Abstract
K+ channels are highly selective for K+ over Na+. Here we present several crystal structures of the MthK K+ channel pore at up to 1.45 Å resolution. The MthK selectivity filter maintains a conductive conformation even in the absence of K+, allowing the channel to conduct Na+. The high resolution structures along with single channel recordings allow for an accurate analysis of how K+ competes with Na+ in a conductive selectivity filter. At high K+ concentrations, two K+ ions equivalently occupy the four sites in the selectivity filter while at low K+/high Na+ a single K+ ion remains bound in the selectivity filter, preferably at site 1 or 3. This single K+ binding at low concentration effectively blocks the permeation of Na+, providing a structural basis for the anomalous mole-fraction effect, a key property of multi-ion pores.
K+ channels are tetrameric cation channels that display very high K+ over Na+ selectivity, despite Na+ being a smaller ion1. This property arises from the presence of the conserved signature sequence of TVGYG, which forms the selectivity filter of the K+ channel2,3. The structural basis for ion selectivity in K+ channels has been extensively studied using KcsA, a prototypic K+ channel, as a model system4,5. The KcsA structure, captured in its closed state, reveals four chemically equivalent K+ binding sites (numbered 1 to 4 from extracellular side) in the selectivity filter, occupied on average by two K+ ions. This has led to the proposal of 1, 3 and 2, 4 configurations of K+ binding in the filter6,7. Interestingly, K+ binding seems requisite to maintaining a conductive KcsA filter, which otherwise collapses to a “nonconductive” state in a low K+/high Na+ environment5. This effect is mitigated by replacing the first conserved glycine residue in the TVGYG signature sequence (underlined) with an unnatural D-alanine (KcsAD-Ala77) 8. In addition to preventing filter collapse in a low K+ environment, this substitution renders the channel, which is still highly K+ selective, permeable to Na+ in the absence of K+. As Na+ conduction in K+ free conditions has been observed in several naturally existing K+ channels9-14, the K+ dependent conformational changes observed in KcsA selectivity filter could well be common only to a subset of K+ channels rather than a general property of all of them, warranting further investigation. To this end, we performed a high resolution structural analysis of the isolated ion conduction pore of MthK15, a Ca2+-gated K+ channel from Methanobacterium thermoautotrophicum, in complex with K+, as well as in low K+/high Na+ concentrations. Our high resolution structures, as well as the ability to carry out detailed ion titration experiments in lipid bilayers, reveal some striking differences between MthK and KcsA, and provide novel insights into ion selectivity in multi-ion K+ channel pores.
Results
The open MthK pore structure
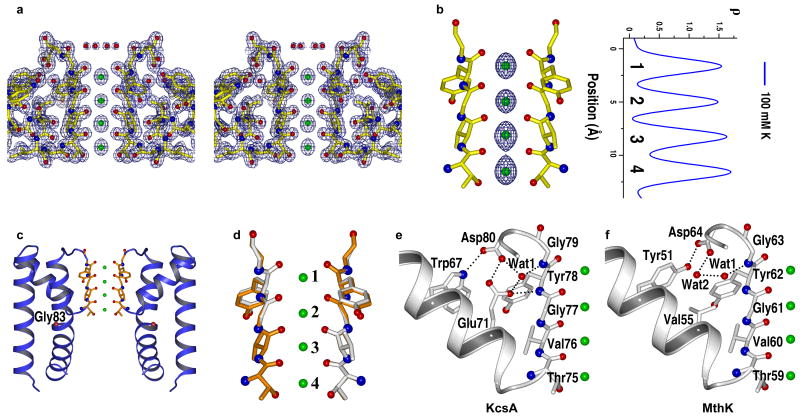
The membrane-spanning ion conduction pore of the MthK channel was obtained by limited trypsin digestion of the full-length channel, resulting in the removal of the intracellular Ca2+ sensing gating ring16. The structure of the MthK pore in complex with K+ was determined to 1.45 Å resolution from a crystal grown in the presence of 100 mM K+ (Fig. 1a, Table 1 and Methods). The Fo-Fc ion omit map of K+ complex reveals four electron density peaks in the filter with equivalent intensity (Fig 1b), indicating a similar K+ occupancy at each site as observed in KcsA.
Figure 1.
