Abstract
Objective
Juvenile Idiopathic Arthritis (JIA) is a heterogeneous group of inflammatory of diseases and there no clinically useful prognostic markers to predict disease outcome in these children. Synovial fluid is likely a reflection of the proteins present in the inflamed synovium. The purpose of this study was to delineate the synovial fluid proteome and determine whether there are differences in the protein expression in subtypes of JIA.
Methods
Synovial fluid from children with oligoarticular, polyarticular and systemic JIA were compared. Two dimensional gel electrophoresis for protein separation and Matrix associated laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS) and Quadripole time of flight mass spectrometry (Q-TOF-MS) for protein identification were used for this study. Synovial fluid cells were analyzed by PCR for the presence of haptoglobin mRNA.
Results
The synovial fluid proteome of the samples was delineated. The majority of proteins showed overexpression in JIA synovial fluid as compared to non-inflammatory controls. There were 24 statistically significant differentially expressed spots (> 2 fold change and p<.05) between the subtypes of JIA. PCR analysis revealed haptoglobin mRNA suggesting that haptoglobin is locally produced in an inflamed joint in JIA.
Conclusions
Despite similar histological appearance of inflamed joints in subtypes of JIA, there are differences in protein expression in the subtypes of JIA. Haptoglobin is differentially expressed between the subtypes of JIA and is locally produced in an inflamed joint in JIA. Haptoglobin and other differentially expressed proteins may be potential biomarkers in JIA.
Juvenile Idiopathic Arthritis (JIA) is a heterogeneous group of inflammatory diseases with varying sex distribution, genetic predisposition, clinical manifestations, disease course and prognosis. At present there are no clinically useful prognostic markers to predict disease outcome in these patients. The International League of Associations for Rheumatology (ILAR) defines three main accepted subtypes of JIA [1]: Oligoarticular JIA, the most frequent subtype, is characterized as arthritis affecting four or fewer joints in the first six months of disease. The outcome is usually good, though some patients may have a more extended course and/or develop uveitis. Polyarticular JIA is defined as arthritis affecting more than four joints during the first six months of disease. In polyarticular JIA, there is an increased frequency of chronic, debilitating disease especially in rheumatoid factor positive children. Systemic JIA (SJIA) refers to children with a documented quotidian fever of at least two weeks duration, arthritis in any number of joints, and typical rash, generalized lymphadenopathy, enlargement of liver or spleen or serositis. The arthritis in SJIA is frequently severe and erosion forming. SJIA is also associated with macrophage activation syndrome (MAS), a severe, potentially life threatening condition where activated macrophages exhibit hemophagocytic activity.
In addition to the various clinical manifestations of the three subgroups, there is also evidence of different cytokine production, gene expression and Human Leukocyte Antigen (HLA) associations [2–5]. With such distinct clinical manifestations, immunoregulation, and genetic background, the subtypes of JIA are likely to have different pathophysiologies and mediators of disease.
Proteomic studies are useful to identify protein profiles and biomarkers of disease. Several studies have evaluated arthritis at the protein level by studying the synovial fluid proteome [6–10]. A study by Liao, et al. used LC/LC-MS/MS to differentiate erosive RA and non-erosive RA and identified 33 potential disease severity biomarkers [7]. Sinz et al used two dimensional electrophoresis (2-DE) along with mass spectrometry demonstrating differential protein expression between rheumatoid arthritis and osteoarthritis [6]. Gibson DS, et al have performed proteomic studies in JIA using 2-DE which demonstrated differential expression of proteins in synovial fluid versus serum, identified specific clusters of proteins which differentiated between subtypes in JIA and also identified proteins differentiating those children with a more persistent course [9, 10]. In our study, we used 2-DE gel techniques and MALDI-TOF MS technology to perform a global identification of the synovial proteome in JIA as well as to identify proteomes specific to the subtypes of JIA. In addition we provide data demonstrating that haptoglobin is locally produced in the inflamed joint of JIA which is a novel finding. We hypothesize that the identified proteins may play a key role in the pathophysiology of the subtypes of disease and are potential biomarkers of disease.
Materials and Methods
Patients and study subjects
Synovial fluid (SF) was collected from patients with active JIA defined according to the criteria established by ILAR. Decision to perform an arthrocentesis was at the discretion of the treating physician. The study patients were recruited from the rheumatology clinic at Children’s Hospital of Pittsburgh. Banked synovial fluid was also obtained from Cincinnati Children’s Hospital JRA Tissue Repository. Synovial fluid was also collected from patients with no history of JIA or inflammatory disease who underwent an orthopedic procedure. These were used as non-inflammatory controls. The study was approved by the Institutional Review Board at the University of Pittsburgh. Informed consent was obtained from all guardians of patients and assent was obtained from the subjects when appropriate.
Synovial fluid collection and storage
The synovial fluid (SF) samples were placed on ice immediately after collection, centrifuged at 1400 rpm for 10 minutes to remove cells and debris and stored at −80°C. Synovial fluid mononuclear cells were separated on a Ficoll gradient at 2050 rpm for 25 minutes. The cells were washed with PBS and centrifuged twice at 1400 rpm for 10 minutes. The cell pellet was resuspended in Trizol (Invitrogen, Carlsbad, CA) for RNA preservation and stored at −80°C.
Sample processing for gel electrophoresis
Samples of SF were pooled by subtype for analysis as this method has been shown to reduce inter-individual differences, and empiric studies show that there is generally high correlation between protein abundance in individual gels and in the pools derived from these individuals[11, 12]. Pooled samples from each subtype were used for the 2-DE comparison study. Aliquots of equal volume (100μl) were taken from all samples and combined to from a pooled internal standard. The samples, along with the pooled internal standard, were then processed using Concanavalin A-sepharose beads (GE-Healthcare, Piscataway, NJ) in macrospin columns (Nest Group Inc., Southborough, MA) in order to deplete high-abundant albumin protein from the synovial fluid. The protein solution was washed with a 1M Sodium phosphate 1M sodium chloride solution and concentrated using a molecular weight column (Millipore, Billerica, MA). Precipitation of proteins was by the Perfect Focus Kit (G-Biosciences, St. Louis, MO). The protein was resuspended in lysis buffer containing 2M thiourea and 7M urea. The protein concentration of each sample was determined by the Bradford Assay protocol.
