagtagaagct |
| GST-CSR1C3b | tgggatccccaggaattccggagaccccggcatcttg/atgcggccgcagccctcctcagtagaagct |
Yeast transformation and library screening
The yeast AH109 competent cell preparation has been described previously (Yu and Luo, 2006). Freshly prepared AH109 competent cells (100 μl) were mixed with the pGBKT7-CSR1c plasmid (0.25–0.50 μg), plus 0.5 μg plasmid DNA from a prostate cDNA library constructed in pACT2, in 0.6 ml of PEG/LiAc buffer, and then incubated at 30 °C for 30 min. Following this initial incubation with plasmid DNA, the cell solution was combined with 70 μl of dimethyl sulfoxide (Sigma, St Louis, MO, USA) and incubated for 15 min at 42 °C. The cells were pelleted, resuspended in 0.5 ml YPDA medium and plated on low, medium and high-stringency SD agar plates, either directly, or from colonies grown on the low (SD-Leu/-Trp)- and medium (SD-Leu/-Trp/-His)-stringency plates to a high-stringency plate (SD-Ade/-His/-Leu/-Trp and X-α-Gal). The resulting colonies were subjected to the colony-lift filter β-galactosidase assay as described previously (Yu et al., 2006). pGBKT7-53 and pGADT7-T antigen were co-transformed into AH109 as a positive control, and pGBKT7-Lamin C with pGADT7-T as a negative control. PCL1 transformed into AH109 was used as a positive control for the galactosidase assay.
Validation of protein interactions in AH109
Plasmid DNA from positive clones (blue colonies on the high-stringency plate) were isolated from yeast, transformed into E. coli, and selected with ampicillin (100 μg/ml) to obtain the gene-ACT2 fusion protein that interacts with the bait-domain fusion protein. The individual purified pACT2/library plasmid DNA was then co-transformed with pGBKT7-CSR1c into AH109 yeast cells and grown in a high-stringency medium (SD-Ade/-His/-Leu/-Trp). The colony-lift filter β-galactosidase activity was assayed on the cells grown in this medium. The positive clones were then sequenced.
Immunoprecipitation
Precleared protein extracts of PC3 cells transformed with pCMV-CSR1 were incubated with CSR1 antibodies at 4 °C for 16 h, then with protein G sepharose for 3 h. The immune complex was pelleted and washed five times with RIPA buffer, and the bound proteins were eluted with SDS–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer.
GST fusion protein pull-down
The cells were grown in 5ml of LB medium (with 100 μg/ml ampicillin) overnight at 37 °C and diluted in 20 × LB medium, then incubated with shaking until the optical density reached 0.6–1.0, then induced by IPTG (final concentration of 1mM) for 3 h. The cells were then pelleted, resuspended in 1 × phosphate-buffered saline (PBS) and sonicated for 2 min. The proteins were solubilized in 1% Triton X-100. The supernatant was collected after centrifugation at 15 000 × g for 5 min. The GST and GST-CSR1c fusion proteins were purified through a Glutathione Sepharose 4B column (Amersham Pharmacia Biotech, Piscataway, NJ, USA). The PC3 cell protein extract was preincubated with the column for 15 min at 4 °C. The flow-through was collected after spinning at 3000 × g for 1 min. The flow-through cell lysates were incubated then with GST fusion protein packed Glutathione Sepharose 4B at 4 °C for 2 h. The column was spun at 3000 × g at room temperature for 1 min, and washed twice more with PBS. The protein was eluted from the column with 40 μl of SDS–PAGE sample buffer. The proteins were separated by SDS–PAGE and detected by western blot analysis.
Immunofluorescent staining
PC3 cells transfected with pCMV-CSR1 were cultured on chamber slides for 24 h. The slides were washed three times with PBS. The cells were fixed with 4% paraformaldehyde for 1 h at room temperature. After washing the slides twice with PBS, the cells were blocked with 10% donkey serum with 0.4% Triton X-100. The cells were then incubated with rabbit antiserum against CSR1 and goat antiserum against CPSF3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at room temperature for 1 h. The slides were washed twice with PBS. Secondary antibodies (fluorescein-conjugated donkey anti-goat and rhodamine-conjugated donkey anti-rabbit) were added and incubated at room temperature for 1 h. The slides were washed twice with PBS before the addition of 4′,6-diamidino-2-phenylindole, dihydrochloride. After additional washes with PBS, slides were mounted with Prolong Gold Antifade Reagent (Invitrogen). Immunofluorescent staining was examined under a confocal microscope.
