Abstract
Gene expression profiling provides tremendous information to help unravel the complexity of cancer. The selection of the most informative genes from huge noise for cancer classification has taken centre stage, along with predicting the function of such identified genes and the construction of direct gene regulatory networks at different system levels with a tuneable parameter. A new study by Wang and Gotoh described a novel Variable Precision Rough Sets-rooted robust soft computing method to successfully address these problems and has yielded some new insights. The significance of this progress and its perspectives will be discussed in this article.
Keywords: α depended degree, cancer, classification, gene expression profiling, network, rough sets, soft computing
Gene expression profiles (GEP) either by microarray or by Serial Analysis of Gene Expression (SAGE) provide us with data of unparalleled wealth, but cancer as a system failure is still mysterious. Many existing methods utilize too many genes to obtain discriminative features associated with cancer, and are unclear or not interpretable at a biological level. Developing simpler rule-based models with as few marker genes as possible is preferable. Ideally, such hub genes could naturally exhibit biological relevance. But good research is never simple and requires hard work: there is no “free lunch” researchers. However, based on a Variable Precision Rough Set (VPRS) core1 with the introduction of α depended degree, Wang and Gotoh recently developed a simple, efficient and straightforward method for accurate cancer classification using single genes or gene pairs and subsequently inferred the direct gene regulatory network.2–4 They first identified hub genes associated with colon cancer using this approach, and subsequently inferred the direct gene regulatory network among the identified genes, and how these are regulated within the genome. Finally, two biologically meaningful findings were obtained.5 This method is not only user-friendly, simple and biologically interpretable, but is cost-effective in a clinical setting with single genes or gene pairs.6 The method also has the advantages of being relatively easy to understand and follow, along with the availability of programming codes with either open access or GNU general public license (GPL).
A Brief Introduction of the α Depended Degree Rough Set Soft Computing Approach
Firstly, rough set theory neds to be understood. In this theory, U is a universe of discourse and R is the equivalent relation. The degree of dependency of a set of attributes Q on another set of attributes P is denoted by γP(Q) and is defined as:
where represents the size of the union of the lower approximation of each equivalence class in U/R(Q) on P in U, and |U| represents the size of U (the set of samples).
If Q is the decision attribute D, and P is a subset of condition attributes, then γP(D) represents the depended degree of the condition attribute subset P by the decision attribute D; i.e the degree to which P can discriminate between the distinct classes of D. In this sense, γP(D) reflects the classification power of the subset P of attributes. The greater γP(D) is, the stronger the classification ability that P possesses. The measure of the depended degree becomes the basis for selecting informative genes.
For some datasets, it is difficult to detect the discriminative features based on the canonical depended degree because of its excessively rigid definition. Therefore, Wang and Gotoh introduced α depended degree, a generalization form of the depended degree sets in their VPRS model,2–5 then utilized the α depended degree as the basis for choosing genes. The α depended degree of the condition subset P by the decision attribute set D is defined by:
where 0≤α≤1, and pos(P,X,α) = ∪{Y ∈ U/R(P) ||Y ∩ X|/|Y|≥ α}.Here |*| denotes the size of set * and U/R(•) denotes the set of equivalence classes induced by the equivalence relation R(•). The depended degree is a specific case of the α depended degree when α = 1. For the selection of high class-discrimination genes, the lower limit of α has been set to 0.7 in practice.2
Wang and Gotoh created classifiers based on decision rules. One decision rule in the form of “A ⇒ B” indicates that “if A, then B”, where A is the description of the condition attributes and B, the description of the decision attributes. The confidence of a decision rule A ⇒ B is defined as follows: confidence where support (A) denotes the proportion of samples satisfying A and support (A ∧ B) denotes the proportion of samples satisfying A and B simultaneously. The confidence of a decision rule indicates the reliability of the rule.
For each determined α value, only the genes with γ P(D,α) = 1 were selected to build decision rules.2 The sufficient reliability of the derived decision rules as ensured by setting a high threshold for α.2–5
User-Friendly Theory, Practical Simplicity and Biological Interpretability
Biologists generally speak different “languages” from mathematicians. Unlike statistical methods, this novel method, the Bimodality Index,7 sought to be interpretable for biological relevance simple for cancer classification in both theory and practice. Importantly, this method allows a straightforward inference of the direct gene regulatory network. All the gene selection, classification and network construction processes in this method correlate with well biologically meaningful decision rules, such as tumor vs. normal cells, up-vs. down-regulation, and positive vs. negative regulation. This contrasts with the process of many other methods, where the classifying power of the gene expression level and the biological importance of that gene are generally only weakly related and thus many biomarker candidates could turn out to be false positives.
