Abstract
Like most intracellular pathogens, Toxoplasma synthesizes and secretes an arsenal of proteins to successfully invade its host cell and hijack host functions for intracellular survival. The rhoptries are key secretory organelles that inject proteins into the host cell where they are positioned to co-opt host processes, although little is known regarding how these proteins exert their functions. We show here that the rhoptry protein ROP13 is synthesized as a pre-pro-protein that is processed in the parasite. Processing occurs at a conserved SϕXE cleavage site as mutagenesis of glutamic acid to alanine at the P1 position disrupts ROP13 maturation. We also demonstrate that processing of the prodomain is not necessary for rhoptry targeting and secretion. While gene disruption reveals that ROP13 is not essential for growth in fibroblasts in vitro or for virulence in vivo, we find that ROP13 is a soluble effector protein that can access the cytoplasm of host cells. Exogenously expressed ROP13 in human cells remains cytosolic but also appears toxic, suggesting that over-expression of this effector protein is disrupting some function within the host cell.
Keywords: Toxoplasma gondii, ROP13, Rhoptry, Effector protein, Host-pathogen
1. Introduction
Toxoplasma gondii is one of the most successful parasites globally in that it is able to infect any warm-blooded animal and is estimated to infect one-third of all humans (Tenter et al., 2000; Kim and Weiss, 2008). This organism is a major cause of human disease as it can lead to retinal scarring, brain damage or abortion following primary maternal infection, and a potentially fatal encephalitic threat to immunocompromised individuals (Montoya and Liesenfeld, 2004). In addition, Toxoplasma is related to an array of other disease-causing apicomplexan parasites, including Cryptosporidium, Eimeria and Plasmodium, the causative agent of malaria. The particular experimental tractability of Toxoplasma makes it well-suited to be used as a model organism for the study of less amenable apicomplexans.
Apicomplexans are named for their apical complex, which includes the specialized secretory organelles termed micronemes and rhoptries. The latter appear to be structurally and functionally divided into two compartments: the more apical rhoptry necks, containing rhoptry neck (RON) proteins, and the more basal rhoptry bodies, home to rhoptry proteins (ROPs) (Bradley and Sibley, 2007; Boothroyd and Dubremetz, 2008). A subset of the RON proteins localize to the moving junction that forms between the invading parasite and the host membrane, and are therefore thought to be involved in parasite invasion and formation of the nascent parasitophorous vacuole (PV). In agreement with the hypothesis that all obligate intracellular descendants of a common ancestor would share proteins required for invasion is the fact that many RONs are shared between different Apicomplexa (e.g. orthologues of RONs 1–5 exist in multiple genera) (Bradley et al., 2005; Straub et al., 2009). In contrast, most ROPs are unique to an individual genus. Some of these proteins have been detected in the host cell, suggesting that many ROPs are effector proteins that modulate the host response to the parasite. ROP2 family proteins are known to be injected into the host cell and localize to the cytoplasmic face of the PV membrane (PVM) where ROP2 may function in interaction with host organelles and ROP18 modulates parasite growth and virulence (Sinai and Joiner, 2001; El Hajj et al., 2007; Reese and Boothroyd, 2009). Protein phosphatase 2C-host nuclear (PP2C-hn) and ROP16 are also secreted and can be detected in the host nucleus where ROP16 activates STAT signaling and IL-12 production (Gilbert et al., 2007; Saeij et al., 2007). ROPs 1, 2, 7, and 18 have been found in evacuoles, membranous whorls that can be detected in the host cytosol following secretion from invasion-arrested parasites (Håkansson et al., 2001; El Hajj et al., 2006, 2007). Prior to this secretion from the rhoptries, these proteins are often processed, removing prodomains that may function as rhoptry-targeting domains and/or as regulators of protein activity.