Structure of open MthK pore in complex with K+ (in 100 mM K+). (a) Stereo view of 2Fo-Fc map (1 σ) at the selectivity filter region. K+ ions are modeled as green spheres. Only two diagonally opposite subunits are shown for clarity. (b) Fo-Fc ion omit map (5 σ) of K+ complex and its 1-D electron density profile along the central axis of the selectivity filter. The ion binding sites are numbered 1 to 4 from the extracellular to intracellular side. (c) Ribbon representation of the overall structure of the MthK pore in an open conformation. The red sphere in the inner helix indicates the position of the Cα atom of glycine gating hinge (Gly83). (d) Superimposition of the selectivity filter of MthK (grey) and KcsA (gold, PDB code 1K4C). (e & f) Comparison of the hydrogen bonding network surrounding the selectivity filter between KcsA (e) and MthK (f). Only the pore helix and the selectivity filter from one subunit are shown.
Table 1. Data collection and refinement statistics.
| Protein | K+ complex | Low-K+ complex | Na+ complex |
|---|---|---|---|
| Mutations | S68H, V77C | S68R, V77C | S68H, V77C |
| KCl/NaCl concentration | 100 mM / 0 mM | 1 mM / 99 mM | 0 mM / 100 mM |
| Data collection | |||
| Space Group | P4212 | P4212 | P4212 |
| Cell dimensions | |||
| a, c (Å) | 63.8, 44.1 | 63.3, 44.0 | 60.8, 44.8 |
| Resolution (Å) | 50-1.45 (1.48-1.45)* | 50-1.45 (1.48-1.45) | 50-2.2 (2.24-2.2) |
| Rmerge | 0.044 (0.75) | 0.047 (0.51) | 0.114 (0.98) |
| I / σI | 51.1 (1.3) | 57.2 (1.2) | 37.4 (3.2) |
| Completeness (%) | 99.3 (98.5) | 97.7 (76.1) | 99.4 (100.0) |
| Redundancy | 8.6 (6.3) | 9.7 (4.2) | 10.4 (9.7) |
| Refinement | |||
| Resolution (Å) | 50.0-1.45 | 50.0-1.45 | 50.0-2.2 |
| No. reflections | 14,863 | 14.425 | 4081 |
| Rwork / Rfree | 18.7 / 20.2 | 19.1 / 21.6 | 20.2 / 20.8 |
| No. atoms | |||
| Protein | 640 | 641 | 644 |
| K+ Ion | 5 | 5 | 0 |
| Na+ Ion | 0 | 0 | 4 |
| Water | 52 | 56 | 27 |
| B-factors | |||
| Protein | 27.0 | 29.7 | 39.7 |
| K+ Ion | 32.6 | 42.2 | |
| Na+ Ion | 39.7 | ||
| Water | 47.9 | 51.4 | 53.5 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.008 | 0.011 |
| Bond angles (°) | 1.26 | 1.11 | 1.11 |
Values in parentheses are for highest-resolution shell
The isolated MthK pore is in an open conformation, similar to the pore region of the full-length MthK structure with a root-mean-square (r.m.s.) deviation of 1.6 Å for all Cα atoms of the tetrameric pore15 (supplementary Fig. 1a). Consequently, the central cavity becomes a wider vestibule that is continuous with the intracellular side, leaving the 12 Å long selectivity filter the only part of the channel that controls K+ permeation (Fig. 1c). The isolated MthK pore structure also superimposes well with the recently determined high resolution structure of non-selective NaK channel in its open conformation17. With a main chain r.m.s. deviation of 0.74 Å for inner, outer, and pore helices (supplementary Fig. 1b), these two structures demonstrate the remarkable conservation of pore opening mechanics in tetrameric cation channels utilizing the conserved glycine hinge (Gly83 in MthK)18.