TUNEL assay
The CSR1-inducible PC3 cells were plated on eight-well chamber slides for 1 day, and were treated with tetracycline (5 μg/ml) for 48 h. The slides were washed twice with PBS. The adherent cells were fixed with 4% formaldehyde. The terminal addition of fluorescein-labeled UTP was obtained by incubating the slides with 50 μl of TUNEL reaction buffer (Roche Applied Science, Nutley, NJ, USA) for 60 min at 37 °C in the dark. Then, the slides were washed three times before the quantification of cell death under an inverted fluorescence microscope as described previously (Wijsman et al., 1993). The experiments were performed in triplicate. PC3 cells not exposed to tetracycline were used as negative control; the same cells treated with DNase I to cleave the DNA before labeling procedure were used as positive control in the assay.
Fluorescence-activated cell sorting analysis of apoptotic cells
Cells with inducible CSR1 clones were plated and treated with tetracycline (5 μg/ml) as described above. These cells were trypsinized and washed twice with cold PBS. The cells were resuspended in 100 μl annexin-binding buffer (Molecular Probes, Eugene, OR, USA), and incubated with 5 μl Alexa Fluor 488-conjugated annexin V and 1 μl 100 μg/ml propidium iodide (PI;both from Molecular Probes) for 15 min in the dark at room temperature. The binding assays were terminated by the addition of 400 μl cold annexin-binding buffer. Fluorescence-activated cell sorting (FACS) analysis was performed using a BD-LSR-II flow cytometer (BD Science Inc., San Jose, CA, USA). The fluorescent-stained cells were analysed at emissions of 530 nm (FL1) and >575 nm (FL3). The negative controls, cells without Alexa Fluor 488 or PI in the incubation, were used to set the background level of fluorescence during the acquisition. Cells stained with only one fluorophore, either Alexa Fluor 488 or PI, were used for calibration and compensation before the acquisition. UV treated cells were used as positive control for the apoptotic cells. For each acquisition, 10 000 to 20 000 cells were sorted based on fluorescence: only green (apoptotic cells); only red or both red and green stained (late apoptotic stage or dead cells); or without any staining (live cells). WinMDI 2.8 software was used to analyse the data.
siRNA directed against CDSF3
For siRNA analysis, 125 pmol siRNA specific for CPSF3 (5′-CGAGAAUAAUACAUGAUCUUCCUACUU/GUAGGAAGAUCAUGUAUUAUUCUCG-3′) or CSR1 (UUGGAUUCCUUCCAGGCUGCRUCCUG/CAGGAGCAGCCUGGAAGGAAUCCAA) or nonspecific control (UAAUGUAUUGGAACGCAUAUU/UAUGCGUUCCAAUACAUUA) were transfected into PWC1 cells using the Lipofectamine 2000 transfection kit (Invitrogen). Immunoblots and FACS analyses were performed 24 h after transfection.
In vitro polyadenylation assay
A β-actin RNA without poly-A tail was used as the target for adenylation in the polyadenylation reaction and was obtained by in vitro transcription detailed as follows. The 737 bp fragment of T7 promoter driven β-actin C terminus was obtained through PCR on the cDNA template by using β-actin primers of GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGAGATCATTGCTCCTCCTGAGC/AAGGTGTGCACTTTTATTCAACTGGTC. For Δβ-actin, the primers are GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGAGATCATTGCTCCTCCTGAGC/CAACTGGTCTCAAGTCAGTGTACA. The PCR products were purified by electrophoresis in 1% agarose gel followed by the GeneClean kit (Bio101, San Diego, CA, USA). An in vitro transcription was performed using the purified PCR product of β-actin as the template at 37 °C for 4 h in a reaction mixture containing 10mM ATP, 10mM CTP, 10mM GTP and 10mM Fam-labeled UTP using the MEGAscript system (Ambion Inc., Austin, TX, USA) to produce Fam-labeled sense RNA (737 bases for β-actin and 719 for Δβ-actin) and further purified through an RNeasy column. The PC3 cells transfected with pCDNA4-CSR1 (PWC1) were induced with tetracycline (5 μg/ml) for 48 h. Then, the cells were treated with 320mM sucrose buffer (3mM CaCl2, 2mM MgAc and 0.1mM EDTA) with 0.5% NP-40 to lyse the cells and the nuclei pelleted by centrifugation at 1500×g for 5 min. In the polyadenylation reaction, the Fam-labeled RNA was incubated with 20 μg of nuclear or cytoplasmic protein from the induced or uninduced PWC1 or PWR1 cells in the presence of 10mM Texas Red-conjugated ATP, 20mM Tris–HCl, pH 7.5, 50mM KCl, 2.5mM MgCl2 and 50 μg/ml BSA at 37 °C for 90 min. The polyadenylation products were purified through a Microcon YM-30 column (Millipore, Temecula, CA, USA) to remove unincorporated nucleotides. The purified RNA was spotted in quadruplicate on a scanning glass side using the Affymetrix 417 arrayer, and Texas Red-labeled polyA and Fam-labeled RNA signals were quantified by scanning in an Affymetrix 428 scanner. The ratio of Texas Red to Fam was analysed using Juguar 2.0 software. The reaction mixture without stain was used to evaluate the background in the scanning.