This novel method is rooted on the rough sets theory (RS) seminally proposed by Pawlak8 for analysis of inconsistent, incomplete, imprecise and precise data. The main advantage of RS is that it does not need any preliminary or additional information about data, e.g. probability in statistics or basic probability assignment in Dempster–Shafer theory. RS has been successfully applied in the areas of medicine and pharmacology.9 Its application in cancer classification and prediction has begun.2–5 As the inhibition of a single molecular target can alter the morphology of tumor cells in lrECM and reduce tumor growth in vivo,10 so a few genes, gene pairs or even a single gene can become biomarkers.2–5,11 Logically, the low complexity classifiers for single genes or gene pairs aids interpretability, i.e. they enhance our ability to interpret the selected (pair of) genes.
This theory itself may be akin to our routine identification (or classification) of objects in the real-world setting. The rationale is first to filter lots of redundant information (i.e. noise) but to retain the critical information (i.e. signal). This is followed by making decision rules based on core information and classifying the whole dataset. In order to extract the hidden meaningful rules, we sometimes need to lose some rigid definitions. Thus Wang and Gotoh introduce the flexible α depended degree under soft computing consideration. This allows some single genes or gene pairs to have strong class discriminatory power, although they would be ignored with the conventional attribute depended degree.2 Interestingly, this also enables us to infer the networks and modules.
In fact, Wang and Gotoh reject the attribute reductions in classic rough set theory due to its high computational expense, uncertainty of predictive performance and non-uniqueness.2 Because of depended degree, they use the entropy-based discretization method12 for discrete gene expression values within datasets.2–5 The stopping point of the recursive step for this algorithm depends on the minimum description length (MDL) principle and the discretization was implemented in the Waikato Environment for Knowledge Analysis (WEKA) package,13 which gives open access to a collection of state-of the-art techniques in machine learning algorithms for data mining tasks; these algorithms can either be applied directly to a dataset or called from user’s own Java code, so it is an excellent unified “workbench” not only for data preprocessing, classification, regression, clustering, association rules and visualization but is also well suited for developing new machine learning schemes.
This process is more or less streamlined. In the discretized decision table, Wang and Gotoh found that most genes were unable to distinguish different classes and were removable, while some genes can distinguish different classes by decision rules.5 They achieved very high leave-one-out-cross-validation (LOOCV) accuracy for an array of datasets.2–5 The reported accuracy is superior to or comparable with other established approaches.2–5
In their new work on the colon cancer dataset, Wand and Gotoh identified 18 discriminative hub genes for cancer. Ten of these (e.g. DES and ACTA2) belong to down-regulated genes in a tumor, while eight other genes (e.g. IL8, HSPD1, SRPK1) belong to up-regulated genes in a tumor. Most, if not al, l of these genes are involved in cancerogenesis, as shown in published literature. Strikingly, IL8 and DES have been identified as cancer hub genes in several independent studies.14
Inference of the Gene Regulatory Network
Obtaining a direct regulatory network of these discriminative hub genes is of particular interest. Functional entities, such as pathways nad signalling networks are more robust descriptors than gene lists.15 The similarity measures, such as Pearson’s correlation and mutual information16 cannot characterize the cause–effect gene regulatory relations in undirected networks very well. In contrast, directed gene regulatory networks, such as Bayesian networks, Boolean networks, Ordinary Differential Equations or IDA17,18 can explore the cause–effect regulatory relations and provide better insights into biological systems than the co-expression relation. Moreover, most previous efforts utilized all gene expression data from microarrays so that the authentic gene interactions were covert due to many genes that were unrelated to cancer. However, it is expected that a few highly class-discriminative hub genes could greatly enhance the authenticity and confidence of computed gene interaction networks.
