Abstract
Human papillomavirus (HPV) gene expression is dramatically altered during cervical carcinogenesis. Because dysregulated genes frequently show abnormal patterns of DNA methylation, we hypothesized that comprehensive mapping of the HPV methylomes in cervical samples at different stages of progression would reveal patterns of clinical significance. To test this hypothesis, thirteen HPV16-positive samples were obtained from women undergoing routine cervical cancer screening. Complete methylation data were obtained for 98.7% of the HPV16 CpGs in all samples by bisulfite-sequencing. Most HPV16 CpGs were unmethylated or methylated in only one sample. The other CpGs were methylated at levels ranging from 11% to 100% of the HPV16 copies per sample. The results showed three major patterns and two variants of one pattern. The patterns showed minimal or no methylation (A), low level methylation in the E1 and E6 genes (B), and high level methylation at many CpGs in the E5/L2/L1 region (C). Generally, pattern A was associated with negative cytology, pattern B with low-grade lesions, and pattern C with high-grade lesions. The severity of the cervical lesions was then ranked by the HPV16 DNA methylation patterns and, independently, by the pathologic diagnoses. Statistical analysis of the two rating methods showed highly significant agreement. In conclusion, analysis of the HPV16 DNA methylomes in clinical samples of cervical cells led to the identification of distinct methylation patterns which, after validation in larger studies, could have potential utility as biomarkers of neoplastic cervical progression.
INTRODUCTION
Persistent high-risk human papillomavirus (HPV) infection is a necessary cause of virtually all cases of cervical cancer. Routine cytologic screening of Papanicolau-stained cervical cells has dramatically reduced the incidence of cervical cancer, but cytology is not an ideal screening tool due to its low sensitivity for high grade lesions (Spitzer, 2002). This is why screening is typically repeated annually and why more than 5% of cervical cytology results without an obvious high-grade lesion (those with “atypical squamous cells of undetermined significance”) require close follow-up. The high cost of cervical cancer prevention is due to the frequency of Pap testing, the large number of cytologic and histologic follow-ups, and the expense of treating high grade CIN. A new HPV prophylactic vaccine is expected to further reduce the incidence of cervical cancer, although not for several years. Cervical cancer will continue to develop in unvaccinated women, the 3% of American women already infected with cancer-associated HPVs (Dunne et al., 2007) and women infected with high-risk types of HPV not in the vaccine, which cause about 30% of cervical cancer. The importance of continued cervical screening cannot be overemphasized, because inadequate screening in the post-vaccination era could negatively impact on the control of cervical cancer (Goldhaber-Fiebert et al., 2008)). At the same time however the sensitivity of cervical cytology will decline (Goldhaber-Fiebert et al., 2008). This situation urgently calls for the discovery of novel biomarkers of cervical oncogenesis (Kiviat, Hawes, and Feng, 2008). The hallmark of carcinogenesis is deregulation of cellular gene expression. A major mechanism controlling gene expression is DNA methylation. DNA methylation is a non-mutational, heritable and reversible epigenetic process. It is critical for normal development and cellular differentiation (Ballestar and Esteller, 2008), including epithelial differentiation (Paradisi et al., 2008). While the basic mechanism of DNA methylation has been known since the 1980s, intensive efforts to understand its role in gene regulation are more recent. Many studies indicate that the absence of DNA methylation in a gene promoter allows full expression of a gene, while its presence is correlated with gene silencing (Esteller, 2002). These studies are only a start though, as the vast majority of CpGs occur outside promoters and have not been surveyed systematically.
To better understand the role of DNA methylation in carcinogenesis, there is great desire to compare normal vs. malignant cells for differences in DNA methylation. Unfortunately the most accurate method for mapping the methylation status of each CpG in a DNA sequence, bisulfite-sequencing, is not cost-effective for this endeavor due to the very large size of the human genome (Bernstein, Meissner, and Lander, 2007). In contrast, bisulfite-sequencing is entirely sufficient for mapping the relatively tiny genomes of viruses. Viruses cause an estimated 15% of all human cancers (zur Hausen, 1991), and their study over the past thirty years has been integral to understanding molecular processes that regulate both normal and transformed cells (DiMaio and Miller, 2006).
Previous studies of HPV16 DNA methylation (Badal et al., 2003; Kalantari et al., 2004) showed that the viral long control region (LCR) and early region were relatively unmethylated in most cervical lesions. In contrast the late gene L1, encoding the major viral capsid protein, was methylated at several CpGs in cervical carcinoma cell lines and most cervical carcinoma tissues, but not in most asymptomatic infections. More variable results were reported for premalignant lesions although low- and high-grade cervical intraepithelial neoplasia were generally not analyzed individually (Badal et al., 2003; Kalantari et al., 2004). Similar results have been reported for HPV18 in cervical lesions (Badal et al., 2004; Kalantari et al., 2008a; Turan et al., 2006; Turan et al., 2007) and for HPV16 in anal intraepithelial neoplasias (Wiley et al., 2005), penile carcinomas (Kalantari et al., 2008b) and oral squamous cell carcinomas (Balderas-Loaeza et al., 2007). These studies established a trend for increasing HPV 16/18 DNA methylation, particularly in the L1 gene, with increasing lesion severity but they did not identify individual CpGs whose methylation status specifically correlated with the pathology. On the other hand, only a small region the HPV 16 or HPV 18 genome was previously mapped by bisulfite-sequencing (Badal et al., 2004; Kalantari et al., 2008a; Turan et al., 2006; Turan et al., 2007). We hypothesized that comprehensive mapping of all 113 sites of potential DNA methylation in the HPV16 genomes contained in patient samples of non-malignant cervical cells at different stages of progression might reveal patterns of diagnostic or prognostic significance. We report here the analysis of thirteen HPV16-positive non-malignant samples from women undergoing routine cervical cancer screening. HPV16 is the most common HPV type in anogenital cancer. The results show three principal HPV16 DNA methylation patterns, the heaviest of which has two variants. Assuming that DNA methylation represses HPV16 gene expression, the patterns are consonant with what is known regarding the biology of HPV-associated malignant progression. Furthermore, they show significant agreement with the cervical diagnoses arrived at by pathologic examination.