The selectivity filter of MthK
The selectivity filter of MthK is identical to that of KcsA with a main chain r.m.s. deviation of 0.2 Å (Fig. 1d). In light of large scale gate-opening conformational changes, this strict structural conservation between K+ channel selectivity filters is a testament to the importance of preserving their structure to catalyze selective K+ transfer across membranes19. However, differences in protein packing in the region surrounding the filter were observed between MthK and KcsA (Fig. 1e&f). In KcsA, an acidic residue (Glu71) from the pore helix forms a hydrogen-bonding network with a buried water molecule (w1) and Asp80 (Fig. 1e). Glu71, shown to be important for the stability of the selectivity filter in KcsA20,21, is not conserved in most K+ channels. In MthK it is replaced by valine, with the hydrogen-bonding network mediated by two buried water molecules: one (Wat1) is in the same position as that in KcsA, whereas the other (Wat2) is positioned similar to the carboxylate oxygen of Glu71 in KcsA (Fig. 1f). Additionally, Tyr51 in MthK forms a short-range hydrogen bond with Asp64 with a distance of 2.6 Å, whereas the equivalent hydrogen bond between Trp67 and Asp80 in KcsA has a distance of 3.0 Å. These differences in protein packing may account for differences in filter stability. As discussed below, while KcsA requires the presence of K+ to stabilize its selectivity filter, the MthK filter maintains a conductive conformation even in the absence of K+.
The MthK filter maintains a conductive conformation in low K+
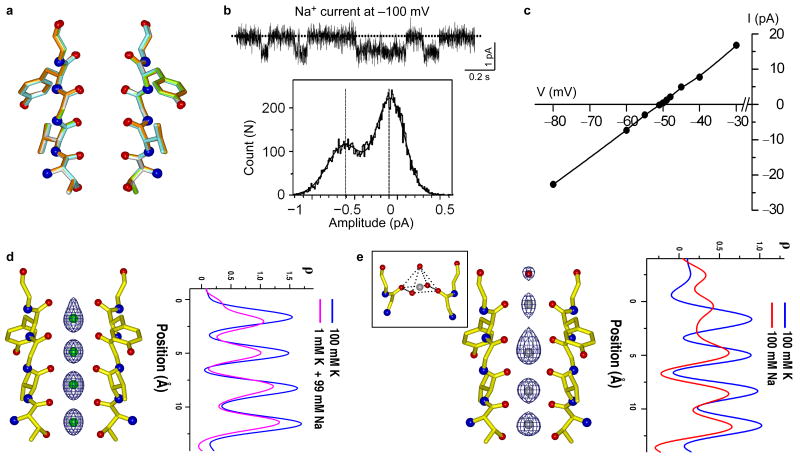
To visualize conformational changes, if any, within the MthK selectivity filter in a low K+ environment, we soaked the K+-complex crystals in a stabilization solution containing 1 mM K+/99mM Na+ (low-K+ complex) or 100 mM Na+ without K+ (Na+ complex), and determined their structures at 1.45 Å and 2.2 Å resolution, respectively. Unlike KcsA, no structural changes were observed in the selectivity filter as shown in the superimposition of the K+ complex with the structures of soaked crystals (Fig. 2a). This is indicative of MthK maintaining a conductive state in a low K+ or Na+ only environment and implies that the channel may be permeable to Na+ as was observed in KcsAD-Ala77 mutant. Indeed, in the absence of K+, MthK can conduct Na+ albeit with a much smaller conductance of ∼5 pS in 150 mM symmetrical NaCl compared to that of ∼220 pS in 150 mM symmetrical KCl (Fig 2b). Nevertheless, the channel is still highly selective for K+ as indicated by a PNa/PK ratio of 0.017 calculated from measured reverse potentials (Fig. 2c). Overall, these results suggest that the K+ concentration dependent conformational changes of the selectivity filter are specific to only a subset of K+ channels. Accordingly, it is likely that other Na+-permeable K+ channels also maintain a conductive filter both in Na+ and K+.
Figure 2.
The selectivity filter of MthK maintains a conductive conformation in low K+ or Na+ only environment. (a) Superimposition of the MthK selectivity filter structures in the presence of 100 mM K+ (gold), 1 mM K+/99 mM Na+ (green) and 100 mM Na+ (cyan). (b) Single channel trace (upper panel) and histogram (lower panel) of Na+ conduction in MthK at -100 mV in the absence of K+. (c) Reverse potential of MthK measured with 100 mM KCl / 170 mM NaCl on the intracellular side and 10 mM KCl / 260 mM NaCl on the extracellular side. (d) Fo-Fc ion omit map (5 σ) of low-K+ complex (in 1 mM K+ / 99 mM Na+) and its 1-D electron density profile (pink) along the central axis of the selectivity filter. The 1-D electron density profile of K+ complex is also shown in blue for comparison. (e) Fo-Fc ion omit map (5 σ) of Na+-complex (in 100 mM Na+) and its 1-D electron density profile (red) along the central axis of the selectivity filter. The inset shows the pyramidal Na+ ion chelation at site 1.