Two-dimensional Difference Gel Electrophoresis (DIGE)
A total of 50 μg of synovial fluid protein from each sample was labeled with Cy3 or Cy5 minimal dyes and the pooled internal standard was labeled with Cy2 in the dark. Lysine was used for the labeling. The labeled protein samples were multiplexed in order to run 2 analytical samples and one internal standard on each gel. In addition, “dye swap” was performed ensuring that differences in protein spots were not due to a specific dye intensity. The labeled protein was brought to a volume of 450 μl in rehydration buffer containing 20mM of dithiotrietol and .05% of (v/v) pH 4–7 carrier ampholytes (GE-Healthcare, Piscataway, NJ). A 24 cm linear ph 4–7 immobilized pH gradient (IPG) strip was immersed each solution. The first dimensional separation of proteins was performed using the IPGphor-3 (GE-Healthcare, Piscataway, NJ) settings as follows: 30V for 12 hours for the rehydration step, then 200v for 1 hour, 500v for 1 hr, 1000V for 1 hr and then a gradient to 8000V over 3 hours to at total of 50,000 Vhrs, according to the manufacturer’s instructions. After isoelectric focusing, the strips were equilibrated in sample buffer containing 100 mg dithiotreitol and then 250 mg iodacetamide. The equilibrated strips were placed onto 12% SDS gels (Jule Gels, Milford, CT). The second dimension was performed using a Ettan DALT 6 (GE-Healthcare, Piscataway, NJ) run at 2W per gel. The samples were assessed in two separate gel runs resulting in 12 gels (36 gel images), and each pooled sample was represented in 6 images.
Image analysis
The 2-D gels were scanned using the Typhoon 9400 Imager (GE-Healthcare, Piscataway, NJ). The resulting gel images were imported into DeCyder v5.02 software (GE-Healthcare, Piscataway, NJ) which outputs a list of statistically significant differences in protein expression including t-test values, using the Cy-2 internal standard. Both Differential In-gel Analysis (DIA) which includes co-detection, background subtraction, normalization, and quantitation of spots in an image pair as well as Biological Variation Analysis (BVA) which matches multiple gels for comparison and statistical analysis of protein abundance changes was used in this analysis. Several studies using 2D gels have utilized these types of analyses [13–15]. A total of 2500 spots per gel protein spot-features were analyzed across all serum sample 2-D spot maps. Spot-features that were significantly differentially expressed (p<0.05 for the unpaired t-test, and ≥2-fold Average Ratio) in each comparison and that were present on 75% of all spot maps, were chosen for further investigation. Each spot identified as significantly differentially expressed was manually assessed to ensure that only true protein spots were picked
Heat map
Expression values of each protein spot were represented as fold change. The data was transferred into Genespring (Agilent Technologies, Santa Clara, CA). The heat map was generated by performing Gene Tree Clustering analysis with default settings.
In-gel protein digestion and Identification
A preparative gel which contained 450 μg of unlabeled pooled internal standard was run using the same running conditions as the analytical gels (as described above), and stained with Deep Purple Protein Stain (GE-Healthcare, Piscataway, NJ), and matched to the analytical gels in BVA. The Ettan Spot Handling Workstation (GE-Healthcare, Piscataway, NJ) was employed for the preparative gel spot picking, tryptic digestion, and spotting onto a MALDI plate that was subsequently analyzed by MALDI-TOF-TOF (ABI 4800). The same spots were also analyzed on the LC/Q-TOF- MS system for peptide sequence information. The MS and MS/MS data were searched against NCBInr and Swissprot human protein database.
RNA extraction and PCR amplification
Frozen samples of synovial fluid cells stored in Trizol (Invitrogen, Carlsbad, CA) were thawed. Total RNA was isolated from cells using the phenol/chloroform extraction technique. To remove possible genomic DNA contamination, RNA was treated with DNase I (Ambion, Austin TX). Complimentary DNA (cDNA) was synthesized with random hexamer oligonucleotides using 1μg of RNA and Invitrogen’s SuperScript™ II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA). PCR was performed in a LightCycler (Mx3000P Stratagene, La Jolla CA) using Brilliant_ SYBR_ Green QPCR Master Mix (Stratagene, La Jolla CA) according to the protocol using oligonucleotide primer sets for human haptoglobin: forward primer 5′-AGAAGTAGGGCGTGTGGGTTATGT-3′, reverse primer 5′-ACTGTGCTGCCTTCATAATGCCT-3′. The 136 base pair product was verified by running a 10% TAE gel.
Results
Identification of the JIA synovial fluid proteome
Table 1 describes the clinical characteristics of the patient populations. Synovial fluid samples were pooled according to subtype (oligoarticular, polyarticular and systemic JIA) in order to decrease inter-individual differences. 2-D gel electrophoresis was used for excellent resolution and identification of proteins. Our previous experience with synovial fluid gel electrophoresis suggests that most of the abundant proteins are in the pH 4–7 range similar to serum (unpublished data). The 2-DE gels in our study encompass this pI range and molecular weight range of 10–200 kD. All distinct protein spots were picked, trypsinized and identified using MALDI-TOF-MS or Q-TOF when no identification was obtained with MALDI-TOF-MS. The global protein identification of the synovial fluid and their known functions are represented in Table 2.
Table 1.