RESULTS
Identification of HPV16-positive cervical samples
To identify samples of cervical cells containing HPV16 DNA, 72 samples collected for routine cervical screening were evaluated by PCR using two primer pairs specific for different regions of the HPV16 genome. Thirteen samples that generated clear bands of the appropriate sizes on agarose gels were selected for further study.
Bisulfite-conversion and primer design
The most accurate method for determining the methylation status of every CpG in a DNA sequence is bisulfite-sequencing. In this method, sodium bisulfite converts all unmethylated cytosines (C) to uracils but leaves methylated Cs intact. PCR is then performed to amplify the bisulfite-treated DNA and convert the uracils to thymines (T). Finally the original status of each C in the PCR product is determined by DNA sequencing, which shows C if the original C was methylated (meCpG) or T if it was not (CpG). Preliminary studies assessed the completeness of bisulfite-conversion using two substrates. One was a plasmid containing the full-length genome of the W12 isolate of HPV16 (Flores et al., 1999), which we methylated in vitro and amplified with primers described below. The other was the Universal Methylated DNA Standard, amplified with its own primers. Both control assays demonstrated complete bisulfite-conversion. As bisulfite-converted DNA contains only adenine, thymine and guanosine, except at methylated cytosines, the HPV16 genome was bisulfite-converted (in silico) for primer design. Twenty primer pairs were designed to collectively amplify all 113 CpGs in the HPV16 genome (Supplementary Table 1). PCR amplification conditions were then optimized for each primer pair using the molecular clone of the W12 isolate of HPV16 (Alazawi et al., 2002) after actual in vitro DNA methylation and bisulfite-conversion.
PCR amplification
The DNAs from the thirteen cervical samples were bisulfite-converted and amplified with each of the first twenty primer pairs (Supplementary Table 1). This work required further optimization of the PCR conditions for some products and some samples. Ultimately, all PCR products were amplified from all samples, indicating that each sample contained all parts of the HPV16 genome (data not shown). The data suggest the presence of episomal HPV16 DNA but do not exclude the possibility of integrated HPV16 DNA in the form of concatamers or partially deleted/rearranged genomes, alone or in combination with episomal copies. Integrated HPV16 DNA is found in about 6% of CIN2 lesions and 19% of CIN3 lesions, while low-grade lesions contain only episomal viral genomes (Vinokurova et al., 2008).
Complete and missing data
To determine which HPV 16 CpGs were methylated, the PCR products from each cervical sample were sequenced in both directions. In some cases, the DNA sequences were not fully readable. In those cases and others, the relevant DNA samples were reamplified and again sequenced in both directions. The data showed that the HPV 16 sequences in all samples were essentially identical to the W12 variant (Flores et al., 1999) and that virtually all cytosines that were not part of CpGs had been converted to thymines, i.e. methylated CpAs, CpTs and CpGs were very rare.
Some data were initially missing, most frequently in PCR product 12 (Supplementary Table 2). The difficulty obtaining readable sequence from product 12 was most likely due to secondary structure resulting from an extraordinarily high concentration of A+T nucleotides, the frequency of which would range from 86 to 88%, depending the methylation frequency. In an attempt to obtain additional data, two new primer pairs were designed that together amplified a region containing the sequence of product 12 plus additional upstream and downstream sequences (primers 12-1 and 12-2, Supplementary Table 1). DNA sequencing of the new PCR products provided new data for 12 CpGs with previously missing data.
A subset of samples still had partially missing data at eight CpGs (Supplementary Table 2). Among the samples with complete data, five of the eight CpGs were unmethylated and three were methylated at low levels, ranging from 4.5 to 8.2% of the HPV16 copies. All together, complete methylation data were obtained for 1450 of the 1469 CpGs in the thirteen samples (98.7%).