Ion occupancy in the selectivity filter
The Electron density in the filter of the low-K+ complex (1 mM K+) is weaker than that of the high-K+ complex (in 100 mM K+) (Fig. 2d), indicating a decrease in K+ occupancy - likely because it is replaced by a Na+ ion, which has a lower number of electrons. Based on ion occupancy studies in KcsA7, it is reasonable to assume that the K+ complex selectivity filter is occupied simultaneously by two water molecules and two K+ ions and that the area under the peaks of the 1-D electron density profile along the central axis of the filter reflects the number of electrons in the filter (56 e- for 2 K+ and 2 H2O). Accordingly, upon comparison of their 1-D electron density profiles, we estimate a reduction of 10 e- in the low-K+ (1 mM) complex of MthK compared to the high-K+ complex (100 mM). This is roughly equivalent to the replacement of a K+ ion with a Na+ ion, suggesting that a single K+ ion remains bound in the filter of the low-K+ complex. The same analysis of the Na+ complex indicates a reduction of about 18 e- compared to the high-K+ complex, suggesting a replacement of both K+ ions by Na+ after crystal soaking in a Na+-only solution (Fig. 2e).
In the low-K+ complex, electron density at site 1 has a shape of a bowling pin with a shoulder peak at the upper edge of the ion binding cage (Fig. 2d). This likely arises from partial Na+ binding since planar coordination is more chemically favorable for the smaller Na+ ions. This is more evident in the structure of the Na+ complex in which the ion at site 1 resides in plane with the four carbonyl oxygen atoms from Tyr62, with ion-ligand distances of 2.4 Å (Fig. 2e), and is consistent with statistics of Na+ coordination commonly observed in small molecules and proteins22. In addition, a water molecule, which is not observed in the K+ complex, participates in Na+ chelation from the extracellular side with a distance of 2.5 Å from the ion (Fig. 2e). A similar pyramidal Na+ coordination scheme was also observed in the Na+ complex of the NaK channel23. The electron density at site 2 is more diffused in the Na+ complex with some density at the upper edge of the site. This can be attributed to an averaged configuration of Na+ ions/water molecules in the center of the cage and Na+ ions binding in plane with the carbonyl oxygen atoms of Gly61. Electron density at sites 3 and 4 also appears to be stronger towards their lower edges, but not as substantial as in sites 1 and 2. Although it is difficult to distinguish Na+ ions from water molecules based on the electron density map since both have an equal number of electrons, it is clear that Na+ can bind in a K+ channel without any structural rearrangements of the selectivity filter.
K+ binding at low concentration
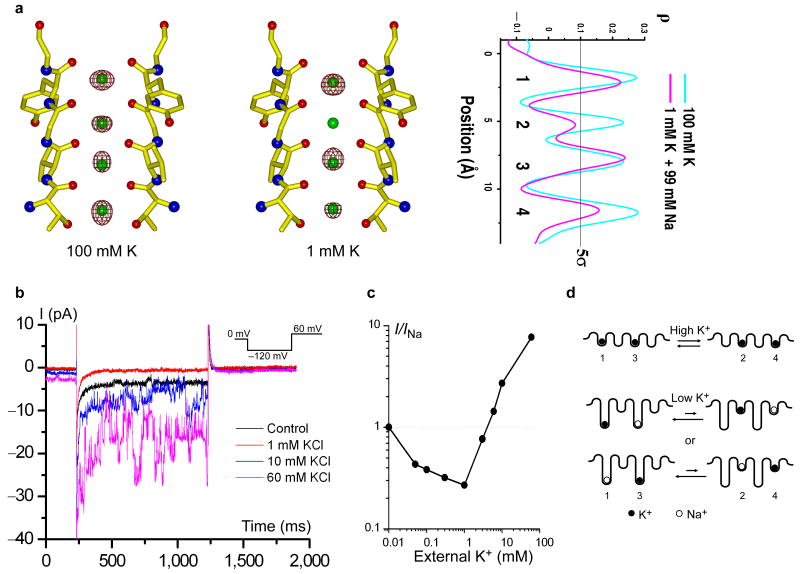
To compare the occupancy of K+ ions at each site in low K+ (in 1 mM K+/99 mM Na+) and high K+ (in 100 mM K+) environments, we calculated anomalous difference Fourier maps of both complexes. At the wavelength used for data collection (0.9794 Å), anomalous scattering is dominated by K+ ions and sulfur atoms in the protein (Supplementary Table 1). Although weak, the high resolution and minimal radiation damage to the crystals allow for sufficient quality data to measure the anomalous difference between Friedel pairs. In the calculated anomalous difference Fourier maps of both complexes (Fig. 3a and supplementary Fig. 2), all major peaks (> 5 sigma) are solely from K+ ions and sulfur atoms. In the high-K+ complex, the anomalous signals from K+ in the filter have similar intensity, confirming the equal occupancy of K+ at each site. However, in the low-K+ complex the anomalous signal at site 1 or 3 is much stronger than that at site 4, with no substantial signal (less than 5 sigma) observed at site 2. This suggests that K+ predominately occupies site 1 or 3 in a low K+/high Na+ environment and has almost no binding at site 2. Furthermore, the overall decrease in intensity of anomalous signals, particularly at sites 2 and 4, confirms the reduction of K+ occupancy in the filter at low K+ concentrations.