Patient characteristics of synovial fluid samples. Control samples were obtained from orthopedic procedures.
| Characteristic | Oligo (n=33) | Poly (n=14) | Systemic (n=11) | Control (n=10) |
|---|---|---|---|---|
| Age, years | 9.6 + 4.4 * | 13.1 + 5.6 | 12.5 + 4.1 | N/A |
| Sex, no (%) | ||||
| Male | 6 (18) | 5 (36)** | 6 (54) | 8 (80) |
| Female | 27 (82) | 9 (64)** | 5 (45) | 2 (20) # |
| Disease Duration, years | 4.1 + 4.7 | 7.7 + 3.5 | 6.9 + 5.7 | N/A |
| ANA status, no (%) | ||||
| positive | 17 (52) | 5 (36) | 1 (9) | N/A |
| negative | 14 (42) | 8 (57) | 8 (73) | N/A |
| unknown | 1 (3) | 1 (7) | 2 (18) | |
| Rf status, no (%) | ||||
| positive | 0 | 1 (7) | 1 (9) | N/A |
| negative | 20 (61) | 7 (50) | 5 (45) | N/A |
| unknown | 13 (39) | 6 (43) | 5 (45) | |
| Treatment at time of procedure, no (%) | N/A | |||
| None | 2 (6) | 3 (21) | 1 (9) | |
| Non-steroidals | 31 (94) | 7 (50) | 4 (36) | |
| Methotrexate | 1 (3) | 2 (14) | 4 (36) | |
| Sulfasalazine | 1 (3) | 4 (28) | 0 | |
| Plaquenil | 2 (6) | 3 (21) | 0 | |
| Prednisone | 0 | 0 | 1 (9) | |
| Leflunomide | 0 | 1 (7) | 0 | |
| Gold | 0 | 1 (7) | 0 | |
| Biologics | 0 | 2 (14) | 1 (9) | |
Each subtype was pooled into a single sample for the analyses. Student’s T-test used for comparison between individual groups. No significant differences between the groups except as indicated:
P<.05 for comparison between oligoarticular vs polyarticular,
P<.05 for comparison between systemic versus oligoarticular,
P<.05 for comparison between oligoarticular versus control.
JIA = Juvenile Idiopathic Arthritis, N/A = data not available.
Table 2.
JIA Synovial Fluid Proteins.
| Protein name | Functional category | ||
|---|---|---|---|
| Acute Phase | Coagulation | Complement | |
| ADP/ATP translocase 3 (P12236)* | |||
| Albumin (P02768) | x | ||
| Alpha-1 antitrypsin precursor (P01009) | x | x | |
| Alpha-1-antichymoptrypsin (P01011) | x | ||
| Alpha 2-macroglobulin precursor (P01023) | x | x | |
| Amyloid P component, serum (P02743) | x | ||
| Apolipoprotein A-1 (P02647) | x | ||
| Complement component 3(P01024) | x | x | |
| Complement component 9 (P02748) | x | x | |
| Complement factor B (P00751) | x | x | |
| Complement factor H (P08603) | x | ||
| Ceruloplasmin (ferroxidase) (P00450) | x | ||
| Fibrinogen beta chain (P02675) | x | x | |
| Fibrinogen gamma chain (P02679) | x | x | |
| Haptoglobin (P00738) | x | ||
| Hemopexin (P02790) | x | ||
| Ig kappa chain C region (P01834)* | |||
| Inter-alpha (globulin) inhibitor H4 (Q14624) | x | ||
| Mitochondrial 28S ribosomal protein (P51398)* | |||
| Pigment epithelium derived factor P36955) | x | ||
| Transferrin (P02787) | x | ||
| Ubiquitone biosynthesis protein (O75208)* | |||
| Zinc-alpha-2-glycoprotein precursor COQ9 (P25311)* | |||
Identification of synovial fluid proteins was performed using MALDI-TOF or Q-TOF. The proteins were grouped into 3 main categories based on the function of the protein.
Proteins not categorized in main functional groups
The three main functional categories of proteins represented are: acute phase response proteins, coagulation system proteins and complement system proteins. The acute phase response proteins can be divided between those that are positive acute phase reactants (those that increase during the inflammatory phase) such as fibrinogen beta and gamma protein, alpha-1 anti-trypsin and haptoglobin family proteins and the negative acute phase reactants (a protein whose level is lowered by more than 25% during the acute phase) such as albumin, apolipoprotein A-1 and transferrin. There were several complement component proteins present in synovial fluid including complement component 3, complement component 9, and complement factors B and H. The last major functional classification is the coagulation protein group which includes alpha-1 anti-trypsin precursor, and fibrinogen family proteins. There are 5 proteins identified whose functions are not encompassed by the 3 main categories, and these are ADP/ATP translocase 3, Ig kappa (κ) chain C region, Mitochondrial 28S ribosomal protein, Ubiquitone biosynthesis protein and Zinc-alpha-2-glycoprotein precursor COQ9
2-D gel analysis identifies multiple differentially expressed proteins between the subtypes of JIA
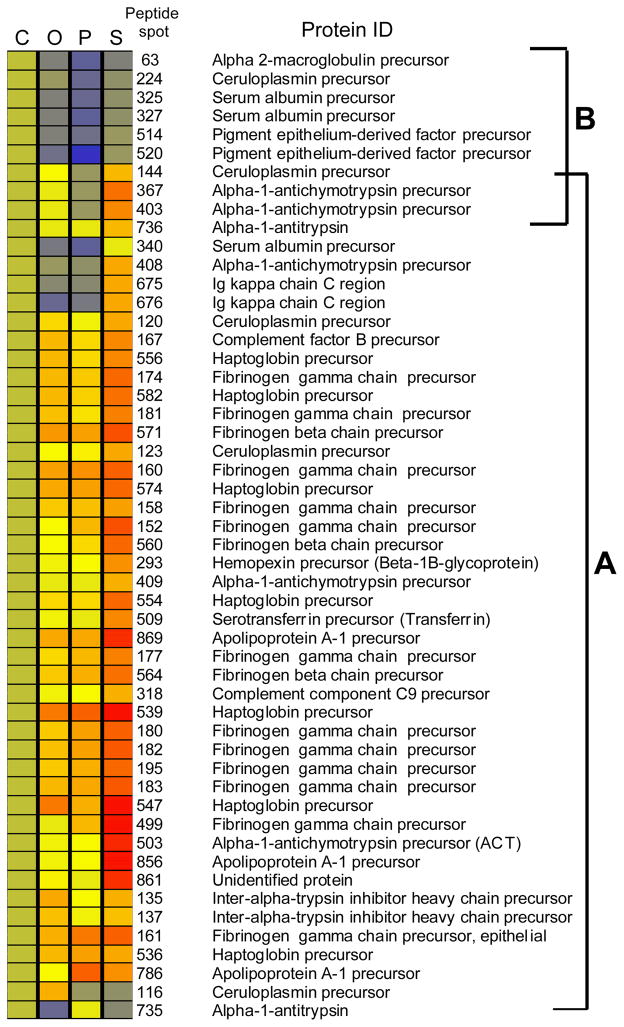
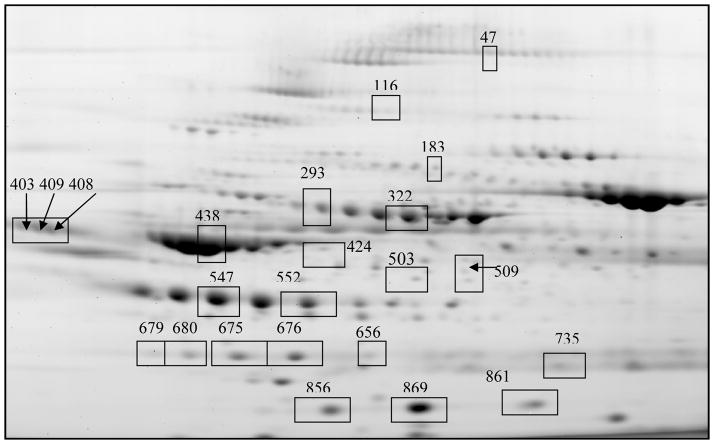