Distribution and methylation frequency of all HPV16 CpGs
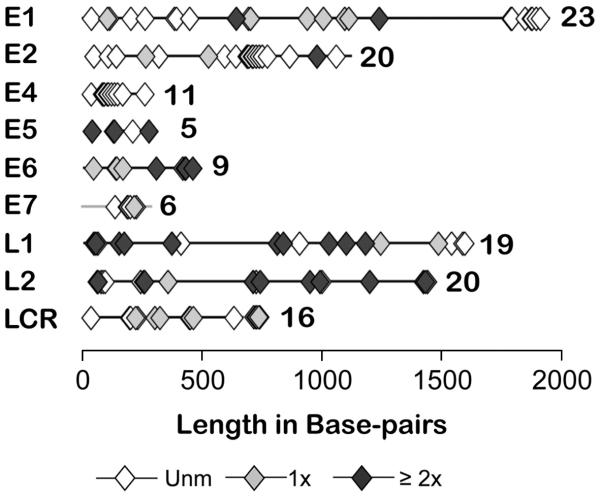
The HPV16 genome contains 113 CpGs, and multiple CpGs occur in each gene as well as the LCR. Direct sequencing of the individual PCR products frequently showed the presence of C and T in different reactions and/or C in one sequencing direction and T in the other. The variation was probably due to the heterogeneity of the population of HPV 16 molecules in the original sample, and hence PCR product, and not to hemimethylation. This interpretation was also supported by peaks that contained both C and T in some chromatograms (data not shown). To estimate the frequencies of methylation among the HPV16 copies per sample, we averaged the mean frequencies per sample, calculated by averaging all the sequence data (2.7 ± 0.1 readable sequences per CpG per sample (mean ± S.E.M.)). The frequency of methylation at the individual meCpGs ranged from a mean of 11.1% (at four CpGs with nine data points) to 100% of the HPV16 genomes per sample. Fifty-two CpGs, including all in the E4 ORF, all but one in the E7 ORF, and most in the E2 ORF were not methylated in any sample (Fig. 1). Thirty-two CpGs were methylated in just one sample, and only 29 were methylated in multiple samples.
Figure 1.
Distribution among cervical samples of methylated and unmethylated CpGs in the HPV16 open reading frames and long control region.
The diamonds represent CpGs that were unmethylated in all samples (◇), in a single sample (grey  ), or in multiple samples (◆). The number of CpGs in each ORF/LCR is listed to the right of each schematic.
), or in multiple samples (◆). The number of CpGs in each ORF/LCR is listed to the right of each schematic.
HPV16 DNA methylation patterns in clinical samples
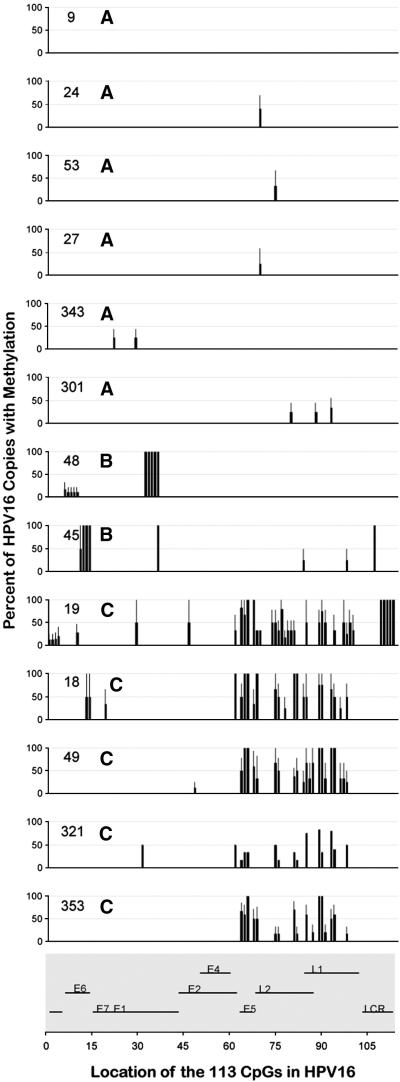
We next examined the profiles of the HPV16 methylomes within each sample. As shown in Fig. 2, most methylated CpGs were heterogeneously methylated, as indicated by levels greater than 0 and less than 100% and by the error bars. From visual inspection of the individual profiles three patterns were deduced (Fig. 2). The first six samples were completely or nearly completely unmethylated (pattern A). The next two were methylated at limited numbers of CpGs, primarily in the early region (pattern B). The last five were heavily methylated at several CpGs, primarily in the late region (pattern C). Overall, the number of CpGs methylated per sample was 1.3 ± 0.4 in pattern A, 9.0 ± 1.0 in pattern B, and 22.2 ± 3. in pattern C (mean ± S.E.M.). While some meCpGs occurred in only one sample per pattern and therefore were not part of the pattern per se, the different levels indicate different degrees of susceptibility to DNA methylation (or demethylation) at different stages of disease.
Figure 2.
HPV16 DNA methylation profiles of the individual cervical samples The HPV16 DNA methylation profile of each sample at each potential methylation site is shown. The numbers inside each plot are the sample numbers and the adjacent letters are the HPV16 DNA methylation pattern (A, B or C). Nucleotides other than the 113 CpGs are not shown. The organization of the HPV16 genome with sequential numbering of the CpGs is shown at the bottom of the figure.
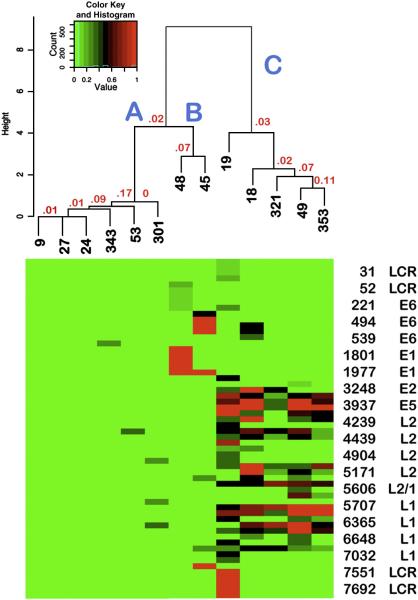
Cluster analysis: identification of related subgroups of HPV16 DNA methylation profiles
To further examine the relationships among the thirteen cervical samples, the data were subjected to cluster analysis. As shown in Fig. 3, the samples with patterns A and B formed one major branch with two closely related subgroups, one consisting of the six samples with pattern A, and the other, the two samples with pattern B. The second major branch contained the five samples with pattern C. As the probabilities of the clusters occurring by chance were small (Fig. 3), our visual impressions were validated.