Prodomains have been found to exist in many Toxoplasma rhoptry proteins: ROPs 1, 2, 4, and 8, TgSUB2, and RONs 2, 4, 5, and 8 (Beckers et al., 1997; Soldati et al., 1998; Sinai and Joiner, 2001; Miller et al., 2003; Bradley et al., 2004; Besteiro et al., 2009). Each of these contain putative cleavage sites with the consensus sequence SϕXE (where ϕ is a hydrophobic amino acid and X is any amino acid) which may serve as the processing site, although experimental evidence has only been shown for ROP1 and TgSUB2 (Bradley et al., 2002; Miller et al., 2003). The glutamic acid at the P1 position is important because processing is completely ablated if it is mutated to methionine or arginine. Mutation to aspartic acid, which retains the negative charge, only partially disrupts processing. However aspartic acid at the P1 position has not been identified in any known or hypothesized rhoptry protein processing sites. The dearth of verified processing sites and mutagenesis studies makes it difficult to predict where processing may occur, and which residues are strictly required for rhoptry protein processing.
We have previously identified ROP13 as a novel rhoptry body protein that shows no homology to any known protein and lacks any identifiable domains (Bradley et al., 2005). Its only recognizable orthologue within any sequenced genome is a protein encoded by Neospora caninum, a parasite that is extremely closely related to Toxoplasma. Here we show that ROP13 is a soluble protein and that it is proteolytically processed en route to the rhoptries. This processing occurs at a site fitting the consensus SϕXE, and mutation of the glutamic acid at the P1 position to alanine ablates processing. No severe parasite phenotype was seen upon disruption of the ROP13 gene. However, this protein’s function may involve host response to the parasite as we are able to detect ROP13 in the host cell in evacuoles. While the parasite-secreted protein is injected at levels too low to determine its destination within the host cell, exogenous expression of mature ROP13 in human cells suggests that its destination is the host cytosol and indicates that over-expression is toxic to the host cell.
2. Materials and methods
2.1. Host cell and parasite cultures
RHΔhpt (parental) and modified strains of T. gondii were maintained in confluent monolayers of human foreskin fibroblast (HFF) cells. HFFs were grown in DMEM supplemented with 10% FBS and 2 mM glutamine, and maintained as previously described (Donald et al., 1996).
2.2. Antibodies
Primary antibodies used in Western blot analyses and immunofluorescence assays (IFAs): polyclonal rabbit anti-ROP13 and the monoclonal mouse anti-PP2C-hn 9D6 (Gilbert et al., 2007), polyclonal mouse anti-ROP13 (Bradley et al., 2005), mouse anti-ROP7 monoclonal antibody 1B10 (Rome et al., 2008), monoclonal mouse anti- hemagglutinin (HA) antibody (Covance, Princeton, NJ), polyclonal rabbit anti-HA (Invitrogen, Carlsbad, CA), rabbit anti-SAG1 (Burg et al., 1988).
2.3. Western blot analyses
Parental and modified strain whole-parasite lysates were separated by 12% or 15% SDS-PAGE as previously described (Bradley et al., 2002). Samples were transferred to nitrocellulose overnight and probed with primary antibodies (see above). For all secondary antibody incubations, horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit antibodies (Sigma-Aldrich, St. Louis, MO) were used at a 1:2000 dilution. Following secondary incubation, an ECL detection kit was used for the detection of HRP activity (GE Amersham, Little Chalfont, United Kingdom).
2.4. IFAs and fluorescence microscopy
For IFAs, wild type and transgenic Toxoplasma strains were used to infect coverslips of HFFs for 24 h. The coverslips were then fixed in 3.7% formaldehyde/PBS for 15 min. The formaldehyde was quenched in PBS with 100 mM glycine for 5 min. The coverslips were washed in PBS and blocked and permeabilized in a solution containing PBS/3% BSA/0.2% Triton X-100 for 30 min. Samples were then incubated with primary antibody diluted in PBS/3% BSA/0.2% Triton X-100 for 1 h. The samples were then washed in PBS and treated with secondary antibodies Alexa 488-conjugated goat anti-mouse and/or Alexa 594-conjugated goat anti-rabbit (Molecular Probes) diluted 1:2,000 in PBS/3% BSA. Following secondary washes, coverslips were mounted onto microscope slides with Vectashield mounting media and the fluorescence was observed using a Zeiss Axio Imager Z1 upright light microscope with a 100× oil immersion objective. All images were rendered using the Zeiss Axiovision software and a Zeiss digital CCD camera (AxioCam MRm).