The synovitis of subtypes of JIA have similar histological appearances despite differences in clinical characteristics. Studies have shown different cytokine profiles in the synovial fluid of subtypes of juvenile idiopathic arthritis[4, 5]. In order to determine if there are differences on the larger protein scale, we performed DiGE analysis of pooled synovial fluid subtypes. A total of 100 protein spots were determined to be differentially expressed (p<.05). Fifty picked spots were determined to be statistically significant in at least one subtype or control comparisons. Figure 1 is a heat map representation of the differentially expressed proteins, where each JIA protein spot is compared to the non-inflammatory control protein spot and illustrated as fold change difference. Several proteins were more highly expressed in the JIA samples as compared to the non-inflammatory controls suggesting markers of disease activity. Isotypes of these proteins include Ig κ chain C region, ceruloplasmin, complement factor B precursor, haptoglobin precursor, fibrinogen beta chain precursor, fibrinogen gamma precursor, hemopexin precursor, complement component 9 precursor, serotransferrrin precursor, inter-alpha-1 trypsin precursor, and apolipoprotein A-1. Several proteins in the JIA synovial fluid showed decreased expression when compared to the controls including alpha-2 macroglobulin, ceruloplasmin, serum albumin and pigment epithelial derived factor (Figure 1A). There are also differentially expressed proteins seen between the subtypes of JIA. The heat map representation shows that the majority of proteins in the JIA synovial fluid are overexpressed in systemic JIA (Figure 1B). The cluster of proteins in Figure 1A which are underexpressed in JIA synovial fluid as compared to controls is also decreased in polyarticular JIA as compared to the other subtypes. There are also a number of proteins that appear to have higher levels in oligoarticular JIA as compared to polyarticular JIA. These include isotypes of antichymotrypsin, ceruloplamin, apolipoprotein A-1 and haptoglobin. The individual statistically significant differentially expressed proteins between subtypes are outlined in the Table 3. Significantly differentially expressed proteins spots between the subtypes of JIA are illustrated in Figure 2 labeled by their spot number. There are 24 statistically significant differentially expressed spots between JIA subtypes. These proteins are: alpha-1 antichymotrypsin, apolipoprotein A-1, ceruloplasmin, fibrinogen gamma chain, haptoglobin, hemopexin, Ig kappa chain C region, transferring, serum albumin and several unidentified proteins. This table mirrors the trends seen in the heat map where the majority of proteins show overexpression in systemic JIA. The proteins that showed the most marked overexpression in systemic JIA include alpha-1 antichymotrypsin precursor (fold change 2.11–5.2), apolipoprotein A-1 precursor (fold change 2.79–6.58), haptoglobin precursor (fold change 2.27–4.11) and Ig kappa chain C region (fold change 3.36–5.5). Other proteins with significant overexpression in systemic JIA included fibrinogen gamma chain precursor, hemopexin precursor, transferrin, serum albumin precursor and ubiquinone biosynthesis protein COQ9. There are several proteins with higher expression in systemic JIA which we were unable to identify (fold change 2.57–6.63). The data in the table also confirm an observation of the heat map that ceruloplasmin precursor (fold change 2.23) and several unidentified peptides (fold change 2.23–2.36) are overexpressed in oligoarticular JIA as compared to polyarticular JIA.
Figure 1.
Heat map representation of differentially expressed proteins. The heat map was generated as described in Materials and Methods. Each column represents the JIA subtype or control sample. Each row represents an individual protein that was identified as being significantly differentially expressed in at least one subtype comparison. Each cell in the matrix represents the relative protein expression level in a pooled sample. The protein spot number is the automated number assigned to a spot on the protein gel. The protein identification for the specific spot is listed. The control group was used as the basis for each individual comparison. The relative amount of a protein is denoted by a color in the spectrum from red to blue, red being the highest amount. (1A) These brackets indicate the proteins that are increased in the systemic JIA (1B) brackets indicate the proteins that are decreased in the polyarticular JIA group. Oligo = Oligoarticular JIA, Poly = Polyarticular JIA, Systemic = Systemic JIA.
Table 3.
Differentially expressed proteins between subtypes of JIA.