Figure 3.
Heat Map: Relationship of the thirteen HPV16 DNA methylation profiles Shown is the heat map derived by Cluster analysis and, above it, the corresponding dendrogram with the sample numbers in black and P-values in red. The original values are shown without scaling. Each branch is labeled with the corresponding HPV16 methylation pattern (A, B or C). The CpGs are shown in the same order as they occur in the HPV16 genome; every fourth CpG is listed on the y-axis. Not shown are the 52 CpGs without any methylation and the eight with partially missing data (see Supplementary Table 2).
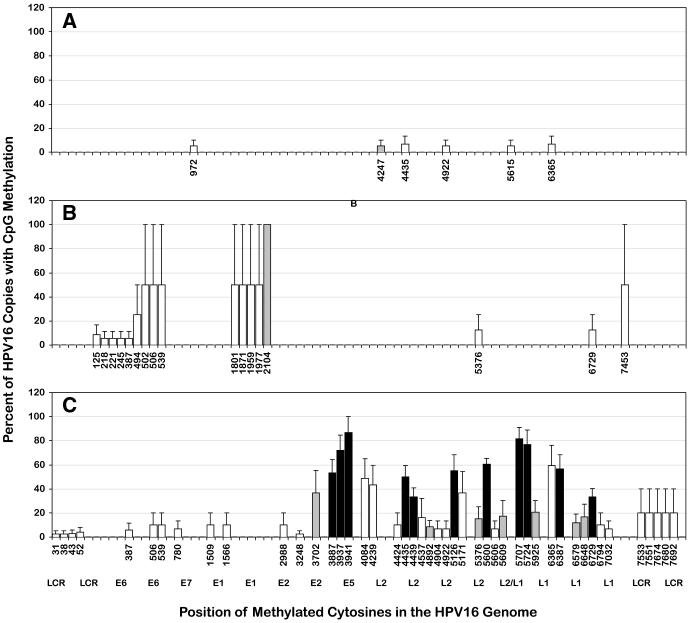
We then plotted the mean data for each pattern to relate its major features to the HPV16 genome. We also distinguished between CpGs that were methylated in multiple samples (potentially part of a pattern) vs. only one. In pattern A, up to three CpGs were methylated, at low frequency, and they were located variously in the E1, L2 or L1 ORFs (Fig. 4A). Only one CpG in pattern A was methylated in two samples (in the L2 gene) and none in more than two. In pattern B, both samples were methylated at four or five contiguous CpGs in the E6 open reading frame (ORF) (from position 125 to 387 in sample #45, or position 494 to 539 in sample #48) (Fig. 4B). One (sample #45) also was methylated in the LCR at the CpG in E2 binding site 4 (E2BS#4). In pattern C, all five samples were heavily methylated at eleven CpGs located in the E5/L2/L1 region, nine of which were pattern C-specific, i.e. not methylated in any other sample. The nine pattern C-specific CpGs included three of five in the E5 ORF, three of twenty in the L2 gene and four of nineteen in the L1 gene (one overlapping the L2 gene) (Fig. 4C) (for positions see Table 1). In summary, pattern A was characterized by minimal methylation, pattern B by methylation in the E1 and E6 ORFs, and pattern C by heavy methylation at nine specific CpGs located at the end of the early region (E5) and in the L2 and L1 ORFs but not including the L1 terminus.
Figure 4.
HPV16 DNA methylation patterns A, B and C
The panels show three HPV16 methylation patterns: A, B and C, respectively. The x-axis is the same for all panels, but only the CpGs with methylation in each pattern are labeled. The nucleotide numbers are those of the W12 isolate of HPV16 (Flores et al., 1999) (Genbank AF125673). The bars represent CpGs that were methylated in only one sample per pattern (□); only two samples (patterns A and B) or two to four samples (pattern C) (grey  ), and all five samples of pattern C (■). One sample in pattern A was missing data at position 1509, and one in pattern C, at position 4247. Not shown are the 52 unmethylated CpGs. The y-axis shows the mean percentage of HPV16 genomes per sample with methylation at each CpG plus the S.E.M.
), and all five samples of pattern C (■). One sample in pattern A was missing data at position 1509, and one in pattern C, at position 4247. Not shown are the 52 unmethylated CpGs. The y-axis shows the mean percentage of HPV16 genomes per sample with methylation at each CpG plus the S.E.M.
Table 1.
Methylated CpGs specific to pattern C and unique to the C-2 variant
| Feature Location | Cytosine a | Methylation Frequency b |
|---|---|---|
| Specific to pattern C (variants C-1 and C-2) c | ||
| E5 | 3887 | 53.3 ± 11.1 |
| E5 | 3937 | 72.0 ± 12.7 |
| E5 | 3941 | 86.7 ± 13.3 |
| L2 | 4439 | 33.3 ± 7.5 |
| L2 | 5126 | 55.0 ± 13.3 |
| L2/L1 | 5600 | 60.3 ± 4.8 |
| L1 | 5707 | 81.7 ± 9.3 |
| L1 | 5724 | 76.7 ± 12.2 |
| L1 | 6387 | 56.7 ± 11.7 |
| Unique to variant C-2 d | ||
| Enhancer | 7533 | 100.0 |
| Enhancer | 7551 | 100.0 |
| Enhancer | 7674 | 100.0 |
| Enhancer e | 7680 | 100.0 |
| Enhancer | 7692 | 100.0 |
| Promoter | 31 | 12.5 |
| Promoter e | 38 | 12.5 |
| Promoter | 43 | 14.3 |
| Promoter e | 52 | 20.0 |
| E2 | 2988 | 50.0 |
| L2 | 4424 | 50.0 |
| L2 | 4537 | 80.0 |
| L2 | 4904 | 33.3 |
| L2 | 5171 | 0.0 f |
| L1 | 6365 | 0.0 g |
| L1 | 6794 | 50.0 |
| L1 | 7032 | 33.3 |
Position of the cytosine in the HPV16 genome.
