Abstract
The chloroplast psaB mRNA encodes one of the reaction centre polypeptides of photosystem I. Protein pulse-labelling profiles indicate that the mutant strain of Chlamydomonas reinhardtii, F14, affected at the nuclear locus TAB2, is deficient in the translation of psaB mRNA and thus deficient in photosystem I activity. Genetic studies reveal that the target site for Tab2 is situated within the psaB 5′UTR. We have used genomic complementation to isolate the nuclear Tab2 gene. The deduced amino acid sequence of Tab2 (358 residues) displays 31–46% sequence identity with several orthologues found only in eukaryotic and prokaryotic organisms performing oxygenic photosynthesis. Directed mutagenesis indicates the importance of a highly conserved C-terminal tripeptide in Tab2 for normal psaB translation. The Tab2 protein is localized in the chloroplast stroma where it is associated with a high molecular mass protein complex containing the psaB mRNA. Gel mobility shift assays reveal a direct and specific interaction between Tab2 and the psaB 5′UTR. We propose that Tab2 plays a key role in the initial steps of PsaB translation and photosystem I assembly.
Keywords: Chlamydomonas/chloroplast/photosynthesis/photosystem I mutant/translation
Introduction
The role of nucleus-encoded factors in the biogenesis of the photosynthetic apparatus has been studied intensively in recent years (reviewed in Barkan and Goldschmidt-Clermont, 2000). Biochemical and genetic approaches in the unicellular green alga Chlamydomonas reinhardtii and in the land plants Zea mays (maize) and Arabidopsis thaliana have revealed the involvement of numerous nuclear genes in post-transcriptional steps of chloroplast gene expression such as RNA processing, RNA stability and translation (for reviews, see Rochaix, 1992, 1996; Mayfield et al., 1995). While most of the nuclear mutations affecting chloroplast post-transcriptional steps in land plants display pleiotropic effects, in C.reinhardtii similar mutations have more specific effects on chloroplast gene expression (Goldschmidt-Clermont, 1998; Leon et al., 1998).
Genetic approaches in Chlamydomonas have identified several nucleus-encoded factors involved in chloroplast translation. A characteristic feature of these factors is that they act specifically on single chloroplast mRNAs. Thus, mutants of Chlamydomonas have been isolated that are specifically deficient in the translation of the mRNA for petA (Wostrikoff et al., 2001), psaB (Stampacchia et al., 1997), psbA (Girard-Bascou et al., 1992; Yohn et al., 1998) and psbC mRNAs (Rochaix et al., 1989; Zerges and Rochaix, 1994; Auchincloss et al., 2002). In most of these cases, the nuclear mutations affect the initiation of translation as shown by reduced expression of chimeric reporter genes driven by the 5′UTRs of these mRNAs in the presence of the nuclear mutant allele. Only one mutant deficient in elongation of psbD mRNA translation has been characterized (Cohen et al., 2001). Mutants affected in chloroplast translation have also been identified in maize. Some of these mutants are specifically deficient in the translation of the atpB/E mRNA or the petD and petA mRNAs (Barkan et al., 1994; McCormac and Barkan, 1999; Fisk et al., 1999).
Here we report the characterization of a nuclear gene and its product involved specifically in the initiation of translation of the psaB mRNA in C.reinhardtii. PsaB and PsaA are the two reaction centre subunits of photosystem I (PSI) that act as ligands of the primary electron donor P700 and the primary acceptors A0, A1 and the 4Fe–4S centre FX. The strain carrying the nuclear mutation F14-tab2 examined here is affected in the synthesis of both PsaA and PsaB. Previous studies showed that mutants deficient in PsaB are unable to synthesize both PsaB and PsaA whereas mutants affected primarily in PsaA synthesis are still able to produce PsaB (Girard-Bascou et al., 1987; Stampacchia et al., 1997). Based on these results it was proposed that PsaB is an anchor protein during PSI assembly, which needs to be synthesized and integrated into the thylakoid membrane before the other PSI subunits are synthesized (Stampacchia et al., 1997). In its absence these polypeptides are no longer synthesized and/or are rapidly degraded. Elucidating how PsaB is translated and inserted into the thylakoid membrane is thus important for understanding the initial steps of PSI assembly. We have characterized Tab2, the gene affected in F14 and we show that it encodes a new type of RNA binding protein that is part of a high molecular weight complex which also includes psaB mRNA. The primary effect of the F14-tab2 mutation is a dramatic decrease in psaB mRNA translation. The predicted amino acid sequence of the Tab2 protein displays similarities with several putative proteins found in both prokaryotic and eukaryotic organisms capable of oxygenic photosynthesis and represents to date the first identified factor specifically involved in the translation and possibly assembly of photosynthetic subunits that is conserved in this broad class of organisms.
Results
The F14 mutant is deficient in the initiation of translation of the chloroplast psaB mRNA
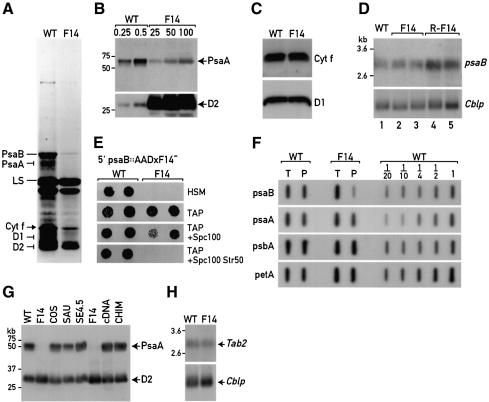
The nuclear mutant F14 was originally described as a high-fluorescence mutant requiring acetate for growth (Bennoun and Levine, 1967) and was classified as a mutant lacking PSI activity. This strain carries a recessive nuclear mutation (Girard et al., 1980). We first tested whether the synthesis of any of the chloroplast-encoded subunits of PSI is impaired in this mutant. Wild-type and mutant cells were pulse-labelled for 45 min with [14C]acetate in the presence of cycloheximide, an inhibitor of cytosolic translation, and total cell proteins were analysed by SDS–PAGE and autoradiography. The labelling of the two reaction centre polypeptides of PSI, PsaA and PsaB, is greatly reduced in the mutant (Figure 1A). Moreover, the bands corresponding to cytochrome (Cyt f) and the PSII D1 polypeptide are significantly weaker in F14 than in the wild type.