2.5. Generation of ROP13HA parasites
To epitope tag ROP13, pROP13HA was generated by PCR-amplifying the entire rop13 coding region plus ~1.5 kb of untranslated region (UTR) upstream of the transcription start site (containing the rop13 promoter) using forward primer ctcgagCCACTACGGGCAGTAGACA and reverse primer gcggccgcCAATAGCCTCAAGGAATTCGC and subcloned using XhoI and NotI sites into the pGRA-HA_HPT vector, which adds a C-terminal HA tag and GRA2 3’ UTR (Rome et al., 2008). The construct was linearized by NotI restriction digest and 50 μg of DNA transfected by electroporation into both RHΔhpt (to generate the ROP13HA line) and Δrop13 (to generate the complemented ROP13HA line, see below) parasites. The transfected parasites were grown in media containing 50 μg/ml mycophenolic acid (MPA) and 50 μg/ml xanthine (Sigma-Aldrich, St. Louis, MO, USA) and selected parasites were cloned by limiting dilution. To confirm proper ROP13 targeting, transgenic parasites were analyzed by IFA with rabbit polyclonal HA antibody and colocalized to the rhoptries using the anti-PP2C-hn monoclonal antibody 9D6 (Gilbert et al., 2007). To confirm that ROP13HA was expressed at similar levels compared with endogenous ROP13 protein, parental lysates and ROP13HA lysates were subjected to Western blot analysis and probed with anti-ROP13 polyclonal antiserum.
2.6. Generation of ROP13 processing mutant parasites
To generate pROP13HA_E66A, E66 at the P1 position of the putative ROP13 processing site was substituted by site-directed mutagenesis and verified by DNA sequence analysis. Two complementary primers (1.25 ng of each; CACTGTCTTTCACTGCAGGCACCAACGAGAC and GTCTCGTTGGTGCCTGCAGTGAAAGACAGTG) containing the desired mutation (underlined) and 50 ng of template (pROP13HA) in 50 μl of reaction buffer were denatured at 95°C for 0.5 min and annealed at 62°C for 1 min, and DNA synthesis was carried out by Pfu polymerase at 68°C for 6.5 min; this cycle was repeated 11 times. The methylated template was removed by incubation with 20 units of DpnI at 37°C for 1 h, with two repetitions. A portion of the resulting product (2 μl )was transformed into chemically competent Mach-1 Escherichia coli and plated overnight on agar containing Luria Broth/1,000 ug/ml ampicillin. DNA was isolated and sequenced to confirm the E66A mutation. The DNA was then used to transfect RHΔhpt parasites as described for generation of ROP13HA parasites above.
2.7. Discovery and alignment of Neospora caninum ROP13 orthologue
A search of the Toxoplasma amino acid sequence against translated DNA of N. caninum using tBLASTn (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/n_caninum) reveals a gene from contig 1049 to have a P-value of 1.1e-70. This gene model has since been identified as NC_LIV_132210 on www.toxodb.org. The alignment was performed using Invitrogen AlignX.
2.8. Gene disruption of rop13
For generation of a ROP13 deletion vector, 4.4 kb of the genomic region upstream of the ROP13 coding region (PCR-amplified with forward primer gcggccgcCTCTCCGTCGCGTTTCTTTC and reverse primer actagtCGGAGAGTACGCCGCTAAAA) and 2.5 kb of the 3’ flank (amplified with the forward primer ctcgagCCGCTAATGTTGCTGCCCAAG and reverse primer gggcccGAGGGAAGGCAAATCTGAGTC) of the sequence encoding ROP13 were amplified from T. gondii RH strain genomic DNA and subcloned into the pMini-GFP.ht vector (at NotI and SpeI sites for the 5’ and XhoI and ApaI sites for the 3’flank) (Karasov et al., 2005). This vector contains the selectable marker hypoxanthine-xanthine-guanine phosphoribosyl transferase (HPT), and a GFP cassette located downstream for negative selection of heterologous recombinants. The final construct, pROP13 KO, was linearized by NotI digestion and 30 μg of DNA was transfected by electroporation into RHΔhpt strain T. gondii. For selection of transformants, the transfected parasites were grown in media containing 50 μg/ml MPA and 50 μg/ml xanthine. After 8 days of selection, parasites were cloned by limiting dilution. GFP negative parasites were then screened by IFA for parasites lacking ROP13 and confirmed as Δrop13_HPT(+) by Western blot.