Percent of HPV16 genomes per sample with methylation (mean ± S.E.M.).
Each CpG was sequenced 3.9 ± 0.3 times per sample (mean ± S.E.M.).
Each CpG was sequenced 4.7 ± 0.4 times (mean ± S.E.M.) in sample #19, with variant pattern C-2.
Cytosine located within an HPV16 E2 protein-binding site (E2BS).
Methylated in all pattern C samples except #19 at 45.8% ± 19.7% of the HPV16 copies per sample (mean ± S.E.M.)
Methylated in all pattern C samples except #19 at 74.2 % ± 10.6% of the HPV16 copies per sample (mean ± S.E.M.)
Correlation of pathology with HPV16 DNA methylation
The cervical cytology results and pathologic diagnoses were reviewed by an expert cytopathologist (M.H.) and two expert histopathologists (M.M. and G.K.H. III). Five samples were diagnosed by cytology alone: two as negative, two as atypical squamous cells of undetermined significance (ASC-US, an equivocal diagnosis), and one as a low-grade squamous intraepithelial lesion (LSIL). Four samples with histologic follow-up were diagnosed as low-grade cervical intraepithelial neoplasia (CIN1), and four others as high grade CIN (CIN2/3). We speculated that the high-grade lesions (CIN2/3) would have pattern C because DNA methylation generally silences gene expression and the heavily methylated HPV genes in pattern C are silenced during malignant progression. We further speculated that the negative samples would have pattern A, i.e. be the most different from pattern C, and that the CIN1 lesions, being intermediate in severity, would have an intermediate but functionally distinct level of methylation, i.e. pattern B. Three samples with ASC-US or LSIL and no follow-up had insufficient data for definitive diagnosis. Of the other samples, two with pattern A had negative cytology, both samples with pattern B showed signs of HPV infection and cervical intraepithelial neoplasia grade 1 (CIN1), i.e. low-grade lesions, and three with pattern C were high-grade lesions (CIN2/3) (Fig. 4). Thus seven of the ten cases with definitive diagnoses had the predicted correlation.
One apparently discordant low-grade lesion with pattern C (#19) was found upon re-review of the bisulfite sequencing data to be methylated at 15 CpGs that were not methylated in any other sample (Table 1). It was also completely unmethylated at two CpGs that were methylated in every other pattern C sample (Table 1). Re-review of the cluster analysis reinforced the importance of the differentially methylated CpGs because it showed that sample #19 was more distantly related to the other pattern C samples than they were to each other, by a factor of approximately two (Fig. 3). Together, the data led to the conclusion that sample #19 had a distinct variant of pattern C. Pattern C was therefore divided into a C-2 variant represented by sample #19 and a C-1 variant represented by the other samples.
The uniquely methylated CpGs in pattern C-2 (sample #19) included nine in the LCR. Five of six in the keratinocyte-specific enhancer were completely methylated and four of the five in the E6/E7 promoter were lightly methylated (Table 1). Since DNA methylation in the 5′ regulatory regions of genes, including the HPV16 LCR (Kim et al., 2003), usually represses expression of the gene(s) it controls, E6/E7 expression may have been compromised in sample #19.
The remaining two cases in which the HPV16 DNA methylation pattern did not agree with the pathologic diagnoses showed no meaningful differences in HPV16 methylation. One sample (#343) clearly had HPV16 DNA methylation pattern A but showed a high grade lesion by both cytology and histology. The other one (sample #353) had the high-risk variant of pattern C but was diagnosed as CIN1. The early region of HPV16 in sample #353 was completely devoid of methylation, while the same region of the other pattern C samples, including sample #19, had one to four methylated CpGs. This tiny difference was however insufficient to consider the pattern of sample #353 as a variant.
Finally, we evaluated the extent of agreement between the HPV16 DNA methylation patterns and the pathologic diagnoses, after classifying pattern C-2 as a low-grade variant and excluding the three samples without definitive diagnoses (Table 2). Statistical analysis showed highly significant concordance between the two methods (P=0.005, Cohen’s Kappa statistic).
Table 2.
Lesion severity ranked by HPV16 DNA methylation pattern vs. pathologicg
| Pap result | HPV16 DNA methylation pattern |
Rank | Pathologic diagnosis | ||
|---|---|---|---|---|---|
| Neg. a | CIN1 | CIN2/3 | |||
| Rank | |||||
| 1 | 2 | 3 | |||
| Neg. a | A | 1 | #27 f | ||
| Neg. | A | 1 | #53 | ||
| HSIL b | A | 1 | #343 | ||
| ASC-US c | B | 2 | #48 | ||
| ASC-H d | B | 2 | #45 | ||
| LSIL e | C-2 | 2 | #19 | ||
| ASC-US | C-1 | 3 | #353 | ||
| ASC-US | C-1 | 3 | #18 | ||
| ASC-US | C-1 | 3 | #49 | ||
| ASC-US | C-1 | 3 | #321 | ||
Cytologically negative for intraepithelial lesion or malignancy.