Figure 3.
K+ binding in the selectivity filter at high and low concentrations. (a) Anomalous difference Fourier maps (5 σ) of K+ (in 100 mM K+) and low-K+ (in 1 mM K+/99mM Na+) complexes of MthK pores and their 1-D electron density profiles at the selectivity filter region. (b) Inward currents of MthK recorded at -120 mV in the presence of various extracellular K+ concentrations. Both sides contain 170 mM NaCl. The control trace was recorded in the absence of extracellular K+. (c) Magnitude of inward current as a function of extracellular K+ concentration. Data points were obtained from an ensemble of 20 traces at each [K+]. Currents were normalized to the control trace (0 mM K+). (d) Schematic representation of K+ binding in the conductive selectivity filter at high (top, two bound K+ ions) and low concentrations (bottom, one bound K+ ion). K+ and Na+ ions are represented by solid and open spheres, respectively.
Discussion
The observation that a single K+ ion remains bound in the conductive MthK filter with preference for sites 1 and 3 is consistent with data from the KcsAD-Ala77 mutant structure in low K+ captured in a conductive conformation8. In that structure, the K+ ion has an occupancy of 1.1 for all four sites, and the electron density distribution in the filter is suggestive of K+ binding at sites 1 and 3 but not definitively due to the resolution limit (2.4 Å). Here, with the extremely high resolution data, analysis of anomalous signals allows us to unambiguously assign the K+ binding sites at both high and low K+ concentrations in a naturally existing K+ channel.
The observation of a single K+ ion binding in the filter with non-equivalent occupancy in the low-K+ structure of MthK allows us to probe another fundamental property of multi-ion pores1. This so called anomalous mole-fraction effect refers to the ratio dependent changes in channel permeability arising from the addition of a second permeable ion species and has been observed in various cation channels including MthK and other Na+-conducting K+ channels as well as Ca2+ channels24-26. As shown in the single channel recording of MthK with 170 mM NaCl on both sides (Fig. 3b&c), addition of 1 mM K+ to the external solution is sufficient to block the inward current carried by Na+ ions. Increasing K+ concentration above 3 mM leads to an increase in current now carried by K+ rather than Na+. The differences in ion occupancy among the Na+, high-K+, and low-K+ complexes provide a plausible explanation. Efficient conduction of K+ necessitates a balance between high affinity binding of multiple ions within the filter and the resulting electrostatic repulsion between them, which destabilizes the ions and lowers this affinity. In the high K+ complex, this is achieved through two K+ ions binding equivalently in the 1, 3 and 2, 4 configurations (Fig. 3d). An additional K+ ion entering the filter effectively expels a bound ion from the opposite end, allowing the filter to maintain two bound ions6. The balance of two K+ ions in energetically equivalent 1, 3 and 2, 4 configurations is disrupted in a low K+ / high Na+ environment as demonstrated in the structure of low-K+ complex in which a K+ ion is replaced by a Na+ ion, leaving a single K+ ion remain bound preferably at site 1 or 3. With K+ predominantly occupying site 1 or 3 in the mixed ion system (1 Na+ and 1 K+), Na+ is likely to reside at site 1 (when K+ is at site 3) or site 3 (when K+ is at site 1). In other words, the two mixed ions have higher occupancy in the 1,3 configuration compared to the 2,4 configuration (Fig. 3d). This imbalance of Na+/K+ occupancy between 1,3 and 2,4 configurations leads to a reduction of ion conductance, reminiscent of what is observed for Rb+ conduction in K+ channels6, and effectively blocks the passage of Na+ ions that would otherwise pass through the pore in the absence of K+. This blockage of Na+ conduction by a single K+ ion in the filter at low K+ concentration virtually prevents the K+ channel from being a leaky valve for Na+ ions, which may be crucial for the biological functioning of K+ channels in light of the high Na+/low K+ extracellular environment. Increasing K+ concentrations works to restore the balance, returning to the favorable 1, 3 and 2, 4 configurations for multiple K+ ion binding and conduction. MthK presents us a first time structural perspective on this classically observed property of multi-ion pores.