Fig. 1. Analysis and rescue of the F14 mutant strain. (A) Chloroplast protein synthesis in the wild-type (WT) and the F14 mutant strain. Cells were pulse-labelled with [14C]acetate for 45 min in the presence of cycloheximide. Thylakoid membrane proteins were separated by gradient 7.5–15% SDS–PAGE. PsaA and PsaB, the two reaction centre polypeptides of PSI; LS, Cyt f, D1 and D2 correspond respectively to the large subunit of ribulose-1,5 bisphosphate carboxylase, Cyt f and the two reaction centre subunits of PSII. (B) Immunoblot analysis of the wild-type and F14 strains. Crude cell extracts were separated on a 6% SDS–PAGE and electroblotted. The membrane was reacted with PsaA and D2 polyclonal antisera. The amounts of protein loaded are expressed in µg of total proteins. (C) Immunoblot analysis of 25 µg of total proteins from wild-type and F14 strains with D1 and Cyt f antisera. (D) RNA blot analysis of the wild-type, F14 and rescued F14 (R-F14) strains. Five micrograms of total RNA from these strains were fractioned on a 1% agarose–formaldehyde gel, blotted and hybridized with a 32P-labelled psaB probe. Equal loading of mRNA was checked by hybridization of the blot with a Cblp probe. Lane 1, WT; lanes 2 and 3, two F14 isolates; lanes 4 and 5, two independently rescued F14 transformants (R-F14) complemented with the cosmid and pKS-SE 4.5, respectively. (E) Growth patterns of strains containing the chloroplast chimeric psaB 5′UTR–aadA gene in wild-type and F14-tab2 mutant nuclear backgrounds. One representative tetrad derived from the cross between F14 (mt–) and the wild-type psaB 5′UTR–aadA (mt+) is displayed (see Materials and methods). WT, progeny with wild-type Tab2 allele; F14, progeny with F14-tab2 allele; HSM, minimal medium; TAP, acetate-containing medium; Spc100, spectinomycin 100 µg/ml; Str50, streptomycin 50 µg/ml. (F) 2 μg of total RNA (T) and 2 µg of polysomal RNA (P) from wild type and F14 was slot blotted and hybridized with probes specific for psaB, psaA, psbA and petA. In each case a wild-type dilution series was used for estimating the amount of RNA. The slot blots used were duplicates of one another. (G) Immunoblot analysis of PsaA polypeptide in wild type, F14 and rescued F14. Total cell protein (5 µg) of the wild type (WT), two replicas of the F14 mutant strain (F14) and the F14 mutant rescued by transformation with the cosmid (COS), the 11 kb Sau3A genomic fragment (SAU), the 4.5 kb SacI–EcoRI minimal genomic fragment (SE4.5), the 2.2 kb Tab2 cDNA driven by the psaD promoter (cDNA) and the chimeric genomic–cDNA Tab2 gene containing a triple HA epitope (CHIM). D2 antiserum was used to check for equal protein loading. Molecular mass markers are indicated in kDa. (H) RNA gel blot analysis of Tab2 mRNA in the wild-type and F14 strains. Five micrograms of total RNA were loaded per lane.
The level of PsaA protein in the mutant is at least 200 times lower than in the wild type as shown by immunoblotting with serial dilutions of crude extracts (Figure 1B). Because we were unable to raise an antiserum against PsaB, we used a PsaA antiserum throughout this study to measure the amount of PsaB. This is possible because the PsaA and PsaB proteins accumulate to the same level in mutants impaired in the synthesis and/or stability of any of these two PSI reaction centre subunits as they form a heterodimer and are stabilized by each other (Girard-Bascou et al., 1987). In contrast to PsaA and PsaB, the apparent reduced synthesis of Cyt f and D1 does not prevent the normal accumulation of these polypeptides in F14 (Figure 1C).
A similar phenotype was observed previously with the non-allelic F15 mutant (Girard et al., 1980) in which a block in PsaB synthesis leads to impaired PsaA synthesis (Stampacchia et al., 1997). In contrast, mutants deficient in psaA gene expression synthesize PsaB protein at the normal rate, although this protein turns over rapidly in the absence of PsaA (Girard-Bascou et al., 1987). From these earlier results we predicted that the primary defect in F14 is in the expression of psaB although the normal accumulation of Cyt f and D1 does not rule out the possibility that the factor deficient in F14 impacts on the translational efficiency of the petA and psbA mRNAs.
Using RNA blot analysis, we first determined that the psaB and psaA mRNAs accumulate to normal levels in the F14 mutant, indicating that the mutation acts at a post-transcriptional step, most likely at the level of translation (Figure 1D, data not shown). To test whether psaB is indeed the target of the mutated factor, we used chimeric genes in which the expression of the aadA reporter gene conferring spectinomycin and streptomycin resistance (Goldschmidt-Clermont, 1991) is driven by the psaB and psaA 5′UTRs. These constructs were integrated at the atpB locus of the Fud50 strain as described previously (Nickelsen et al., 1994; Zerges and Rochaix, 1994). The transformants were first selected for growth on minimal medium and then tested for their ability to grow on spectinomycin-containing medium. Once homoplasmicity was obtained, the strains were subjected to DNA gel blot analysis and confirmed to contain only copies of the chimeric genes (data not shown). These transformants (mating type +) containing the psaB 5′UTR–aadA or psaA 5′UTR–aadA were crossed to the F14 mutant (mating type –). Because of the uniparental inheritance of the chloroplast genome of Chlamydomonas during crosses, all the progeny contain the chimeric chloroplast gene, whereas the nuclear mutation F14-tab2 segregates 2:2. The progeny of both crosses were scored for phototrophic growth and spectinomycin and/or streptomycin resistance. In the crosses involving the psaB 5′UTR–aadA-containing strains, we observed cosegregation between the ability to grow on minimal medium and antibiotic resistance in seven tetrads tested. One illustrative tetrad is shown in Figure 1E. In contrast no effect of the F14-tab2 mutation was detected on the expression of the chimeric psaA 5′UTR–aadA gene (data not shown). These results indicate that a target site of the Tab2 factor is comprised within the psaB 5′UTR and that this factor acts most likely at the level of translation initiation. This was tested further by purifying polysomes by sedimentation through a sucrose cushion and by hybridizing total and polysomal RNA with 32P-labelled probes specific for psaB, psaA, psbA (D1) and petA (Cyt f) (Figure 1F). As expected the amount of psaB RNA associated with polysomes is vastly decreased, <5% of wild-type levels, in the F14 mutant. In contrast, the level of psaA mRNA bound to polysomes in the mutant is reduced only slightly (70% of wild-type levels) and the amount of polysomal psbA and petA mRNA is the same in the mutant and wild type. To test that the polysomes were intact, their fast sedimentation on sucrose gradients was verified to be sensitive to EDTA which is known to dissociate polysomes (data not shown). Furthermore the bulk of psbA mRNA, a representative chloroplast mRNA, was associated with the polysome fraction in the sucrose gradient (data not shown). We thus conclude that the F14-tab2 mutation impacts principally on translation initiation of psaB mRNA. It is likely that other processes are responsible for the decreased signals of PsaA, D1 and Cyt f observed during protein pulse-labelling (Figure 1A).