| Spot no | Protein Identification | Poly vs Oligo | S vs Oligo | S vs Poly | |||
|---|---|---|---|---|---|---|---|
| FC | p | FC | p | FC | p | ||
| 408 | Alpha-1-antichymotrypsin | 2.45 | .00019 | 2.57 | .015 | ||
| 409 | Alpha-1-antichmotrypsin | 2.31 | .011 | 2.17 | .011 | ||
| 438 | Alpha-1-antichmotrypsin | 2.11 | .037 | ||||
| 503 | Alpha-1-antichmotrypsin | 5.2 | .0081 | 4.73 | .046 | ||
| 735 | Alpha-1 anti-trypsin | 2.24 | .0055 | ||||
| 856 | Apolipoprotein A-1 precursor | 6.58 | .05 | ||||
| 869 | Apolipoprotein A-1 precursor | 2.79 | .017 | 2.9 | .012 | ||
| 116 | Ceruloplasmin precursor | −2.23 | .021 | ||||
| 183 | Fibrinogen gamma chain precursor | 2.33 | .034 | ||||
| 547 | Haptoglobin precursor | 3.46 | .017 | ||||
| 552 | Haptoglobin precursor | 4.11 | .032 | ||||
| 656 | Haptoglobin precursor | 2.27 | .045 | ||||
| 293 | Hemopexin precursor | 2.28 | .048 | ||||
| 322 | Hemopexin precursor | 2.32 | .0066 | ||||
| 675 | Ig kappa chain C region | 3.36 | .012 | 3.39 | .0024 | ||
| 676 | Ig kappa chain C region | 5.5 | .025 | 3.73 | .0031 | ||
| 403 | Serum albumin precursor | 2.92 | .025 | 3.73 | .026 | ||
| 509 | Transferrin | 2.58 | .048 | 2.92 | .011 | ||
| 539 | Ubiquitone biosynthesis protein COQ9 | 2.26 | .028 | ||||
| 47 | Unidentified peptide | −2.23 | .029 | ||||
| 424 | Unidentified peptide | −2.36 | .0033 | ||||
| 680 | Unidentified peptide | 3.72 | .041 | ||||
| 861 | Unidentified peptide | 6.63 | .03 | ||||
| 679 | Unidentified peptide | 2.57 | .048 | ||||
Gel protein spots were compared using DeCyder image analysis. Any spot comparison that showed a 2 fold or greater difference with a p value of .05 and was present in at least 75% of the images was determined to be statistically significant. These spots were then visually inspected to verify validity of the comparison. The resulting spots are outlined and their fold change difference is listed. C=Control, O= oligoarticular JIA, P=polyarticular JIA, S=systemic JIA, FC= fold change
Figure 2.
Gel image of significantly differentiated proteins. The proteins are separated vertically by molecular weight and horizontally by pH. The gel image represents the internal standard, the combination of all the sample groups. The differentially expressed proteins spots are identified by spot number.
Haptoglobin is significantly differentially expressed between the subtypes of JIA and haptoglobin mRNA is detected in the joints
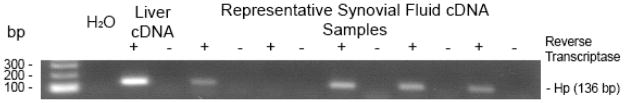
Haptoglobin, a protein known to be synthesized by the liver and which functions as an acute phase response protein is significantly overexpressed in the systemic and polyarticular JIA synovial fluid (Table 3). Haptoglobin in its full form is in the 86 kD range, and is formed by the disulfide bonding of 2 alpha and 2 beta chains. The molecular weight range of the haptoglobin identified in our study is a 17–22 kD isoform suggesting that it is a cleaved portion. The alpha chain of haptoglobin is 17 kD. We wanted to determine whether the difference we observed in haptoglobin was due to local production in the inflamed joint or whether it represented overflow from the plasma. We used PCR analysis of synovial fluid cells to amplify the alpha chain of haptoglobin. Figure 3 is the gel image of the PCR product of 5 polyarticular JIA samples tested for haptoglobin mRNA. In this representation, the majority (4/5) of the polyarticular samples shown were positive for haptoglobin mRNA. This is, to our knowledge, the first time that haptoglobin has been shown to be produced in a human inflamed joint.
Figure 3. Detection of synovial fluid haptoglobin by PCR.
Gel image of polymerase chain reaction products of 5 representative JIA synovial fluid cDNA samples. The primer identified haptoglobin mRNA. Hp = haptoglobin, bp=base pair
Discussion
In this study, we explored the proteomic profiles of synovial fluid in juvenile idiopathic arthritis to determine if there is differential protein expression between oligoarticular JIA, polyarticular JIA and systemic JIA.
Our studies indicate that there is differential protein expression in synovial fluid between JIA and non-inflammatory joints. Most of these proteins are known to be normal constituents of synovial fluid, and the differential expression may clue into the pathogenesis of disease. The majority of the differentially expressed proteins are acute phase reactant proteins which are elevated in JIA synovial fluid. Our data also show a cluster of proteins that have increased expression in non - JIA synovial fluids as compared to JIA synovial fluids. One of these proteins is Pigment Epithelium –Derived factor (PEDF). PEDF is an effective neutrotrophic factor and has potent anti-angiogenic activity [16] [17]. Furthermore, PEDF has been implicated in the pathogenesis of various conditions, including chronic inflammatory disease, atherosclerosis, diabetic complications and cancer [18]. There are no published studies of the role of PEDF in arthritis to date, but studies in uveitis show that retinal and plasma PEDF levels were drastically decreased endotoxin-induced uveitis (EIU), which suggests that PEDF functions as a negative acute-phase protein [19]. It is possible that it plays a similar role in the arthritis of JIA either locally or systemically and the decreased levels in JIA synovial fluid represent consumption or clearance of this protein.
Alpha-2-macroglobulin, an important inhibitor of cartilage-degrading proteinases, is another protein that was found to have decreased expression in JIA synovial fluid. Cartilage oligomeric matrix protein(COMP) is a glycoprotein found in cartilage [20], and fragments of this glycoprotein have been found in cartilage, synovial fluid and serum of patients with knee injuries, osteoarthritis, rheumatoid arthritis and JIA [21–23]. Members of the ADMATS (a disintegrin and metalloproteinase with thrombospondin motifs), specifically ADAMTS-7 and ADAMTS-12, family cleaves COMP in vitro and the size of the resulting fragments are similar to that found in arthritis [24]. Alpha-2 macroglobulin inhibits members of the ADAMTS family and protects COMP degradation by these enzymes [24, 25]. The differential expression of alpha-2 macroglobulin in non-JIA synovial fluid versus JIA synovial fluid may represent consumption of the protein in attempts to prevent COMP degradation in the diseased joint of JIA.
Although it did not reach statistical significance by BVA, there was decreased PEDF and alpha-2 macroglobulin in the synovial fluid of polyarticular JIA as compared to the other subtypes. Decreased amounts of PEDF and alpha-2 macroglobulin in JIA may have a role in extension of joint involvement in polyarticular JIA.