Methods
Protein purification
The MthK channel was expressed and purified in n-Decyl-β-D-Maltoside (DM) as described before15. The membrane-spanning portion of the MthK channel which forms the ion conduction pore was obtained by limited trypsin digestion of the MthK channel16 and further purified on a superdex 200 gel filtration column in a buffer of 5 mM N,N-dimethyldodecylamine-N-oxide (LDAO), 20 mM Tris.HCl, pH 8.0, and 100 mM KCl.
Crystallization
Purified MthK pores in LDAO were concentrated to about 8 mg ml-1 and crystallized using the sitting drop vapor diffusion method by mixing equal volumes of concentrated protein and well solution containing 3.0-3.5 M 1,6-Hexandiol, and 100 mM HEPES, pH 7.5. Initial crystals of the wild type MthK pore were of space group I4 and displayed anisotropic diffraction with 2.3 Å resolution in the best direction. Two mutations, Val77 to Cys and Ser68 to Arg or His, were introduced into the pore region of MthK to improve the protein packing in the crystal. Both mutations have no effect on channel function but yield much better diffracting crystals that were used in structure determination. Crystals of mutant MthK pore were of a different space group (P4212) with unit cell dimensions around a=b=63.8 Å, c=44.0 Å, α=β=γ=90° and contained one molecules per asymmetric unit.
Data collection and structure determination
X-ray data were collected at the Advanced Photon Source (APS) Beamlines 19-ID/BM and 23-ID. Data processing and scaling was performed using the HKL2000 software27. The structures were determined by molecular replacement using AMoRe in the CCP4 suite28 using KcsA structure (PDB code 1K4C) as an initial search model followed by repeated cycles of model building with XtalView29 and refinement with REFMAC28. Detailed data collection and refinement statistics are listed in Table 1.
One dimensional electron density profiles
One dimensional electron density profiles of bound ions in the selectivity filter were obtained as described6. All data were scaled against the K+ complex crystal (100 mM K+) before map calculation. 1-D profiles were obtained by sampling the Fo-Fc difference maps, with ions and selectivity filter residues omitted, along the central axis of the filter using MAPMAN30.
Electrophysiological study
The MthK channel, purified in DM detergent, was reconstituted into lipid vesicles composed of 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPE, 7.5 mg ml-1) and 1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG, 2.5 mg ml-1) and its activity was recorded in a vertical lipid bilayer setup using the same method as described16. Na+ current was recorded with 150 mM symmetrical NaCl. Reverse potentials were measured with 100 mM KCl / 170 mM NaCl on the intracellular side and 10 mM KCl / 260 mM NaCl on the extracellular side. The permeation probability (PNa/PK) was calculated using the following equation:
Where [X]o and [X]i are ion concentrations on extracellular and intracellular sides. The K+ blocking effect on Na+ current was measured with 170 mM NaCl, 10 mM CaCl2 and 10 mM HEPES, pH 7.6 on intracellular side, and 170 mM NaCl, 0-60 mM KCl and 10 mM HEPES, pH 7.4 on the extracellular side. Membrane voltage was controlled and current recorded using an Axopatch 200B amplifier with a Digidata 1322A analogue-to-digital converter (Axon Instruments). Currents were low-pass filtered at 1 kHz and sampled at 10 kHz.