Tab2 is conserved in oxygenic photosynthetic organisms
The mutant strain F14 was rescued by transformation with an ordered cosmid library and selection for phototrophic growth as described (Purton and Rochaix, 1994; Zhang et al., 1994; Vaistij et al., 2000). A cosmid that rescued F14 was isolated (for details see Supplementary data). Subcloning of this cosmid gave rise to a 4.5 kb SacI–EcoRI fragment (pKS-SE 4.5) that was still able to restore the wild-type phenotype by transformation. This fragment was used to screen a C.reinhardtii cDNA library. A 2.2 kb cDNA was identified, which, when fused to the PsaD promoter and 5′UTR, was able to rescue the mutant. The gene was named Tab2 for translation of psaB mRNA. The efficiency of transformation of F14 with the different DNA fragments is summarized in Table I. The PsaA protein accumulated to wild-type levels in the various complemented strains as estimated by immunoblotting using an anti-PsaA polyclonal antibody (Figure 1G). As mentioned above, the amount of PsaA protein detected in this blot provides a measure of the amount of PsaB.
Table I. Complementation of the F14 mutant strain.
| Plasmid | Size (kb) | Complementation |
|---|---|---|
| Cosmid | 30 | + |
| pKS-SE4.5 | 4.5 | + |
| pKS-cDNA | 2.2 | – |
| PsaD 5′UTR cDNA | 2.2 | + |
| Chimeric gDNA–cDNA | 3.6 | + |
| Chimeric gDNA–cDNA::HA | 3.7 | +/– |
Rescue is indicated by ‘+’, lack of rescue by ‘–’, lower rescue efficiency by +/–. pKS-SE4.5 refers to the 4.5 kb SacI–EcoRI fragments cloned in pKS-Bluescript; pKS-cDNA, Tab2 cDNA with partial 5′UTR cloned directly in pKS without Chlamydomonas promoter; PsaD 5′UTR cDNA, cDNA of Tab2 cloned in pSL18 driven by the PsaD promoter. The chimeric genes result from the fusion of the 4.5 SacI–EcoRI genomic DNA containing the Tab2 5′UTR and the complete Tab2 ORF except for the end of the last exon, which has been replaced with the corresponding Tab2 cDNA sequence.
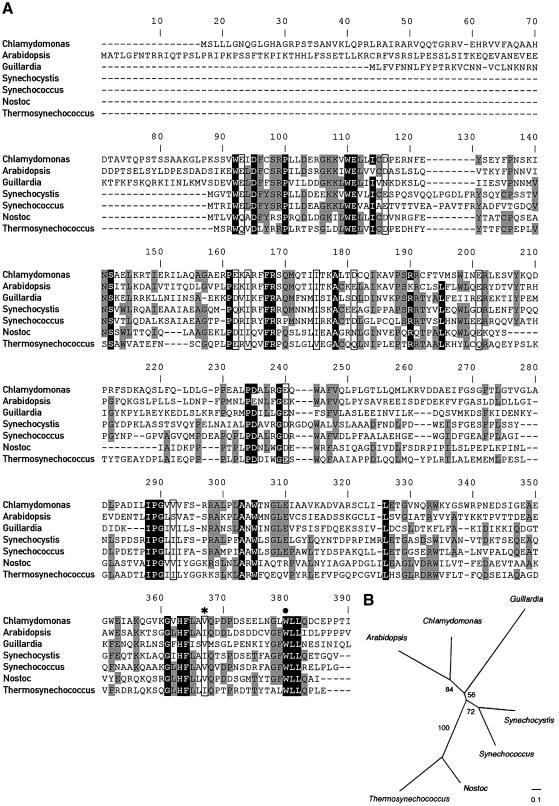
Sequencing of the genomic and cDNA fragments revealed that the Tab2 gene consists of six exons with sizes ranging from 90 to 355 bp and five introns with sizes comprised between 73 and 478 bp. Several stop codons in the same reading frame as Tab2 are present within the 68 bp sequence upstream of the first ATG detected in the cDNA, indicating that it contains the entire Tab2 coding sequence. The deduced Tab2 open reading frame (ORF) encodes a protein of 358 amino acids of 39.9 kDa. A putative chloroplast transit sequence of 50 amino acids rich in arginine and serine residues is predicted by ChloroP. The predicted mature protein consists of 308 amino acids with a molecular mass of 34.3 kDa and a pI of 5.2. A Blast search did not reveal any similarity with proteins known to be involved in chloroplast translation or with known protein motifs. However, sequence similarity with several proteins of unknown function in both prokaryotic and eukaryotic organisms capable of oxygenic photosynthesis was observed. Figure 2A displays the alignment between the C.reinhardtii Tab2 protein and its orthologues in the land plant A.thaliana, the cryptophyte Guillardia theta and in the cyanobacteria Synechocystis sp., Nostoc punctiforme, Synechococcus sp. and Thermosynechococcus elongatus. The phylogenetic tree derived from these alignments is shown in Figure 2B. The eukaryotic sequences are weakly monophyletic with respect to the prokaryotic homologues, suggesting a single origin from the cyanobacterial ancestor of plastids.
Fig. 2. Conservation of Tab2 protein. (A) Sequence alignment of the C.reinhardtii Tab2 protein and its orthologues in the land plant A.thaliana (At3g08010), the cryptophyte G.theta (NP_113515) and several cyanobacteria including Synechocystis sp. (NP_441046), Synechococcus sp. (BAA92865), N.punctiforme (ZP_00112111) and T.elongatus (NP_682776). Sequences were aligned with ClustalW (Thompson et al., 1994). (B) The tree was constructed using protein maximum likelihood with the Molphy program (Adachi and Hasegawa, 1996). Numbers next to branches indicate RELL bootstrap proportions (Adachi and Hasegawa, 1996). The scale bar indicates 0.1 substitution per site.
Of particular interest is the tripeptide motif WLL near the C-terminal end that is conserved in all Tab2 orthologues. Sequencing of the Tab2 gene in the original mutant F14-tab2 revealed a 14 nucleotide insertion 72 bp upstream of the stop codon that resulted in a frameshift. This mutation is expected to give rise to a truncated form of the Tab2 protein in which the 24 C-terminal residues of Tab2 are replaced by a stretch of 14 unrelated amino acids that lacks the conserved tripeptide. RNA blot analysis indicated that the size of the Tab2 mRNA is 2.8 kb and that the level of this mRNA is similar in the wild-type and F14 mutant strains (Figure 1H).
Tab2 is a chloroplast stromal protein
Because we were unable to raise an antiserum against Tab2, we constructed a chimeric genomic–cDNA gene containing a triple HA epitope at the 3′ end of the coding sequence. This construct was obtained by fusing the 5′ part of the 4.5 kb genomic fragment described above and the cDNA in frame with the triple HA epitope by using a unique BglII restriction site in the last exon of the gene. This construct, which is driven by the endogenous promoter and 5′UTR region of the Tab2 gene, was able to restore phototrophic growth of the mutant after transformation. However, the efficiency of transformation with this HA construct was lower (see Table I). Whole-cell proteins from three photosynthetic transformants were probed by immunoblot analysis with the anti-HA monoclonal antibody (Figure 3A). A 38 kDa protein with the size expected for the HA-tagged Tab2 protein without its transit peptide was detected in all transformants (lanes 1–3) but not in a wild-type strain (lane 4) or in the untransformed F14 mutant strain (lane 5). Moreover, these transformants containing the HA-tagged Tab2 protein accumulate wild-type levels of PsaA protein (Figure 1G, lane CHIM).