There are several proteins which are significantly differentially expressed between the subtypes of JIA. Apolipoprotein-A1 (apoA-1) showed differential expression in systemic JIA versus oligoarticular and polyarticular JIA, and similarly, Gibson, et al showed this protein to be increased in polyarticular versus oligoarticular fluid [10]. In the absence of inflammation, high-density lipoprotein cholesterol (HDL-C) has a complement of antioxidant enzymes that work to maintain an anti-inflammatory state. In the presence of systemic inflammation, these antioxidant enzymes such as ApoA-1 can be inactivated, and HDL can accumulate oxidized lipids and proteins that make it proinflammatory [26]. When not activated by inflammation, ApoA-1 has anti-inflammatory properties and has been shown to block contact-mediated activation of monocytes in vitro causing inhibition of TNF-α and IL-1β production [27] and c-reactive protein[28]. Localization of apoA-1 in inflamed synovium can inhibit the production of proinflammatory cytokines by macrophages upon direct contact with stimulated T cells [29]. In an inflamed joint where joint integrity and lipid homeostasis is compromised the apoA-1 may become reactive and proinflammatory. Further studies will need to be done to determine the role of ApoA-1 in inflammatory arthritis.
Another identified protein of interest is Ig kappa (κ) chain C region. Children with JIA have been shown to produce increased levels of serum circulating immune complexes (CIC’s) that correlates with disease activity [30, 31]. A recent study by Low, et al delineate the CIC proteome in JIA [32]. The authors demonstrated several disease associated proteins that are present in the CIC’s in active and erosive polyarticular JIA including Ig kappa chain regions. In our study there was increased amounts of the immunoglobulin κ chain in the synovial fluid of systemic JIA which may suggest that immunoglobulins in synovial fluid in sJIA have a different antibody response.
Haptoglobin was significantly differentiated between subtypes and increased levels seen in both systemic JIA and polyarticular JIA. During hemolysis, free hemoglobin which is toxic and inflammatory is released. Haptoglobin binds to hemoglobin and inhibits the ability of hemoglobin to serve as an oxidant [33]. The deactivation and clearance of free hemoglobin is facilitated by the hemoglobin-haptoglobin (Hb-Hp) complex which activates monoctyes and macrophages via the scavenger receptor, CD163 [34].
Systemic juvenile arthritis is associated with macrophage activation syndrome (MAS). MAS is a severe, potentially life threatening complication characterized by activation of well-differentiated macrophages and is clinically manifested by fever, hepatosplenomegaly, lymphadenopathy, severe cytopenias and intravascular coagulation [35]. There is an uncontrolled and persistent expansion of activated T lymphocytes and hemophagocytic macrophages. The macrophages in MAS express CD163 [36, 37]. The genes for haptoglobin were shown to be one of the most highly overexpressed genes in early systemic JIA and especially those with subclinical MAS [38]. The synovium is highly vascular and there is presumably a significant degree of hemolysis occurring which leads to increased increased RBC turnover and initiation of this cascade.
Contrary to its role as an anti-oxidant, haptoglobin may have pro-inflammatory effects on the joint. It functions as an acute phase reactant and its synthesis is induced by various cytokines including IL-1 and IL-6 [39]. It also appears to play a role in the inflammatory process of bone destruction via bradykinin and thrombin stimulation on prostaglandin E2 formation leading to bone resorption [40, 41]. Haptoglobin was identified as an essential factor for cell migration and may play a direct role in arthritis [42]. Recent data suggests an immune modulating role of haptoglobin in modulating Th1 versus Th2 balance by promoting Th1 cellular response [43].
Haptoglobin is primarily produced in the liver. However, there is evidence that haptoglobin is expressed in extra hepatic tissues such as lung, kidney, skin, heart and arteries [42, 44–48]. Haptoglobin has been shown to be a normal constituent of synovial fluid [49]. A previous study by Smeets, et al showed that haptoglobin is locally produced in arthritic rats [50]. Our data indicate that haptoglobin is locally produced in the inflamed joint in JIA and to our knowledge the only data showing this in human synovial cells. Further studies will be needed to localize haptoglobin to a specific cell population and identify the role of this protein in JIA.
There are several limitations of the study. The first is that the “control” synovial fluid originates from joints with traumatic injury and is therefore not completely normal. However, we used these samples as they do not originate from children with JIA. There were some differences in patient characteristics in the pooled samples between the subtypes of JIA that reached statistical significance including a difference in the disease duration between the oligoarticular and polyarticular group. Disease duration as well as differences in medication treatment could be confounding factors and may alter proteomic profiles. However, all of these patients had active disease at the time of arthrocentesis, so the proteins most likely represent the proteome of active JIA as reflected in the number of acute phase proteins.
There were also collective differences in the gender and age between the groups, but these differences most likely did not alter the resulting data of the differential proteins identified.
Pooling of synovial fluid by subtype may mask inter-individual differences, and this might be important if the proteins affected are the ones of particular interest for further study. However, given the low throughput of 2D gels, pooling is an efficient method to find global differences between patient subsets as there is generally a high correlation between protein abundance in individual gels and in the pools derived from these individuals [11, 12]. Similar studies in synovial fluid proteomics have not been performed.
A final limitation to our study is that DiGE analysis for protein identification will only detect those proteins of high abundance, so low abundance proteins are not identified in our study. We used affinity based techniques for depleting our samples of albumin and immunoglobulin for improving detection of low abundance proteins, but this technique is non-specific and may remove other wanted proteins from the fluid. There may be important low molecular weight proteins that are bound to albumin (albuminone) which might include additional differentially-expressed proteins not identified here. Further studies analyzing those fractions may identify other proteins to expand the repertoire of the protein profiles.
Despite advances in our understanding of the molecular basis of JIA, substantial gaps remain both in our understanding of disease pathogenesis, and in the development of effective strategies for early diagnosis and for treatment. Proteomic analysis of biological fluids provides an opportunity for better understanding of disease processes. Our study has identified proteins that are differentially expressed and potential biomarkers in JIA. Our study demonstrates how proteomic platforms can be used for further targeted discovery in the understanding the specific roles of proteins in inflammatory arthritis of JIA and other inflammatory arthritides.
Acknowledgments
We would like to thank Dr. Robert Boudreau, PhD, (University of Pittsburgh, Department of Biostatistics), for his statistical support.