Following the identification of hub genes, Wang and Gotoh investigated the gene regulatory network by employing the method described above. The details of this method are as follows: one gene instead of a class is used as the decision attribute. If “GENEI” is substituted for “Class label” in a decision table, GENE-I is regarded as the decision attribute with two distinct values: up-regulation and down-regulation, and a new derivative table can be obtained. Likewise, Wang and Gotoh implement the discretization of this derivative table to obtain another newly derived table. Applying the same learning algorithm to this latest derived table, they can induce the decision rules linking GENE-I to GENE-II: if the expression level of GENE-I in one sample is not greater than value A, then GENE-II is down-regulated; otherwise, GENEII is up-regulated. In other words, if GENE-I is down-regulated, then Gene-II is down-regulated; if Gene-I is up-regulated, then Gene-II is up-regulated. They are not necessarily true in reverse. Therefore, a directed regulatory relation of GENE-I to GENE-II, a positive one, is established.5
Similarly, Wang and Gotoh regard each of the 18 identified genes as the decision attribute in turn, and examine the regulatory relations that the other genes exert on them. They constructed all their network graphs using Cytoscape software.19 They analyzed one network containing only these 18 genes, and another containing genes other than these 18. The first networke one orchestrates the core of the latter in the genome. Modules constitute the “building blocks” of molecular networks. To explore the modularity of networks, Wang and Gotoh use the Cytoscape plugin MCODE19 to analyze the network constructed and detected two significant modules, one of which forms a feed-forward loop. They conclude that the co-regulation of multiple activators could be at least partly responsible for the occurrence of tumors. Further, they chose the Cytoscape plugin BiNGO20 to perform a Gene Ontology (GO) based enrichment analysis of the two modules. Other gene functional analysis, such as Gene Set Enrichment Analysis (GSEA), could also be useful. Finally, they observed that in colon cancer, the gene regulatory network, the up-regulated genes are regulated by more genes than down-regulated ones, while the down-regulated genes regulate more genes than up-regulated ones; secondly, tumor suppressors inhibit tumor activators and activate as many other tumor suppressors as possible. In contrast, tumor activators activate other tumor activators and inhibit as few tumor suppressors as possible.5 A fascinating question: is it true for other cancers and how about its validation of wet-lab experiments?
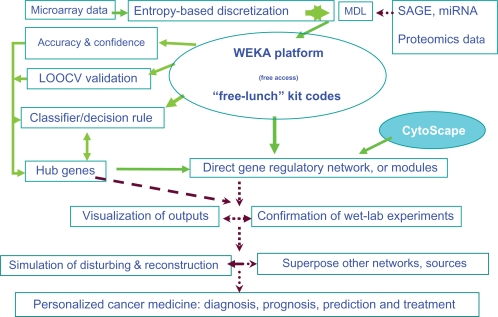
This method is a new option for cancer classification and direct gene regulatory network inference. For these processes, it exhibits its inherent biological relevance. Finally, this method out-performs or at least matches other approaches, though LOOCV may have a large variance of accuracy.2–5 Taking into account its other merits, especially its simplicity, this is a great way to explore the cancerogenesis according to Occam’s Razor: the simple theory is preferable to the complex one. A scheme of a “free-lunch” toolkit for cancer classification and networks is shown in Figure 1.
Figure 1.
Scheme of the “free lunch” toolkit for cancer classification at the network level and beyond. Arrow: executed Dash arrow: being executed “Free lunch” kit codes: the programming codes for cancer classification, hub gene identification and inference of gene regulatory network under GNU GPL.
Future Directions
This kind of cause–effect inference could have practical value in the prioritization and design of perturbation experiments. Of course, only verification via follow-up wet-lab studies rather than published literature could prove that the conclusions from this new study are perfectly valid and reliable, though, theoretically, the process always demonstrates biological relevance, which may have already sparked the curiosity and passion of biologists and clinicians.
In the near future, a wide variety of datasets, such as subtype or multi-class cancer microarray data, microRNA array data, Serial Analysis of Gene Expression (SAGE) data and proteomic data could challenge the “free-lunch” toolkit. Thus far, we have identified seven highly discriminative (hub) genes in the SAGE breast cancer dataset,21 which has approximately 2.7 million tags and which has 27 samples, each of which are described as lymph node [LN(+)] and [LN(−)] primary breast tumors. All identified genes have high classification accuracy using this method under α = 0.8 (Results are presented in Table 1). These seven hub genes are very interesting and informative for their biological relevance. First, it is well known that the role of the ATF2/AP1 complex and its network is at the hub of tumorigenesis22,23 and this has been reflected by a high classification accuracy of 88.89%. ATF2 communicates with an array of cell signalling pathways that are important for mammary tumors, e.g. TGFbeta. This emphasizes that comprehensive understanding of how ATF2 functions promises to provide new avenues for therapeutic intervention in breast cancer. CARD10/CARMA3 has a physical and functional interaction with IkappaKgamma-NEMO in lymphoid and non-lymphoid cells, is required for GPCR-induced NF-kappaB activation24 and is important in LPA-induced cancer cell in vitro invasion. Secondly, this hub gene list includes master regulators in angiogenesis (ATF2, CARD10 and VG5Q/AGGF1), the age-related neurodegenerative disease (MGRN1 and CARD10; cancer is one disease associated with ageing) and the main cell signalling pathways for breast cancer, such as the NF-KappaB (CARD10) pathway, the IL-6 (PKD1-like) pathway, the TGFbeta/STAT3/p38alphaMAPK/ATF-2 pathway, ATM/DNA repair, and the PGE(2)/PKA/PKC signalling pathways (ATF2). Thirdly, novel proteins like CGI-41 and UBLCP1 (MGC10067 nad the ubiquitin-like domain containing CTD phosphatase 1) may point us in a new direction for future breast cancer study because CTD phosphatase, UBLCP1, has a relatively lower level of expression in most normal adult tissues and at a higher level in tumor tissues, and it could play a major role in polymerase recycling.25
Table 1.