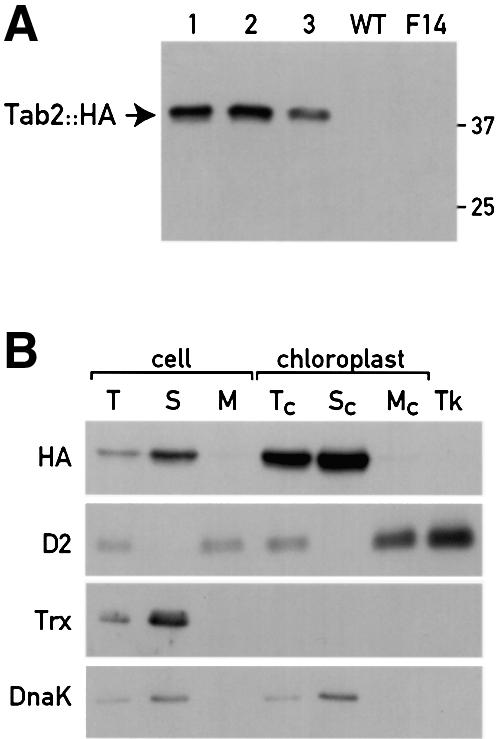
Fig. 3. Localization of the Tab2::HA protein. (A) Immunoblot analysis of total cell proteins (20 µg) of the wild-type, the F14 and three independent F14 strains complemented with the HA-tagged chimeric genomic–cDNA Tab2 gene (lanes 1–3). (B) Subcellular localization of the Tab2::HA protein. Immunoblot analysis of Tab2::HA in whole cells and cellular fractions of F14 complemented with the Tab2::HA construct. T (total), S (soluble) and M (membrane) extracts; Tc, Sc and Mc, total, soluble and membrane fractions from isolated chloroplasts, respectively. The analysis was also performed on purified thylakoids (Tk). The same fractions were analysed with antibodies directed against the thylakoid D2 subunit of PSII, the cytosolic soluble protein thioredoxin h (Trx) and the chloroplast stromal chaperone DnaK.
The HA-tagged strains were used to determine the subcellular localization of the Tab2 protein (Figure 3B). The Tab2::HA protein detected in total extract (lane T) was enriched in the soluble fraction (lane S). This protein was undetectable in the membrane fraction (lane M). The Tab2::HA protein was observed in isolated chloroplasts (lane Tc). The lack of cytosolic thioredoxin h (Trx) in all chloroplast-derived fractions (lanes Tc, Sc, Mc and Tk) demonstrates that our chloroplast preparations were not contaminated by cytosolic proteins. Separation of extracts from isolated chloroplasts into soluble and insoluble fractions revealed that the 38 kDa protein is in the soluble fraction (lane Sc). No significant amount of Tab2::HA protein was detected in the chloroplast membrane fraction (lane Mc) or in the purified thylakoids (lane Tk). The soluble chloroplast fraction was not contaminated with thylakoid membranes as indicated by the absence of the PSII D2 protein in lanes S and Sc. The chloroplast stromal DnaK chaperone has a similar distribution to the Tab2::HA protein confirming that the Tab2 protein is present in the same compartment.
Tab2 is part of a high molecular weight complex containing psaB RNA
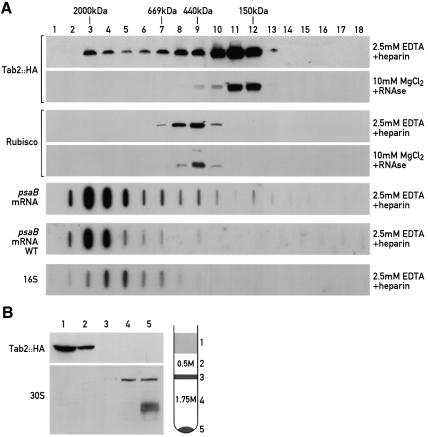
To test whether Tab2 is stably associated with other proteins, the soluble fraction of cells from the F14 strain complemented with Tab2::HA was subjected to size exclusion chromatography (Figure 4A). The bulk of Tab2::HA protein was detected in fractions corresponding to a size of 300 kDa. However, a significant part of Tab2::HA was also found in the heavier fractions with a size range up to 2000 kDa (Figure 4A). These large complexes were stable in the presence of 2.5 mM EDTA, but destabilized in the presence of 10 mM EDTA (data not shown). In the presence of RNase and MgCl2, the higher molecular weight complexes completely disappeared. As a control we verified that the elution of Rubisco was unaffected by the RNase treatment (Figure 4A). These results together with the fact that Tab2 is specifically required for translation of the psaB mRNA raised the possibility that the complex contains psaB mRNA. To test this, total RNA from each chromatography fraction was extracted, slot blotted and hybridized with a 32P-labelled psaB probe. The bulk of psaB mRNA was found in the high molecular weight fractions containing Tab2::HA, which disappear when the sample is treated with RNase. The same distribution of psaB mRNA was found in the wild-type control strain in fractions corresponding to a slightly larger size as compared with the small chloroplast ribosomal subunit containing 16S rRNA (Figure 4A). The Tab2 protein was undetectable in the polysome fraction (Figure 4B, fraction 5).
Fig. 4. Tab2 is part of a high molecular weight complex and is not associated with polysomes. (A) Size exclusion chromatography and immunoblot analysis of a cell extract of F14 complemented with the HA-tagged Tab2 gene. A soluble fraction was prepared in the presence of either 0.5 mg/ml heparin and 2.5 mM EDTA or 100 µg/ml RNase and 10 mM MgCl2, and subjected to size exclusion chromatography. Part of each fraction (20%) was separated by 6% SDS–PAGE, blotted and reacted with HA antiserum. As control the protein blots were performed with Rubisco antiserum. Molecular mass standards are included. The remaining part of the fractions (80%) was used to extract the RNA and analyse the psaB mRNA and 16S rRNA distribution by slot blotting. In the case of the psaB RNA hybridizations, the slot blots were performed on both the F14 strain complemented with the HA-tagged Tab2 gene or a wild-type reference strain. (B) Polysome fractions were immunoblotted with HA antiserum or an antiserum against the chloroplast 30S ribosomal subunit. The drawing corresponds to the classical profile obtained after centrifugation of a soluble crude extract on a discontinouous sucrose gradient (1.75 and 0.5 M). Most of the polysomes are contained in the pellet.
To confirm the specific association of psaB mRNA with the Tab2 complex, a crude extract from the strain expressing the Tab2::HA protein was immunoprecipitated with HA antiserum. RNA was extracted from the immunoprecipitated material and subjected to slot blot hybridization with the labelled psaB probe (Figure 5). A signal was readily detected with this probe, but not with a psbD probe. Both probes detected their corresponding RNA in the supernatant fraction from the immunoprecipitation (Figure 5). Moreover, when the same experiment was performed with a strain containing the IscU::HA protein, a protein involved in iron–sulfur cluster assembly, no signal was detected with either the psaB or the psbD probe, indicating that the immunoprecipitation with the HA antiserum was specific (Figure 5). Taken together these results show that psaB RNA is specifically associated with Tab2 in a high molecular weight complex.