To remove the selectable marker HPT, the pROP13 KO vector was digested with SpeI and XbaI, blunted by treatment with Klenow enzyme (New England Biolabs, Ipswich, MA, USA), and recircularized by ligation. The resulting pHPT-KO vector was linearized by NotI digestion and transfected into Δrop13_HPT(+) parasites. Selection for the absence of HPT was achieved using 200 μg/ml 6-thioxanthine (Sigma-Aldrich, St. Louis, MO, USA) for 4 weeks, after which Δrop13_HPT(−) (hereafter referred to as Δrop13) parasites were cloned by limiting dilution. Clones were then screened for the absence of GFP and tested for the inability to survive in media containing 50 μg/ml MPA and 50 μg/ml xanthine.
2.9. Competitive growth rate assay
RHΔhpt (parental) and Δrop13, or Δrop13_HPT(+) and complemented ROP13HA parasites were mixed in equal numbers (105 each) and allowed to infect a confluent HFF monolayer. Upon lysis, the resulting parasites were used to infect a new monolayer at low multiplicity of infection (~1 parasite per HFF) as well as used to infect HFF-coated coverslips for IFA. Rabbit anti-SAG1 was used to stain all parasites, while mouse anti-ROP13 was used to detect only parental or complemented parasites. The percentage of each strain in the population was assessed by counting two coverslips two times each, counting at least 200 vacuoles for each timepoint. Complementation was attempted multiple times with tagged or untagged versions of the protein, with or without the selectable marker hypoxanthine-xanthine-guanine phosphoribosyl transferase (HPT).
2.10. In vivo infections in mice
Virulence of parental and Δrop13 strains was assessed by injecting parental (RHΔhpt) or Δrop13 parasites i.p. into BALB/c mice at a low dose (~30 parasites). Groups of four mice were used for each experiment and the parasite dose was confirmed by plaque assay. Mice that were moribund at day 8–9 were euthanized.
2.11. Triton X-114 phase partitioning
To assess whether ROP13 partitions with membrane or soluble fractions, 4 × 107 parasites were subjected to Triton X-114 (TX-114) phase partitioning. Extracellular parasites were pelleted at 2,500 g for 10 min, washed in PBS, and lysed in 1% Triton X-114/10 mM Tris (pH 7.5)/5 mM NaCl for 30 min at 4°C. Following a low speed spin at 2,500 g to remove insoluble material, lysates were partitioned as previously described (Seeber et al., 1998).
2.12. Detection of ROP13HA in evacuoles
Evacuoles were detected as previously described (Håkansson et al., 2001). Briefly, ~ 4 × 106 parasites were syringe-lysed from an HFF monolayer, washed in PBS and resuspended in 1 mL pre-chilled media with 1 μM cytochalasin D (Sigma-Aldrich, St. Louis, MO, USA), then incubated on ice for 10 min. Parasites (200 μl)were added directly to pre-chilled HFF-coated coverslips, allowed to settle for 20 min, and subsequently washed with ice-cold PBS. After removing the PBS, 37°C media containing 1 μM cytochalasin D was added to the coverslips, which were then incubated for 20 min in a 37°C water bath. After a PBS wash, coverslips were fixed for 15 min in 3.7% formaldehyde and IFAs were conducted as described above, although 0.1% saponin was substituted for Triton X-100 in both primary and secondary antibody incubations, and primary antibodies were used at a five-fold higher concentration.
2.13. Exogenous expression of ROP13 and ROP5 in human cells
Rop13ha from base 199 (which encodes the P1’ position of the processing site) was PCR-amplified from pROP13HA using forward primer gatatcGGCACCAACGAGACAAACCCA and reverse primer ctcgagTCATGCGTAGTCGGGGACG and cloned into a pCDNA3 vector in frame with an N-terminally encoded PTP (Protein C-TEV protease site-Protein A) tag (Schimanski et al., 2005). PP2C-hn cDNA was also cloned into pCDNA3 for use as a positive control. HeLa cells were seeded at 60% confluence 24 h prior to transfection in a 24-well plate. Vector (0.55 μg) was diluted with 73 μl serum-free medium (Invitrogen, Carlsbad, CA, USA) prior to addition of 1.65 μg polyethyleneimine (PEI) and vortexing. After a 15 min room temperature incubation during which the cells were washed with complete medium three times, the DNA/PEI mix was added to the cells for 16–22 h. For parasite studies, transfected mammalian cells were then infected with 8 × 104 RH parasites. Cells were fixed 24 h p.i. and subjected to IFA.