Detection of polyadenylation by RNase protection assay
To generate DNA templates for in vitro transcription of GAPDH and β-actin, PCR was performed using primers specific to GAPDH (ACTCAGTCCCCCACCACACTGAATCTCCCCTC/GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24) and β-actin (CCCTTTTTTGTCCCCCAACTTGAGATGTATG/GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24) from a cDNA library from the prostate gland (BD Bioscience). PCR products were purified and transcribed in vitro to generate complementary RNA (cRNA) similarly to the procedure described above (polyadenylation assay) except that both CTP and UTP are biotin-labeled. These biotin-labeled RNA probes (148 bases for GAPDH and 143 for β-actin; 20 ng per sample), which include a stretch of 24 thymidine residues at the 5′end of the RNA, were hybridized with 10 μg total RNA extracted from PWC1 or PWR1 cells in hybridization buffer from the Multi-Probe RNase Protection Assay kit (BD Bioscience) at 56 °C for 16 h. The hybridization products were subsequently incubated with 100U RNase A at room temperature for 45 min. The reaction was terminated by adding 10U of proteinase K for 15 min at 37 °C. Then, the RNase digested products were precipitated using 4M LiCl in 80% ethanol. The products were centrifuged, washed with 90% ethanol and air-dried. The purified RNA products, the protected double-stranded RNA (GAPDH or β-actin), were separated by electrophoresis in a 4.75% polyacrylamide gel, electrotransferred onto a Nylon membrane, exposed to horseradish peroxidase-conjugated streptavidin and detected using a chemiluminescent probe detection kit (BD Bioscience).
Acknowledgements
This study was supported by grants from the National Cancer Institute to JHL (RO1 CA098249), the development fund from the Department of Urology, University of Pittsburgh and the John Rangos Foundation for Enhancement of Research in Pathology.
References
- Coon SW, Savera AT, Zarbo RJ, Benninger MS, Chase GA, Rybicki BA, et al. Prognostic implications of loss of heterozygosity at 8p21 and 9p21 in head and neck squamous cell carcinoma. Int J Cancer. 2004;111:206–212. doi: 10.1002/ijc.20254. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor cpsf-73 is involved in histone-pre-mrna processing. Cell. 2005a;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. A cpsf-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of cpsf-100. Mol Cell Biol. 2005b;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci M, Merola R, Farsetti A, Orlandi G, Sentinelli S, De Carli P, et al. Cytogenetic profiles as additional markers to pathological features in clinically localized prostate carcinoma. Cancer Lett. 2006;237:76–82. doi: 10.1016/j.canlet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Han HJ, Tokino T, Nakamura Y. Csr, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Hum Mol Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- Isaacs JT. Molecular markers for prostate cancer metastasis. Developing diagnostic methods for predicting the aggressiveness of prostate cancer. Am J Pathol. 1997;150:1511–1521. [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Kurimoto F, Gemma A, Hosoya Y, Seike M, Takenaka K, Uematsu K, et al. Unchanged frequency of loss of heterozygosity and size of the deleted region at 8p21–23 during metastasis of lung cancer. Int J Mol Med. 2001;8:89–93. doi: 10.3892/ijmm.8.1.89. [DOI] [PubMed] [Google Scholar]
- Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, et al. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- Ohata H, Emi M, Fujiwara Y, Higashino K, Nakagawa K, Futagami R, et al. Deletion mapping of the short arm of chromosome 8 in non-small cell lung carcinoma. Genes Chromosomes Cancer. 1993;7:85–88. doi: 10.1002/gcc.2870070204. [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor cpsf-73 is the mrna 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman JH, Jonker RR, Keijzer R, van de Velde CJ, Cornelisse CJ, van Dierendonck JH. A new method to detect apoptosis in paraffin sections: In situ end-labeling of fragmented DNA. J Histochem Cytochem. 1993;41:7–12. doi: 10.1177/41.1.7678025. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, Milchgrub S, Syed S, Lam S, et al. Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59:1973–1979. [PubMed] [Google Scholar]
- Yu G, Tseng GC, Yu YP, Gavel T, Nelson J, Wells A, et al. Csr1 suppresses tumor growth and metastasis of prostate cancer. Am J Pathol. 2006;168:597–607. doi: 10.2353/ajpath.2006.050620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Yu YP, Luo JH. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Research. 2006;66:7414–7419. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]