We would like to thank Dr. Manimalha Balasubramani, Ph.D. (Genomics and Proteomics Core Laboratories, University of Pittsburgh) for her support with the MALDI-TOF-MS data.
Supported by NIH (National Human Genome Research Institute K23-HG-003978-01) Cincinnati Rheumatic Diseases Core Center (NIAMS P30 AR47363)
References
- 1.Brewer EJ, Jr, et al. Current proposed revision of JRA Criteria. JRA Criteria Subcommittee of the Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Section of The Arthritis Foundation. Arthritis Rheum. 1977;20(2 Suppl):195–9. [PubMed] [Google Scholar]
- 2.De Benedetti F, Ravelli A, Martini A. Cytokines in juvenile rheumatoid arthritis. Curr Opin Rheumatol. 1997;9(5):428–33. doi: 10.1097/00002281-199709000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Glass DN, Giannini EH. Juvenile rheumatoid arthritis as a complex genetic trait. Arthritis Rheum. 1999;42(11):2261–8. doi: 10.1002/1529-0131(199911)42:11<2261::AID-ANR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Murray KJ, et al. Contrasting cytokine profiles in the synovium of different forms of juvenile rheumatoid arthritis and juvenile spondyloarthropathy: prominence of interleukin 4 in restricted disease. J Rheumatol. 1998;25(7):1388–98. [PubMed] [Google Scholar]
- 5.Thompson SD, et al. Chemokine receptor CCR4 on CD4+ T cells in juvenile rheumatoid arthritis synovial fluid defines a subset of cells with increased IL-4:IFN-gamma mRNA ratios. J Immunol. 2001;166(11):6899–906. doi: 10.4049/jimmunol.166.11.6899. [DOI] [PubMed] [Google Scholar]
- 6.Sinz A, et al. Mass spectrometric proteome analyses of synovial fluids and plasmas from patients suffering from rheumatoid arthritis and comparison to reactive arthritis or osteoarthritis. Electrophoresis. 2002;23(19):3445–56. doi: 10.1002/1522-2683(200210)23:19<3445::AID-ELPS3445>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 7.Liao H, et al. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50(12):3792–803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 8.Tilleman K, et al. Chronically inflamed synovium from spondyloarthropathy and rheumatoid arthritis investigated by protein expression profiling followed by tandem mass spectrometry. Proteomics. 2005;5(8):2247–57. doi: 10.1002/pmic.200401109. [DOI] [PubMed] [Google Scholar]
- 9.Gibson DS, et al. Proteomic analysis of recurrent joint inflammation in juvenile idiopathic arthritis. J Proteome Res. 2006;5(8):1988–95. doi: 10.1021/pr060129t. [DOI] [PubMed] [Google Scholar]
- 10.Gibson DS, et al. Comparative analysis of synovial fluid and plasma proteomes in juvenile arthritis--proteomic patterns of joint inflammation in early stage disease. J Proteomics. 2009;72(4):656–76. doi: 10.1016/j.jprot.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Proteomic analysis of individual variation in normal livers of human beings using difference gel electrophoresis. Proteomics. 2006;6(19):5260–8. doi: 10.1002/pmic.200600006. [DOI] [PubMed] [Google Scholar]
- 12.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30(17):2967–75. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 13.Ryu OH, et al. Identification of parotid salivary biomarkers in Sjogren’s syndrome by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry and two-dimensional difference gel electrophoresis. Rheumatology (Oxford) 2006;45(9):1077–86. doi: 10.1093/rheumatology/kei212. [DOI] [PubMed] [Google Scholar]
- 14.Brown LM, et al. Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Mol Carcinog. 2006;45(8):613–26. doi: 10.1002/mc.20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandra Rao SP, et al. Profiling of human mesangial cell subproteomes reveals a role for calmodulin in glucose uptake. Am J Physiol Renal Physiol. 2007;292(4):F1182–9. doi: 10.1152/ajprenal.00268.2006. [DOI] [PubMed] [Google Scholar]
- 16.Tombran-Tink J, Chader GG, Johnson LV. PEDF: a pigment epithelium-derived factor with potent neuronal differentiative activity. Exp Eye Res. 1991;53(3):411–4. doi: 10.1016/0014-4835(91)90248-d. [DOI] [PubMed] [Google Scholar]
- 17.Dawson DW, et al. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285(5425):245–8. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 18.Ek ET, Dass CR, Choong PF. PEDF: a potential molecular therapeutic target with multiple anti-cancer activities. Trends Mol Med. 2006;12(10):497–502. doi: 10.1016/j.molmed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SX, et al. Pigment epithelium-derived factor (PEDF) is an endogenous antiinflammatory factor. Faseb J. 2006;20(2):323–5. doi: 10.1096/fj.05-4313fje. [DOI] [PubMed] [Google Scholar]
- 20.Hedbom E, et al. Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J Biol Chem. 1992;267(9):6132–6. [PubMed] [Google Scholar]
- 21.Neidhart M, et al. Small fragments of cartilage oligomeric matrix protein in synovial fluid and serum as markers for cartilage degradation. Br J Rheumatol. 1997;36(11):1151–60. doi: 10.1093/rheumatology/36.11.1151. [DOI] [PubMed] [Google Scholar]
- 22.Saxne T, Heinegard D. Cartilage oligomeric matrix protein: a novel marker of cartilage turnover detectable in synovial fluid and blood. Br J Rheumatol. 1992;31(9):583–91. doi: 10.1093/rheumatology/31.9.583. [DOI] [PubMed] [Google Scholar]
- 23.Gilliam BE, et al. Measurement of biomarkers in juvenile idiopathic arthritis patients and their significant association with disease severity: a comparative study. Clin Exp Rheumatol. 2008;26(3):492–7. [PubMed] [Google Scholar]