High-grade squamous intraepithelial lesion.
Atyical squamous cells of undetermined significance.
ASC-US cannot rule out HSIL.
Low-grade squamous intraepithelial lesion (diagnosed as ASC-US before expert review).
Number of the cervical sample.
DISCUSSION
While it has been known for 25 years that papillomavirus genomes are highly methylated in carcinomas (Wettstein and Stevens, 1983), mapping the methylation status of specific sites began only recently. Previous studies of HPV16 DNA methylation mapped up to 19 CpGs at the 3′ end of the L1 ORF and the LCR (Badal et al., 2004; Kalantari et al., 2004). We report here the precise mapping of 98.7% of all CpGs in the HPV16 genomes contained in thirteen cellular samples of cervical cells with pathologic diagnoses ranging from negative to CIN3. To our knowledge, this is the first report of comprehensive DNA methylation mapping for the entire genome of any virus. It is also the first to identify unique HPV16 DNA methylation marks that distinguish high-grade lesions from low-grade lesions and asymptomatic infections.
Most of the 113 CpGs in the HPV 16 genome were unmethylated or methylated in only one sample. The methylated CpGs were located mainly in the bodies of HPV16 genes. CpG methylation within gene bodies has been previously reported, but little is known about the prevalence of such events nor their biologic significance. Methylation within a gene body might repress expression of the gene in which it resides, another gene(s) via regulatory elements contained within the first gene, or merely reflect the stage of malignant progression in neoplastic cells.
Repeat sequencing of the HPV 16 PCR products revealed heterogeneous methylation at most methylated CpGs, as previously reported for smaller genomic regions by molecular cloning (Kalantari et al., 2004; Turan et al., 2006; Turan et al., 2007). Despite the heterogeneity, the HPV16 methylomes in each sample showed one of three distinct patterns of HPV16 DNA methylation or a variant of the most highly methylated pattern. The existence of a limited number of patterns indicates that transfer and/or removal of methyl groups to CpGs in the HPV16 genome is not a random process, that cells with methylation (or not) at particular HPV16 CpGs have a selective growth advantage, and/or that the methylation of certain CpGs is incompatible with continued infection.
Pathologically, the samples with almost no HPV16 DNA methylation (pattern A) were the least severe, those with several methylated CpGs in the E1 and E6 ORFs were intermediate in severity (pattern B), and those with high frequency methylation, particularly in the E5/L2/L1 region (pattern C), the most severe. Excluding three samples with insufficient pathology, the HPV16 DNA methylation patterns ranked the severity of eight of ten lesions identically to the pathologic diagnoses. Moreover the agreement was statistically significant (P=0.005). Thus neoplastic progression was generally associated with increasing numbers of methylated CpGs and increasing proportions of methylated HPV16 molecules, in agreement with previous findings (Badal et al., 2004; Badal et al., 2003; Kalantari et al., 2004; Turan et al., 2006; Turan et al., 2007). Our results also expand previous findings by identifying nine specific CpGs in the E5 ORF, L2 ORF and 5′ two-thirds of the L1 gene that were highly methylated in high-grade lesions (Table 2).
High levels of HPV16 CpG methylation do not however necessarily indicate a high grade lesion, because the sample with by far the most methylation was a low-grade lesion with the C-2 variant pattern. In this lesion expression of the E6/E7 oncogenes may have been repressed due to the methylation of more than 80% of the CpGs in the enhancer/promoter region. Interestingly, the lesion spontaneously regressed as shown by follow-up cytology one year after biopsy. As the growth of transformed cervical cells relies on continued E6/E7 expression (DeFilippis et al., 2003), it is tempting to speculate that methylation of the HPV 16 enhancer/promoter region was mechanistically involved in mediating regression. It is worth noting in this context that complete methylation of the HPV 16 promoter/enhancer region has previously been reported in a subset of asymptomatic infections (Badal et al., 2003) that might have been regressing.
The 3′ terminus of the L1 ORF was completely unmethylated in all our samples. Previous bisulfite-sequencing studies have similarly reported the absence of methylation at the L1 terminus in most asymptomatic and low-grade cervical lesions as well as some high-grade cervical lesions (Kalantari et al., 2004; Turan et al., 2006; Turan et al., 2007). Other high-grade lesions however were highly methylated at the L1 terminus (Kalantari et al., 2004; Turan et al., 2006; Turan et al., 2007). Since DNA methylation and HPV gene expression both change dramatically during epithelial differentiation (Paradisi et al., 2008; Zheng and Baker, 2006), and the L1 terminus of episomal HPV16 transits from a hypermethylated state in undifferentiated cervical cells to a hypomethylated state upon the induction of differentiation (Kalantari et al., 2008a), the absence of methylation at the L1 terminus in at least some of our high grade lesions may reflect the absence of undifferentiated keratinocytes in cytology samples (this study) vs. tissues samples (previous studies). The relative uniformity of epithelial differentiation in our cervical samples may also have facilitated the identification of specific methylation differences between lesions with different diagnoses.