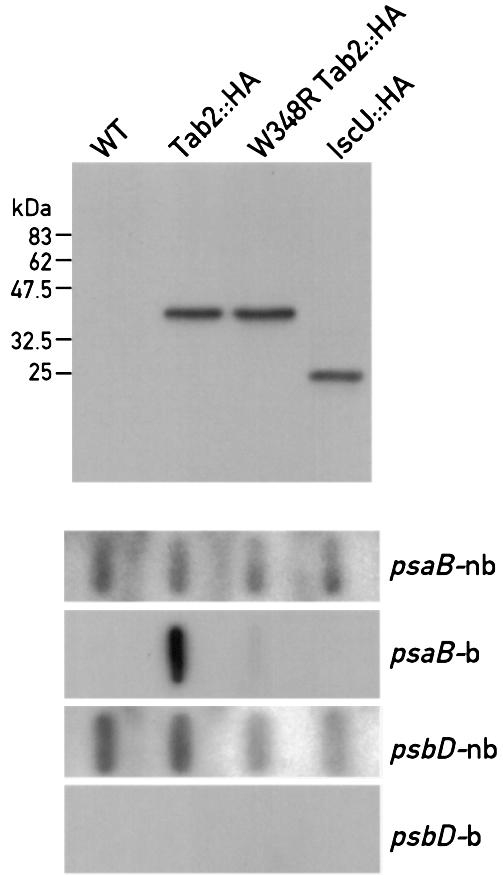
Fig. 5. psaB RNA is contained within the Tab2 complex. Immunoprecipitation of Tab2::HA and psaB mRNA detection by slot blot hybridization. Total soluble extracts were prepared from the F14 strain complemented with the HA-tagged wild-type Tab2 gene (Tab2::HA), with the mutant HA-tagged W348R Tab2 gene, and from a strain carrying an HA-tagged IscU gene or from a wild-type strain (control). After incubating the extracts for 1 h at 4°C with HA antiserum bound to beads, the beads were washed and an aliquot (10%) was used to detect the HA-tagged proteins by immunoblotting. The remaining bound material was used for RNA extraction and RNA slot blot analysis with psaB and psbD probes (psaB-b, psaD-b). RNA from the non-bound fractions were hybridized with the same probes (psaB-nb, psbD-nb). The hybridizations with the bound material were overexposed ∼40-fold relative to the non-bound material.
The W348R mutation abolishes the function of Tab2
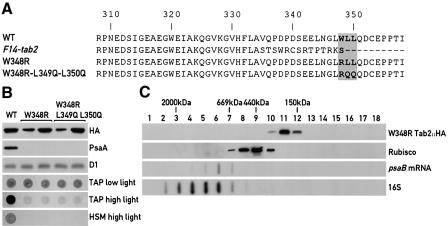
The only difference between the wild-type and the mutant F14-tab2 gene is a 14 nucleotide insertion in the last exon. This insertion creates a premature stop codon and leads to the loss of the C-terminal conserved WLL motif (Figures 2 and 6A). As the mutant strain is not able to grow photoautotrophically, it is possible that this tripeptide motif is essential for the in vivo function of Tab2. This was tested by transforming the F14 strain with mutated versions of the HA-tagged Tab2 gene. The two constructs tested differ from the wild-type copy by a substitution of W348 by R or by changing the WLL motif to RQQ (Figure 6A). The mutated genomic versions of Tab2::HA were inserted into the plasmid pSL18 and the transformants were selected for resistance to paromomycin (conferred by the AphVIII gene present in the plasmid) in low light on acetate-containing medium. Transformants were subjected to immunoblot analysis using the monoclonal anti-HA antibody and strains expressing the mutated Tab2 protein were examined. Genomic DNA from two transformants for each construct was isolated and the presence of the mutation was verified by sequencing. These strains were then tested for photoautotrophic growth on minimal medium (Figure 6B). The selected transformants express the mutated HA-tagged Tab2 protein at levels similar to the wild type. However, both types of transformant containing the two mutant forms of Tab2 were unable to grow photoautotrophically and grew poorly on acetate-containing medium under high light (60 µE/m2/s). Immunoblot analysis revealed that PsaA does not accumulate to detectable levels in the W348R mutant implying that PsaB expression is affected (Figure 6B). Taken together these results confirm that the deficiency in psaB mRNA translation in the mutant is due to the 14 nucleotide insertion near the 3′ end of the gene and point to the importance of the highly conserved W348 for Tab2 protein function.
Fig 6. The WLL motif in Tab2 is essential for its function. (A) Protein sequence comparison between the wild-type Tab2 polypeptide (WT), the putative translated ORF in the F14-tab2 mutant and the two Tab2 mutants (W348R and W348R L349Q L350Q). Only the 50 C-terminal amino acids of the proteins are displayed. The complete protein sequence is shown in Figure 2A. (B) Growth patterns of F14 mutant strains expressing the mutated Tab2 proteins. The F14 mutant strain was transformed using the HA-tagged chimeric genomic–cDNA described above but containing the amino acid substitutions indicated in (A). The transformants were selected on acetate-containing medium under low light in the presence of paromomycin. Total crude extracts from these strains were used to detect the mutated Tab2::HA protein by immunoblotting. Two independently obtained transformants with each of the two Tab2 mutations were tested for growth on TAP (acetate-containing medium allowing heterotrophic growth) under low and high light and on HSM (minimal medium requiring photoautotrophic growth). The control strain (WT) corresponds to the F14 mutant strain complemented with the wild-type HA-tagged Tab2 gene. (C) Gel filtration chromatography of a total soluble extract from the F14 mutant strain expressing the HA-tagged W348R Tab2 gene. Immunoblot analysis using the HA and Rubisco antisera and slot blot detection of psaB mRNA and 16S rRNA were performed as in Figure 4.
Size exclusion chromatography of a soluble extract from the F14 mutant strain transformed with a W348R HA-tagged Tab2 construct revealed that the high molecular weight complex containing the psaB mRNA is no longer present and that psaB RNA is found in fractions corresponding to smaller sizes (Figure 6C). Moreover psaB mRNA was nearly undetectable after immunoprecipitation of extracts from the strain containing the W348R substitution with HA antiserum (Figure 5) further confirming the importance of W348 for the formation of the psaB mRNA–Tab2 complex.