Supplementary Material
Acknowledgments
We thank Dr. Amer Alam for discussion and critical review of the manuscript. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy, Office of Energy Research. We thank the beamline staff for assistance in data collection. This work was supported by Howard Hughes Medical Institute and by grants from the NIH/NIGMS (RO1 GM071621), David and Lucile Packard Foundation and Welch Foundation.
Footnotes
Accession codes The atomic coordinates and the structure factors for the K+, low-K+ and Na+ complexes of the MthK ion conduction pore have been deposited with the accession codes of 3LDC, 3LDD and 3LDE, respectively.
Author Contributions. S.Y determined the structures. Y.L performed the electrophysiological analysis. S.Y, Y.L and Y.J designed the research, analyzed data, and wrote the manuscript.
Competing interests statement. The authors declared no competing interests.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- 2.Heginbotham L, Abramson T, MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- 3.Heginbotham L, Lu Z, Abramson T, MacKinnon R. Mutations in the K+ channel signature sequence. Biophys J. 1994;66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 6.Morais-Cabral JH, Zhou Y, MacKinnon R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, MacKinnon R. The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J Mol Biol. 2003;333:965–975. doi: 10.1016/j.jmb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Valiyaveetil FI, Leonetti M, Muir TW, Mackinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 2006;314:1004–1007. doi: 10.1126/science.1133415. [DOI] [PubMed] [Google Scholar]
- 9.Callahan MJ, Korn SJ. Permeation of Na+ through a delayed rectifier K+ channel in chick dorsal root ganglion neurons. J Gen Physiol. 1994;104:747–771. doi: 10.1085/jgp.104.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korn SJ, Ikeda SR. Permeation selectivity by competition in a delayed rectifier potassium channel. Science. 1995;269:410–412. doi: 10.1126/science.7618108. [DOI] [PubMed] [Google Scholar]
- 11.Ogielska EM, Aldrich RW. A mutation in S6 of Shaker potassium channels decreases the K+ affinity of an ion binding site revealing ion-ion interactions in the pore. J Gen Physiol. 1998;112:243–257. doi: 10.1085/jgp.112.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starkus JG, Kuschel L, Rayner MD, Heinemann SH. Ion conduction through C-type inactivated Shaker channels. J Gen Physiol. 1997;110:539–550. doi: 10.1085/jgp.110.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Starkus JG, Kuschel L, Rayner MD, Heinemann SH. Macroscopic Na+ currents in the “Nonconducting” Shaker potassium channel mutant W434F. J Gen Physiol. 1998;112:85–93. doi: 10.1085/jgp.112.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Hesketh JC, Fedida D. A high-Na(+) conduction state during recovery from inactivation in the K(+) channel Kv1.5. Biophys J. 2000;79:2416–2433. doi: 10.1016/S0006-3495(00)76486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Berke I, Chen L, Jiang Y. Gating and inward rectifying properties of the MthK K+ channel with and without the gating ring. The Journal of general physiology. 2007;129:109–120. doi: 10.1085/jgp.200609655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alam A, Jiang Y. High-resolution structure of the open NaK channel. Nat Struct Mol Biol. 2009;16:30–34. doi: 10.1038/nsmb.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 19.Nishida M, Cadene M, Chait BT, MacKinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. Embo J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 21.Cordero-Morales JF, et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol. 2007;14:1062–1069. doi: 10.1038/nsmb1309. [DOI] [PubMed] [Google Scholar]
- 22.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 23.Alam A, Jiang Y. Structural analysis of ion selectivity in the NaK channel. Nat Struct Mol Biol. 2009;16:35–41. doi: 10.1038/nsmb.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss L, Immke D, LoTurco J, Korn SJ. The interaction of Na+ and K+ in voltage-gated potassium channels. Evidence for cation binding sites of different affinity. J Gen Physiol. 1998;111:195–206. doi: 10.1085/jgp.111.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almers W, McCleskey EW. Non-selective conductance in calcium channels of frog muscle: calcium selectivity in a single-file pore. J Physiol. 1984;353:585–608. doi: 10.1113/jphysiol.1984.sp015352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess P, Tsien RW. Mechanism of ion permeation through calcium channels. Nature. 1984;309:453–456. doi: 10.1038/309453a0. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 30.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.