The seven hub genes identified in the breast cancer SAGE dataset.
| Tag | Gene symbol | Classification accuracy | Alpha value | Classification rules |
|---|---|---|---|---|
| TATATGCCTA | CGI-41 | 85.19% | 0.8 | If gene expression > 1.5, then LN(−);LN(+) otherwise |
| GGCGGGTCGG | MGRN1 | 85.29% | 0.8 | If gene expression > 1.5, then LN(−); LN(+) otherwise |
| GATGTCTTGT | MGC10067 | 81.48% | 0.8 | If gene expression > 6.5, then LN(−);LN(+) otherwise |
| GACTGTTAAT | VG5Q | 85.19% | 0.8 | If gene expression > 5.5, then LN (−);LN(+) otherwise |
| GTGGATTCAT | ATF2 | 88.89% | 0.8 | If gene expression > 4.5, then LN(−);LN(+) otherwise |
| TACTGGAGTA | CARD10 | 81.48% | 0.8 | If gene expression > 4.5, then LN(−);LN(+) otherwise |
| TTGACACTTT | PKD1-like | 81.48% | 0.8 | If gene expression > 31.5, then LN(−);LN(+) otherwise |
Importantly, the ENCODE project tells us that at least 93% of the analyzed human genome is transcribed in different cells into biologically meaningful RNAs that could greatly exceed the ∼ 1.2% encoding proteins.26 More and more attention is being given to RNA, especially Linc RNAs, microRNAs and antisense RNAs. However, the protein levels and IHC staining have a greater variety of available assays in the clinical setting. Archimedes once said, “Give me a lever long enough and a fulcrum on which to place it, and I shall move the world”. Recent advances in deep-sequencing application in ChIP-seq, SAGEseq, HITS-CLIP27 and MALDI-TOF mass-spec in proteomics and the exponential increase of available profiling datasets may act as a metaphorical fulcrum. The method of Wang and Gotoh, together with others, e.g. the Bimodality Index7,29 have made advances in the direction of being the lever. Simple, yet powerful and reliable techniques like the “free-lunch” toolkit could pave the way to unveiling the mystery of cancer.
Another direction is to dissect cancerogenesis in silico in conjunction with software such as Sorting Intolerant From Tolerant (SIFT),28 Polymorphism Phenotyping (PolyPhen)(http://coot.embl.de/PolyPhen) and Function Analysis and selection tool for single nucleotide polymorphisms (FASTSNP) (http://FASTSNP.ibms.sinica.edu.tw), or platforms such as GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/) and Metacore,29 as the mutational load and sequential functional module change could generally cause cancer. Most importantly, this method could further integrate the protein–protein interaction data, published literature information, siRNA library screen or knockout data, and thus construct comprehensive function-oriented gene, genetic and protein networks.30–33 A web-server and visualization module for displaying results in the clinical setting could make this toolkit even more popular.
The perturbations of gene regulatory networks could be essentially responsible for cancinogenesis5 and the therapeutic recovery could reflect the flexibility and robustness of biological system. It will be exciting to perform in silico simulation of perturbation of interaction networks and recovery with this toolkit as in10,34 as well as the in vivo confirmation of biomedical experiments with drug treatment.35
Acknowledgments
The author is deeply grateful to Dr.Wang for programming the code for analysis of SAGE data and for giving his opinion.
Footnotes
Disclosure
This manuscript has been read and approved by the author. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The author and peer reviewers of this paper report no conflicts of interest. The author confirms that they have permission to reproduce any copyrighted material.