There were two discordant cases. The low-grade lesion (CIN1) with the C-1 methylation pattern was persistent as shown by follow-up cytology (LSIL) seven months after the biopsy (sample #353). This outcome suggests the possibility that the C-1 variant in a patient with CIN1 could indicate persistence. Persistent lesions are at greatly increased risk of malignant progression (Schiffman et al., 2005). The other case was a high-grade lesion (HSIL and CIN2) with pattern A (#343). In this case we suspect that the high grade pathology was caused by co-infection with a high-risk HPV type(s) other than HPV 16. Since concurrent HPV infections are common (Trottier et al., 2006), the evaluation of HPV co-infection and the mapping of additional HPV methylomes will be important in future studies.
In summary, HPV16 DNA methylation patterns have the potential to provide useful biomarkers of cervical carcinogenesis. Future validation studies will map the HPV methylomes in larger numbers of cervical samples, infected (and co-infected) with various types of HPV, and collected together with clinical follow-up data at multiple time points. Such studies may identify specific combinations of HPV methylation marks with definitive prognostic and/or diagnostic value that could be easily be incorporated into routine screening assays.
MATERIALS AND METHODS
Origin of samples
Seventy-two residual samples of exfoliated cervical cells were obtained from patients being routinely screened for cervical cancer by the Department of Pathology at Yale University. The cervical cells were collected in either PreservCyt® solution and held at room temperature for approximately one month prior to use. The samples were collected between March and October of 2007. The samples were obtained with approval from the Yale Human Investigation Committee.
DNA purification
High molecular weight DNAs were extracted from cervical samples using MasterPure™ DNA purification kits (EPICENTRE®Biotechnologies, Madison, WI 85201).
HPV16 DNA screening
Sample DNAs were screened for HPV16 DNA by PCR using two primer pairs that amplified fragments containing nucleotides (nts) 79 to 559 or nts 1800 to 1942. PCR reactions were performed using Taq PCR Master Mix Kits (Qiagen Inc., Valencia, CA 91355) with primers at 10 μm concentration. The PCR profile was: 94°C × 5 minutes, followed by 35 cycles of 94°C × 20 seconds, 55°C × 45 seconds × 72°C for 1 minute, with a final incubation at 72°C for 10 minutes.
Bisulfite modification
Sample DNAs were modified using the DNA Methylation-Gold Kit™ (catalog number D5006, Zymo Research Corp., Orange, CA 92867) according to the manufacturer’s instructions. To control for complete bisulfite-conversion, we methylated a plasmid containing the complete genome of the W12 isolate of HPV16 in vitro using the CpG Methyltransferase SssI (New England Biolabs, Ipswich, MA) and used it as a substrate. Other control reactions used the Universal Methylated DNA Standard (catalog number D5010, (ZYMO Research, Orange, CA 92867).
Amplification of bisulfite-treated DNAs
Twenty-two pairs of primers were designed to amplify bisulfite-modified HPV16 DNA using the MethPrimer Design program (urogene.org/methprimer/index1.html) (Li). The primers are listed in Table 1 without M13 tails, which were added to facilitate DNA sequencing. The PCR amplification conditions for each primer set were optimized for MgCl2 concentration (1.5 to 4.0 μm) and annealing and elongation temperature (50°C to 68°C). Each PCR reaction contained 1.25 units of AmpliTaq Gold (catalog number 808-0241) (Roche Applied Science, Indianapolis, IN 46250) and 0.5 units of PfuTurbo® polymerase (Stratagene, La Jolla, CA). The standard optimized PCR profile was 95°C × 10 minutes, followed by five cycles of 95°C × 1 minute, 54°C to 60°C × 2 minutes, 72°C × 3 minutes, and 35 cycles at 95°C × 1 minute, 60°C × 2 minutes, 72°C × 2 minutes, with a final incubation at 72°C for 10 minutes. PCR reactions were performed in a MasterCycler® Gradient (Eppendorf Scientific Inc., Westbury, NY 11590). The production of each PCR product was confirmed by electrophoresis in ethidium-bromide stained agarose gels. Further optimization was required to amplify some PCR products from several patient samples.
DNA sequencing
PCR products were purified and sequenced by Agencourt Bioscience Corporation (Beverly, MA), and the DNA sequencing data were analyzed using the multiple sequence alignment program Clustal W (http://www.ebi.ac.uk/Tools/clustalw/).
Statistical analysis
Hierarchical clustering was performed using the hclust library in the R statistical package (Team, 2007) using Ward’s minimum variance method with Euclidean distance metric. Above-average expression is in red, whereas below-average expression is in green. The dendrograms were generated as defined for hierarchical clustering. Cluster stability was evaluated and permutation-based cluster stability P-values calculated using the multi-scale permutation clustering (R package ‘pvclust’ (Suzuki and Shimodaira, 2006)).