The Tab2 protein interacts directly with the 5′UTR of psaB mRNA in vitro
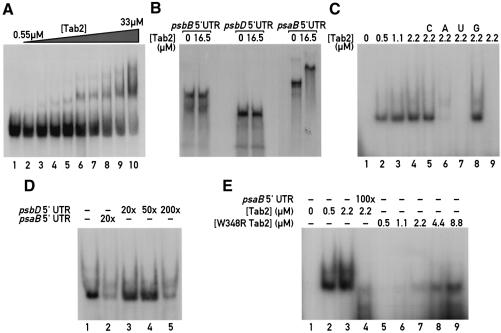
The dependency of the expression of the chimeric psaB 5′UTR–aadA gene on the Tab2 gene and the identification of a high molecular weight Tab2–psaB mRNA complex indicate that the Tab2 factor interacts either directly or indirectly with the psaB 5′UTR. To distinguish between these two possibilities, electrophoretic mobility shift assays (EMSAs) were performed. The Tab2 protein was expressed in Escherichia coli fused with GST and purified using a standard procedure and the GST epitope was removed by thrombin-mediated cleavage. The protein preparation was then incubated with radiolabelled in vitro transcribed RNAs corresponding to either the 3′ terminal 100 nucleotides of the psaB 5′UTR or to the complete psaB 5′UTR (495 bases). Formation of an RNA–protein complex with reduced mobility in a non-denaturing polyacrylamide gel was observed with the –100/+1 region of psaB 5′UTR with increasing concentrations of purified Tab2 protein (Figure 7A). The first experiments were performed with this sub-fragment of the psaB 5′UTR because we expected that the target region of the Tab2 complex would be close to the ribosome binding site. This interaction was confirmed by competition experiments with a 200-fold excess of cold psaB RNA (data not shown). Large amounts of recombinant purified Tab2 protein were necessary to completely shift the psaB 5′UTR fragment (Figure 7A). We estimate the Kd of this interaction as ∼18 µM based on quantitative measurements of the bands in Figure 7A with a phosphorimager (see Material and methods). This value appears to be surprisingly high. However, it is possible that the RNA region chosen does not contain the entire RNA binding site. Alternatively it is possible that the recombinant Tab2 protein is not fully refolded or that it needs additional factors for efficient RNA binding. We therefore performed additional mobility shift assays with the complete in vitro transcribed radiolabelled psaB 5′UTR. In the presence of large amounts of Tab2 (16.5 µM pure recombinant protein per lane) a complete shift could be readily observed with the psaB 5′UTR, but not with the psbB and psbD 5′UTRs (Figure 7B). At this concentration of Tab2 protein (16.5 µM) only half of the –100/+1 region of psaB 5′UTR was shifted (Figure 7A, lane 8). The large size of the psaB 5′UTR probe prevented clean assays. Further gel shift analysis was therefore performed by treating the samples with T1 RNase prior to electrophoresis (Figure 7C). The interaction between Tab2 and the complete psaB 5′UTR was significantly stronger than with the 3′-terminal subfragment (Figure 7A). Competition was tested with a 200-fold excess of cold psaB 5′UTR and a 500-fold excess of poly(riboA), poly(riboU), poly(riboC) and poly(riboG). Full competition was achieved with the psaB 5′UTR (Figure 7C, lane 9) and with poly(riboU) (lane 7) and to a lesser extent with poly(riboA) (lane 6). The specificity of this interaction was tested further by competition with the psbD 5′UTR. In this case competition was observed only with a 200-fold excess of psbD competitor (Figure 7D, lane 5) whereas it occurred with a 20-fold excess of psaB competitor (Figure 7D, lane 2).
Fig 7. Tab2 interacts directly with the psaB 5′UTR region. (A) EMSA with recombinant purified Tab2 protein and the 100 3′-terminal nucleotides of the psaB 5′UTR. Three femtomoles of in vitro transcribed RNA were incubated with increasing concentrations of purified Tab2 polypeptide (0, 0.55, 1.1, 2.2, 3.3, 8.25, 12.4, 16.5, 24.8, 33µM for lanes 1–10, respectively). (B) The Tab2 protein interacts specifically with psaB 5′UTR. EMSA of Tab2 with complete 5′UTRs of psaB, psbB and psbD. Three femtomoles of each probe were incubated without or with 16.5 µM purified Tab2 recombinant polypeptide prior to electrophoresis. (C) EMSA with RNase T1 digestion. After incubation of 3 fmol of in vitro transcribed complete psaB 5′UTR with Tab2 protein, the samples were treated with T1 RNase prior to electrophoresis (see Materials and methods). Under these conditions, the RNA probe alone is completely degraded (lane 1). In the presence of purified Tab2 protein, a part of the RNA molecule is protected from T1-mediated cleavage. Lanes 2–4 correspond to incubations with 0.55, 1.1 and 2.2 µM Tab2 protein, respectively. Lanes 5–9, competition assays with a 500-fold excess of poly(riboC) (C), poly(riboA) (A), poly(riboU) (U) and poly(riboG) (G) or with 200-fold excess of cold in vitro transcribed complete psaB 5′UTR (lane 9). (D). T1-EMSA in the presence of cold competitor psbD and psaB 5′UTR. One femtomole of complete psaB 5′UTR was incubated with 2.2 µM purified Tab2 recombinant protein prior to T1 digestion. The competitors RNAs were preincubated with the proteins for 5 min before the labelled transcript was added. The fold excess of each competitor is indicated in the figure. (E) T1-EMSA with wild-type and W348R recombinant Tab2 proteins. Three femtomoles of in vitro transcribed complete psaB 5′UTR were incubated with different concentrations of purified mutant Tab2 proteins followed by T1 RNase digestion. Lane 1, psaB 5′UTR probe without protein; lanes 2 and 3, psaB 5′UTR with 0.55 and 2.2 µM wild-type Tab2 recombinant protein, respectively. Lane 4, competition with a 100-fold excess of cold psaB 5′UTR. Lanes 5–9, psaB 5′UTR with 0.55, 1.1, 2.2, 4.4 and 8.8 µM purified recombinant mutant W348R Tab2 polypeptide.
A purified recombinant Tab2 protein containing the W348R substitution was used in mobility shift assay experiments with the complete psaB 5′UTR (Figure 7E). Although the mutated Tab2 protein was still able to interact with the 5′UTR, this interaction was considerably weaker than with wild-type Tab2 (Figure 7E). This result suggests that W348 of Tab2 is involved in the binding of Tab2 to psaB mRNA.
Discussion
Tab2 is an activator of translation initation of psaB mRNA
We have characterized the Tab2 factor that is affected in the PSI-deficient mutant F14. Protein pulse-labelling of F14 mutant cells indicates that the synthesis of the two PSI reaction centre subunits PsaA and PsaB is strongly reduced. The finding that the chimeric gene consisting of the psaB 5′UTR fused to the aadA coding sequence is expressed in the chloroplast in the presence of the wild-type Tab2 allele, but not in the presence of the F14-tab2 allele, shows that the F14-tab2 mutation interferes with psaB expression and that at least one target site of Tab2 is comprised within the psaB 5′UTR. These results, together with the observation that the fraction of psaB mRNA associated with polysomes is strongly diminished in F14 indicate that the Tab2 factor is involved in the initiation of translation and acts as a translational activator. Interestingly although the protein pulse-labelling results indicate that PsaA synthesis is strongly reduced in F14 (Figure 1A), a large portion of psaA mRNA remains associated with polysomes in this mutant suggesting that translation initiation is only moderately affected and that accelerated protein turnover of PsaA may occur in the absence of PsaB. However, this finding does not agree with results obtained with a chimeric psaA 5′UTR–petA construct whose expression was shown to be reduced in the absence of PsaB (Choquet et al., 2001). Further studies are required to settle this point.