References
- 1.Ziarko W. Variable precision rough set model. J Comput Syst Sci. 1993;46(1):39–59. [Google Scholar]
- 2.Wang X, Gotoh O. Microarray-based cancer prediction using soft computing approach. Cancer Inform. 2009;7:123–39. doi: 10.4137/cin.s2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Gotoh O. Accurate molecular classification of cancer using simple rules. BMC Med Genomics. 2009;2:64. doi: 10.1186/1755-8794-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Gotoh O. A robust gene selection method for microarray-based cancer classification. Cancer Inform. 2010;9:15–30. doi: 10.4137/cin.s3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Gotoh O. Inference of cancer-specific gene regulatory networks using soft computing rules. Gene Regul Syst Biol. 2010;4:19–34. doi: 10.4137/grsb.s4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 2009;7:199–216. doi: 10.4137/cin.s2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlak Z. Rough set theory. International J of Information and Computer Science. 1982;11:341–56. [Google Scholar]
- 9.Thangavela K, Pethalakshmib A. Dimensionality reduction based on rough set theory: A review. Appl Soft Comp. 2008;9(1):1–12. [Google Scholar]
- 10.Zhang X, Fournier MV, Ware JL, Bissell MJ, Yacoub A, Zehner ZE. Inhibition of vimentin or beta1 integrin reverts morphology of prostate tumor cells grown in laminin-rich extracellular matrix gels and reduces tumor growth in vivo. Mol Cancer Ther. 2009;(3):499–508. doi: 10.1158/1535-7163.MCT-08-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grate LR. Many accurate small-discriminatory feature subsets exist in microarray transcript data: biomarker discovery. BMC Bioinformatics. 2005;6:97. doi: 10.1186/1471-2105-6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fayyad UM, Irani KB. Proceedings of the 13th International Joint Conference of Artificial Intelligence: August 28–September 3 1993. Chambéry, France: Morgan Kaufmann; 1993. Multi-interval discretization of continuous-valued attributes for classification learning; pp. 1022–7. [Google Scholar]
- 13.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update; SIGKDD Explorations. 2009;11(1):2009. [Google Scholar]
- 14.Jiang W, Li X, Rao S, et al. Constructing disease-specific gene networks using pair-wise relevance metric: application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Syst Biol. 2008;2:72. doi: 10.1186/1752-0509-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–90. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 17.Maathuis MH, Colombo D, Kalisch M, Bühlmann P. Predicting causal effects in large-scale systems from observational data. Nat Methods. 2010;7(4):247–8. doi: 10.1038/nmeth0410-247. [DOI] [PubMed] [Google Scholar]
- 18.Xing B, van der Laan MJ. A causal inference approach for constructing transcriptional regulatory networks. Bioinformatics. 2005;21(21):4007–13. doi: 10.1093/bioinformatics/bti648. [DOI] [PubMed] [Google Scholar]
- 19.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 21.Abba MC, Sun H, Hawkins KA, et al. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5(9):1–10. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer. 2010 Jan;10(1):65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhoumik A, Ronai Z. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle. 2008 Aug;7(15):2341–5. doi: 10.4161/cc.6388. [DOI] [PubMed] [Google Scholar]
- 24.Fraser CC. G protein-coupled receptor connectivity to NF-kappaB in inflammation and cancer. Int Rev Immunol. 2008;27(5):320–50. doi: 10.1080/08830180802262765. [DOI] [PubMed] [Google Scholar]
- 25.Zheng H, Ji C, Gu S, et al. Cloning and characterization of a novel RNA polymerase II C-terminal domain phosphatase. Biochem Biophys Res Commun. 2005 Jun 17;331(4):1401–7. doi: 10.1016/j.bbrc.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 26.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319(5871):1787–9. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. ETS-FUSions networking, triggering and beyond. Genet Epigenet. 2010;3:1–4. [Google Scholar]
- 28.Zaghloul NA, Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 2010;26(4):168–76. doi: 10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bessarabova M, Kirillov E, Shi W, Bugrim A, Nikolsky Y, Nikolskaya T. Bimodal gene expression patterns in breast cancer. BMC Genomics. 2010;11(Suppl 1):S8. doi: 10.1186/1471-2164-11-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ourfali O, Shlomi T, Ideker T, Ruppin E, Sharan R. SPINE: a framework for signaling-regulatory pathway inference from cause–effect experiments. Bioinformatics. 2007;23(13):i359–66. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- 31.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311(5766):1481–4. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 32.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in C. elegans. Nat Genet. 2008;40(2):181–8. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 33.Yip KY, Alexander RP, Yan KK, Gerstein M. Improved reconstruction of in silico gene regulatory networks by integrating knockout and perturbation data. PLoS One. 2010;5(1):e8121. doi: 10.1371/journal.pone.0008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Wong L. Identifying good diagnostic gene groups from gene expression profiles using the concept of emerging patterns. Bioinformatics. 2002;(5):725–34. doi: 10.1093/bioinformatics/18.5.725. [DOI] [PubMed] [Google Scholar]
- 35.Geva-Zatorsky N, Dekel E, Cohen AA, Danon T, Cohen L, Alon U. Protein dynamics in drug combinations: a linear superposition of individual drug responses. Cell. 2010;140(5):643–51. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]