- 24.Luan Y, et al. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthritis Cartilage. 2008;16(11):1413–20. doi: 10.1016/j.joca.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tortorella MD, et al. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J Biol Chem. 2004;279(17):17554–61. doi: 10.1074/jbc.M313041200. [DOI] [PubMed] [Google Scholar]
- 26.Navab M, et al. The oxidation hypothesis of atherogenesis: the role of oxidized phospholipids and HDL. J Lipid Res. 2004;45(6):993–1007. doi: 10.1194/jlr.R400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Hyka N, et al. Apolipoprotein A-I inhibits the production of interleukin-1beta and tumor necrosis factor-alpha by blocking contact-mediated activation of monocytes by T lymphocytes. Blood. 2001;97(8):2381–9. doi: 10.1182/blood.v97.8.2381. [DOI] [PubMed] [Google Scholar]
- 28.Wadham C, et al. High-density lipoproteins neutralize C-reactive protein proinflammatory activity. Circulation. 2004;109(17):2116–22. doi: 10.1161/01.CIR.0000127419.45975.26. [DOI] [PubMed] [Google Scholar]
- 29.Bresnihan B, et al. Apolipoprotein A-I infiltration in rheumatoid arthritis synovial tissue: a control mechanism of cytokine production? Arthritis Res Ther. 2004;6(6):R563–6. doi: 10.1186/ar1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melsom RD, et al. Anti-C1q affinity isolated circulating immune complexes correlate with extra-articular rheumatoid disease. Rheumatol Int. 1986;6(5):227–31. doi: 10.1007/BF00541372. [DOI] [PubMed] [Google Scholar]
- 31.Moore TL, et al. Immune complexes in juvenile rheumatoid arthritis: a comparison of four methods. J Rheumatol. 1982;9(3):395–401. [PubMed] [Google Scholar]
- 32.Low J, et al. Proteomic analysis of circulating immune complexes in juvenile idiopathic arthritis reveals disease associated proteins. Proteomics Clin Appl. 2009;3:829–840. doi: 10.1002/prca.200800073. [DOI] [PubMed] [Google Scholar]
- 33.Melamed-Frank M, et al. Structure-function analysis of the antioxidant properties of haptoglobin. Blood. 2001;98(13):3693–8. doi: 10.1182/blood.v98.13.3693. [DOI] [PubMed] [Google Scholar]
- 34.Kristiansen M, et al. Identification of the haemoglobin scavenger receptor. Nature. 2001;409(6817):198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 35.Hadchouel M, Prieur AM, Griscelli C. Acute hemorrhagic, hepatic, and neurologic manifestations in juvenile rheumatoid arthritis: possible relationship to drugs or infection. J Pediatr. 1985;106(4):561–6. doi: 10.1016/s0022-3476(85)80072-x. [DOI] [PubMed] [Google Scholar]
- 36.Bleesing J, et al. The diagnostic significance of soluble CD163 and soluble interleukin-2 receptor alpha-chain in macrophage activation syndrome and untreated new-onset systemic juvenile idiopathic arthritis. Arthritis Rheum. 2007;56(3):965–71. doi: 10.1002/art.22416. [DOI] [PubMed] [Google Scholar]
- 37.Schaer DJ, et al. Soluble hemoglobin-haptoglobin scavenger receptor CD163 as a lineage-specific marker in the reactive hemophagocytic syndrome. Eur J Haematol. 2005;74(1):6–10. doi: 10.1111/j.1600-0609.2004.00318.x. [DOI] [PubMed] [Google Scholar]
- 38.Fall N, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56(11):3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 39.Oliviero S, Cortese R. The human haptoglobin gene promoter: interleukin-6-responsive elements interact with a DNA-binding protein induced by interleukin-6. Embo J. 1989;8(4):1145–51. doi: 10.1002/j.1460-2075.1989.tb03485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frohlander N, Ljunggren O, Lerner UH. Haptoglobin synergistically potentiates bradykinin and thrombin induced prostaglandin biosynthesis in isolated osteoblasts. Biochem Biophys Res Commun. 1991;178(1):343–51. doi: 10.1016/0006-291x(91)91820-3. [DOI] [PubMed] [Google Scholar]
- 41.Lerner UH, Frohlander N. Haptoglobin-stimulated bone resorption in neonatal mouse calvarial bones in vitro. Arthritis Rheum. 1992;35(5):587–91. doi: 10.1002/art.1780350517. [DOI] [PubMed] [Google Scholar]
- 42.de Kleijn DP, et al. Acute-phase protein haptoglobin is a cell migration factor involved in arterial restructuring. Faseb J. 2002;16(9):1123–5. doi: 10.1096/fj.02-0019fje. [DOI] [PubMed] [Google Scholar]
- 43.Guetta J, et al. Haptoglobin genotype modulates the balance of Th1/Th2 cytokines produced by macrophages exposed to free hemoglobin. Atherosclerosis. 2007;191(1):48–53. doi: 10.1016/j.atherosclerosis.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 44.Friedrichs WE, et al. Expression and inflammatory regulation of haptoglobin gene in adipocytes. Biochem Biophys Res Commun. 1995;209(1):250–6. doi: 10.1006/bbrc.1995.1496. [DOI] [PubMed] [Google Scholar]
- 45.Yang F, et al. Pulmonary expression of the human haptoglobin gene. Am J Respir Cell Mol Biol. 2000;23(3):277–82. doi: 10.1165/ajrcmb.23.3.4069. [DOI] [PubMed] [Google Scholar]
- 46.Yang F, et al. Cell type-specific and inflammatory-induced expression of haptoglobin gene in lung. Lab Invest. 1995;73(3):433–40. [PubMed] [Google Scholar]
- 47.D’Armiento J, Dalal SS, Chada K. Tissue, temporal and inducible expression pattern of haptoglobin in mice. Gene. 1997;195(1):19–27. doi: 10.1016/s0378-1119(97)00123-6. [DOI] [PubMed] [Google Scholar]
- 48.Li P, et al. Localization of haptoglobin in normal human skin and some skin diseases. Int J Dermatol. 2005;44(4):280–4. doi: 10.1111/j.1365-4632.2005.02088.x. [DOI] [PubMed] [Google Scholar]
- 49.Neuhaus OW, Sogoian VP. Presence of haptoglobin in synovial fluid. Nature. 1961;192:558–9. doi: 10.1038/192558a0. [DOI] [PubMed] [Google Scholar]
- 50.Smeets MB, et al. The acute phase protein haptoglobin is locally expressed in arthritic and oncological tissues. Int J Exp Pathol. 2003;84(2):69–74. doi: 10.1046/j.1365-2613.2003.00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]