Tab2 is part of a high molecular weight RNA–protein complex
Size exclusion chromatography indicates that Tab2 is part of a large complex with a molecular mass close to 2000 kDa. This complex contains psaB RNA based on several observations. First, Tab2 and psaB RNA cofractionate after size exclusion chromatography. Second, treatment of the complex with RNase A induces a shift in size from 2000 to 350 kDa. Third, psaB RNA can be specifically co-immunoprecipitated from cell extracts containing the HA-tagged Tab2 protein with HA antiserum. Fourth, a single amino acid change in Tab2 abrogates the formation of this RNA–protein complex.
The picture that emerges from several recent studies in Chlamydomonas and maize is that most of the chloroplast trans-acting factors involved in plastid gene expression are part of distinct RNA–protein complexes. Nac2 and Mbb1, which are involved in psbD and psbB mRNA processing and stability, respectively, are associated with RNA–protein complexes with sizes ranging from 600 to 1800 kDa. Both of these proteins contain several tetratrico peptide (TPR)-like domains (Boudreau et al., 2000; Vaistij et al., 2000). Raa3, which is required for the trans-splicing of the first intron of the psaA precursor RNAs, is part of a 1700 kDa RNA–protein complex (Rivier et al., 2001). The Crs1 and Crs2 splicing factors of maize are also part of high molecular weight RNA–protein complexes (Jenkins and Barkan, 2001; Till et al., 2001).
Tab2 is an RNA binding protein conserved in oxygenic photosynthetic organisms
A rather unique feature of Tab2 is that amongst the ancillary factors involved in plastid gene expression, it is the first to be conserved in both prokaryotic and eukaryotic organisms capable of oxygenic photosynthesis. Our data indicate that Tab2 interacts directly with the psaB 5′UTR and thus has to recognize this sequence specifically. However, 5′UTRs are usually poorly conserved between cyanobacteria, algae and vascular plants. If the Tab2 orthologues share the same RNA binding specificity as the C.reinhardtii protein, one would have to assume that there is a specific match between each Tab2 orthologue and its RNA binding sequence. It is unlikely that Tab2 is required for translation elongation because it is not associated with polysomes. Another possibility is that the C.reinhardtii protein has a dual function by being involved both in the recognition and binding of the psaB 5′UTR and in an early step of PSI assembly. Perhaps only the latter role is shared with the Tab2 orthologues from the other organisms. Two specific PSI assembly factors, Ycf3 and Ycf4, have previously been identified in land plants (Ruf et al., 1997), algae (Boudreau et al., 1997) and cyanobacteria (Wilde et al., 1995). However, in contrast to Tab2, these factors act at a post-translational level and are firmly associated with the thylakoid membrane and therefore cannot be part of the stromal Tab2 complex.
How does Tab2 function?
We propose that Tab2 acts early in the PSI assembly pathway by binding to psaB mRNA, perhaps already to the nascent RNA chain. Tab2 could possibly be involved in targeting this RNA to the thylakoid membrane where, together with the thylakoid membrane protein Tab1 (Stampacchia et al., 1997; F.Laroche and J.D.Rochaix, unpublished results), it promotes the initiation of translation and perhaps also the insertion of the nascent PsaB polypeptide chain into the membrane. Shortly before or after initiation of translation, Tab2 must be released from the psaB mRNA since it is undetectable in the polysome fraction. At this stage it may recruit a new psaB mRNA for translation. Upon insertion of PsaB in the thylakoid membrane, newly synthesized PsaA protein would be stabilized and inserted into the membrane followed by the assembly of the PsaA–PsaB heterodimer that forms the core of the PSI reaction centre.
Materials and methods
Strains and media
Chlamydomonas reinhardtii wild-type and mutant strains were grown as described (Harris, 1989; see Supplementary data).
Genomic complementation
The mutant strain F14 was rescued by transformation with an ordered cosmid library and selection for phototrophic growth as described (Purton and Rochaix, 1994; Zhang et al., 1994; Vaistij et al., 2000; see Supplementary data).
Nucleic acid techniques
The pSL17 plasmid (S.Lemaire and J.D.Rochaix, unpublished results) containing the AphVIII gene conferring paromomycin resistance (Sizova et al., 2001) was used to clone the mutated version of Tab2::HA. The pSL18 plasmid (S.Lemaire and J.D.Rochaix, unpublished results) containing both psaD 5′ and 3′UTRs bordering the multicloning site, and the AphVIII gene was used to clone the Tab2 cDNA. Procedures for standard molecular techniques were performed as described (Ausubel et al., 1998; see Supplementary data). The Tab2 cDNA and genomic sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AJ577849 and AJ577850, respectively.
EMSA
For EMSA, between 200 ng and 12 µg of purified Tab2 recombinant protein (corresponding to concentrations of 0.55–33 µM) were incubated with 32P-labelled transcript (3 fmol in vitro transcribed RNA cloned under control of the T7 promoter in pUC18) in 20 µl of 100 mM KCl, 20% glycerol (v/v), 2 mM EDTA, 20 mM HEPES–NaOH pH 8 and 10 µg of yeast tRNA for 20 min on ice. For competition studies, the competitor RNAs were preincubated with the proteins for 5 min before the labelled transcript was added. In the experiments involving the complete psaB 5′UTR, 10 U of T1 RNase (Roche, Mannheim, Germany) were added and the incubation continued for 20 min. Samples were then subjected to electrophoresis (3 h at 200 V and 4°C) in 4% (w/v) native polyacrylamide gel (29:1) using Tris–borate–EDTA as electrophoresis buffer. Gels were dried and exposed at –80°C overnight. Kd was estimated as the concentration of Tab2 at which the free and Tab2-bound RNA are equal (Figure 7A).
Protein extracts, size exclusion chromatography and immunoblots
Protein extracts and size exclusion chromatography were performed as described (Zerges and Rochaix, 1998; Rivier et al., 2001; see Supplementary data).
For immunoblot analysis, proteins were separated by SDS–PAGE using an acrylamide concentration of 6% (w/v) and electroblotted to nitrocellulose membranes (Protran, 0.45 µm; Schleicher & Schuell, Inc., Keen, NH). Primary antisera used were mouse α-HA (1:1000, Eurogentec), rabbit α-thioredoxin-h (1:10000; gift of S.Lemaire), α-PsbD (D2) (1:10000; gift of P.Nixon), α-PsaA (1:5000; Redding et al., 1998), α-DnaK (1:1000; H.Naver, K.Wilson and J.D.Rochaix, unpublished results) and α-30S (1:10000, gift of A.Boschetti). The signals were visualized by enhanced chemiluminescence (Durrant, 1990).