Supplementary Material
Acknowledgments
We thank Daniel DiMaio for helpful comments on the manuscript. We thank Xueguang Sun and Andrew Dyer for helpful technical suggestions. The study was funded by a pilot grant from the Yale Comprehensive Cancer Center, a generous gift from Laurel Schwartz, and in part by the Yale Center of Excellence in Molecular Hematology, DK072442.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Cancer Society . Cancer facts & figures 2007. ACS; 2007. [Google Scholar]
- Alazawi W, Pett M, Arch B, Scott L, Freeman T, Stanley MA, Coleman N. Changes in cervical keratinocyte gene expression associated with integration of human papillomavirus 16. Cancer Research. 2002;62(23):6959–65. [PubMed] [Google Scholar]
- Badal S, Badal V, Calleja-Macias IE, Kalantari M, Chuang LSH, Li BFL, Bernard H-U. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324(2):483–92. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Badal V, Chuang LSH, Tan EH-H, Badal S, Villa LL, Wheeler CM, Li BFL, Bernard H-U. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. Journal of Virology. 2003;77(11):6227–34. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderas-Loaeza A, Anaya-Saavedra G, Ramirez-Amador VA, Guido-Jimenez MC, Kalantari M, Calleja-Macias IE, Bernard H-U, Garcia-Carranca A. Human papillomavirus-16 DNA methylation patterns support a causal association of the virus with oral squamous cell carcinomas. International Journal of Cancer. 2007;120(10):2165–9. doi: 10.1002/ijc.22563. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Esteller M. Epigenetic gene regulation in cancer. Advances in Genetics. 2008;61:247–67. doi: 10.1016/S0065-2660(07)00009-0. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- DeFilippis R, Goodwin E, Wu L, DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. Journal of Virology. 2003;77(2):1551–63. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Miller G. Thirty years into the genomics era: tumor viruses led the way. Yale Journal of Biology & Medicine. 2006;79(3-4):179–85. [PMC free article] [PubMed] [Google Scholar]
- Dunne EF, Unger ER, Sternberg M, McQuillan G, Swan DC, Patel SS, Markowitz LE. Prevalence of HPV infection among females in the United States. JAMA. 2007;297(8):813–9. doi: 10.1001/jama.297.8.813. [see comment] [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–40. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Flores ER, Allen-Hoffmann BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262(2):344–54. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- Goldhaber-Fiebert JD, Stout NK, Salomon JA, Kuntz KM, Goldie SJ. Cost-effectiveness of cervical cancer screening with human papillomavirus DNA testing and HPV-16,18 vaccination. Journal of the National Cancer Institute. 2008;100(5):308–20. doi: 10.1093/jnci/djn019. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Calleja-Macias IE, Tewari D, Hagmar B, Lie K, Barrera-Saldana HA, Wiley DJ, Bernard H-U. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. Journal of Virology. 2004;78(23):12762–72. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Lee D, Calleja-Macias IE, Lambert PF, Bernard H-U. Effects of cellular differentiation, chromosomal integration and 5-aza-2′-deoxycytidine treatment on human papillomavirus-16 DNA methylation in cultured cell lines. Virology. 2008a;374(2):292–303. doi: 10.1016/j.virol.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari M, Villa LL, Calleja-Macias IE, Bernard H-U. Human papillomavirus-16 and -18 in penile carcinomas: DNA methylation, chromosomal recombination and genomic variation. International Journal of Cancer. 2008b;123(8):1832–40. doi: 10.1002/ijc.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Garner-Hamrick PA, Fisher C, Lee D, Lambert PF. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. Journal of Virology. 2003;77(23):12450–9. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviat NB, Hawes SE, Feng Q. Screening for cervical cancer in the era of the HPV vaccine--the urgent need for both new screening guidelines and new biomarkers. Journal of the National Cancer Institute. 2008;100(5):290–1. doi: 10.1093/jnci/djn038. [comment] [DOI] [PubMed] [Google Scholar]
- Li L-C. MethPrimer - Design Primers for Methylation PCRs. [DOI] [PubMed]
- Paradisi A, Pasquariello N, Barcaroli D, Maccarrone M. Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. Journal of Biological Chemistry. 2008;283(10):6005–12. doi: 10.1074/jbc.M707964200. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Herrero R, Desalle R, Hildesheim A, Wacholder S, Rodriguez AC, Bratti MC, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Wright TC, Solomon D, Chen Z, Schussler J, Castle PE, Burk RD. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337(1):76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Spitzer M. In vitro conventional cytology historical strengths and current limitations. Obstetrics & Gynecology Clinics of North America. 2002;29(4):673–83. doi: 10.1016/s0889-8545(02)00025-6. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22(12):1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Team RDC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- Trottier H, Mahmud S, Costa MC, Sobrinho JP, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL, Franco EL. Human papillomavirus infections with multiple types and risk of cervical neoplasia. Cancer Epidemiology, Biomarkers & Prevention. 2006;15(7):1274–80. doi: 10.1158/1055-9965.EPI-06-0129. [DOI] [PubMed] [Google Scholar]
- Turan T, Kalantari M, Calleja-Macias IE, Cubie HA, Cuschieri K, Villa LL, Skomedal H, Barrera-Saldana HA, Bernard H-U. Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression? Virology. 2006;349(1):175–83. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Turan T, Kalantari M, Cuschieri K, Cubie HA, Skomedal H, Bernard H-U. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology. 2007;361(1):185–93. doi: 10.1016/j.virol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Durst M, Schneider A, von Knebel Doeberitz M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Research. 2008;68(1):307–13. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- Wettstein FO, Stevens JG. Shope papilloma virus DNA is extensively methylated in non-virus-producing neoplasms. Virology. 1983;126(2):493–504. doi: 10.1016/s0042-6822(83)80007-5. [DOI] [PubMed] [Google Scholar]
- Wiley DJ, Huh J, Rao JY, Chang C, Goetz M, Poulter M, Masongsong E, Chang CI, Bernard H-U. Methylation of human papillomavirus genomes in cells of anal epithelia of HIV-infected men. Journal of Acquired Immune Deficiency Syndromes: JAIDS. 2005;39(2):143–51. [PubMed] [Google Scholar]
- Zheng Z-M, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Frontiers in Bioscience. 2006;11:2286–302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Viruses in human cancers. Science. 1991;254(5035):1167–73. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.