Immunoprecipitations of HA-tagged proteins were performed with Anti-HA Affinity Matrix (Roche Diagnostics Corp., IN) following a slightly modified procedure. One hundred micrograms of affinity matrix were first blocked by incubation for 1 h at 4°C with 3 mg of a wild-type total protein extract. The beads were then washed five times with 1 ml of 50 mM Tris–HCl pH 7.5, 150 mM NaCl and 10 mM MgCl2 and resuspended in 100 µl of the same buffer except that the NaCl concentration was lowered to 100 mM. Subsequently, 3 mg of total protein extract obtained from the HA-tagged transformants were incubated at 4°C for 1 h. The beads were washed five times and resuspended in 200 µl of the same buffer, 20 µl were used for immunoblotting with the monoclonal α-HA antibody and RNA was extracted from the remaining 180 µl and used for slot blot analysis.
Polysome purification
The polysome fraction was purified as described (Barkan, 1988; Yohn et al., 1996; see Supplementary data).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank N.Roggli for preparing the figures, W.Martin for assistance with phylogenetic analysis, J.Van Dillewijn for skilled technical assistance, F.Barneche, M.Goldschmidt-Clermont and L.Merendino for critical reading of the manuscript, and A.Boschetti, S.Lemaire and P.Nixon for antisera. D.D. was supported by a long-term EMBO fellowship ALTF 300-2001. This work was supported by grant 3100-0667763.02 from the Swiss National Foundation.
References
- Adachi J. and Hasegawa,M. (1996) Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol., 42, 459–468. [DOI] [PubMed] [Google Scholar]
- Auchincloss A.H., Zerges,W., Perron,K., Girard-Bascou,J. and Rochaix,J.D. (2002) Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol., 157, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,G.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & sons, New York, NY. [Google Scholar]
- Barkan A. (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J., 7, 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. and Goldschmidt-Clermont,M. (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie, 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Barkan A., Walker,M., Nolasco,M. and Johnson,D. (1994) A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J., 13, 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P. and Levine,R.P. (1967) Detecting mutants that have impaired photosynthesis by their increased level of fluorescence. Plant Physiol., 42, 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi,Y., Lemieux,C., Turmel,M. and Rochaix,J.D. (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J., 16, 6095–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Nickelsen,J., Lemaire,S.D., Ossenbuhl,F. and Rochaix,J.D. (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J., 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wostrikoff,K., Rimbault,B., Zito,F., Girard-Bascou,J., Drapier,D. and Wollman,F.A. (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans., 29, 421–426. [DOI] [PubMed] [Google Scholar]
- Cohen A., Yohn,C. and Mayfield,S.P. (2001) Translation of the chloroplast-encoded psbD mRNA is arrested post-initiation in a nuclear mutant of Chlamydomonas reinhardtii. Plant Physiol., 158, 1069–1075. [Google Scholar]
- Durrant I. (1990) Light-based detection of biomolecules. Nature, 346, 297–298. [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker,M.B. and Barkan,A. (1999) Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J., 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J., Chua,N.H., Bennoun,P., Schmidt,G. and Delosme,M. (1980) Studies on mutants deficient in the photosystem I reaction centers in Chlamydomonas reinhardtii.Curr. Genet., 2, 215–221. [DOI] [PubMed] [Google Scholar]
- Girard-Bascou J., Choquet,Y., Schneider,M., Delosme,M. and Dron,M. (1987) Characterization of a chloroplast mutation in the psaA2 gene of Chlamydomonas reinhardtii. Curr. Genet., 12, 489–495. [DOI] [PubMed] [Google Scholar]
- Girard-Bascou J., Pierre,Y. and Drapier,D. (1992) A nuclear mutation affects the synthesis of the chloroplast psbA gene production in Chlamydomonas reinhardtii. Curr. Genet., 22, 47–52. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1991) Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res., 19, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol., 177, 115–180. [DOI] [PubMed] [Google Scholar]
- Jenkins B.D. and Barkan,A. (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J., 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989) The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Leon P., Arroyo,A. and Mackenzie,S. (1998) Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 453–480. [DOI] [PubMed] [Google Scholar]
- Mayfield S.P., Christopher,B.Y., Cohen,A. and Danon,A. (1995) Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol., 46, 147–166. [Google Scholar]
- McCormac D.J. and Barkan,A. (1999) A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell, 11, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., van Dillewijn,J., Rahire,M. and Rochaix,J.D. (1994) Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J., 13, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton S. and Rochaix,J.D. (1994) Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol. Biol., 24, 533–537. [DOI] [PubMed] [Google Scholar]
- Redding K., Redding,K., MacMillan,F., Leibl,W., Brettel, K, Hanley,J., Rutherford,A.W., Breton,J. and Rochaix,J.-D. (1998). A systematic survey of conserved histidines in the PsaA and PsaB proteins of Chlamydomonas reinhardtii reveals the likely axial ligands of P700. EMBO J., 17, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier C., Goldschmidt-Clermont,M. and Rochaix,J.D. (2001) Identification of an RNA–protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J., 20, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D. (1992) Post-transcriptional steps in the expression of chloroplast genes. Annu. Rev. Cell Biol., 8, 1–28. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D. (1996) Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol., 32, 327–341. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D., Kuchka,M., Mayfield,S., Schirmer-Rahire,M., Girard-Bascou,J. and Bennoun,P. (1989) Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J., 8, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel,H. and Bock,R. (1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol., 139, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I., Fuhrmann,M. and Hegemann,P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene, 277, 221–229. [DOI] [PubMed] [Google Scholar]
- Stampacchia O., Girard-Bascou,J., Zanasco,J.L., Zerges,W., Bennoun,P. and Rochaix,J.D. (1997) A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell, 9, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B., Schmitz-Linneweber,C., Williams-Carrier,R. and Barkan,A. (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA, 7, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F.E., Boudreau,E., Lemaire,S.D., Goldschmidt-Clermont,M. and Rochaix,J.D. (2000) Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 97, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Hartel,H., Hubschmann,T., Hoffmann,P., Shestakov,S.V. and Borner,T. (1995) Inactivation of a Synechocystis sp. strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell, 7, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Choquet,Y., Wollman,F.A. and Girard-Bascou,J. (2001) TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics, 159, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn C.B., Cohen,A., Danon,A. and Mayfield,S.P. (1996) Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Mol. Cell. Biol., 16, 3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn C.B., Cohen,A., Rosch,C., Kuchka,M.R. and Mayfield,S.P. (1998) Translation of the chloroplast psbA mRNA requires the nuclear-encoded poly(A)-binding protein, RB47. J. Cell Biol., 142, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W. and Rochaix,J.D. (1994) The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus-encoded functions in Chlamydomonas reinhardtii. Mol. Cell. Biol., 14, 5268–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W. and Rochaix,J.D. (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol., 140, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Herman,P.L. and Weeks,D.P. (1994) Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol., 24, 663–672. [DOI] [PubMed] [Google Scholar]