Rop5 minus its signal peptide was PCR-amplified from genomic DNA (isolated from RH strain T. gondii by a Promega Wizard genomic DNA purification kit) using forward primer gatatcGCCACCatgCGGGCAGGGGCAGTTCAGCTC (including Kozak sequence GCCACC and introduced start codon atg) and reverse primer gcggccgcAGCGACTGAGGGCGCAGCAGT and cloned into the ROP13HA in pCDNA vector from which ROP13 had been removed by digestion with NotI and EcoRV.
3. Results
3.1. Western blot analysis of ROP13 suggests a processing event
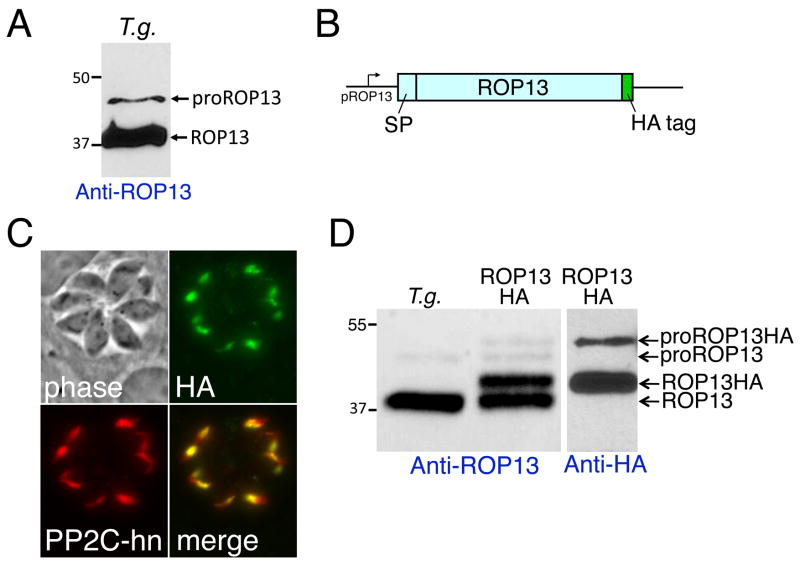
We initially identified ROP13 by mass spectrometric analysis of a purified rhoptry fraction and verified its localization using antibodies against the recombinant protein (Bradley et al., 2005; Gilbert et al., 2007). When these antibodies were used by Western blot analysis of Toxoplasma lysates, we observed two immunoreactive bands: a minor band migrating approximately at the size of the protein lacking its predicted signal peptide (~42 kDa) and a major band at ~38 kDa (Fig. 1A). This pattern is consistent with the processing of a propeptide as seen in several other rhoptry proteins. Each of these contain the consensus sequence SϕXE (where ϕ is a hydrophobic amino acid and X is any amino acid) at the predicted processing site. This consensus has only been verified for two ROPs (Bradley et al., 2002; Miller et al., 2003). To assess processing of ROP13, we expressed an HA-tagged copy of the gene driven from its own promoter (present on an ~1.5 kb region upstream of the start codon) in T. gondii (Fig. 1B). Stably transfected parasite clones were isolated and localization of the tagged protein assessed by IFA. Staining with anti-HA antibodies shows a classic club-like rhoptry staining pattern that colocalizes with the rhoptry body protein PP2C-hn, verifying correct organellar targeting (Fig. 1C). The HA-tagged copy can be distinguished from the endogenous ROP13 by Western blot, which shows a shift of both the pro- and mature forms of the protein consistent with the size of the tag as assessed by staining with anti-ROP13 and anti-HA antibodies (Fig. 1D). Thus, HA-tagged ROP13 is trafficked and processed similarly to endogenous ROP13. Because the epitope tag is positioned at the C-terminus, this data also shows that ROP13 processing occurs in the N-terminal region of the protein.
Fig. 1.
Western blot analysis of rhoptry protein 13 (ROP13) suggests processing which is supported by hemagglutinin (HA) -tagged protein. (A) Rabbit anti-ROP13 antiserum detects a 38 kDa major band and a 42 kDa minor band in Toxoplasma gondii lysate by Western blot analysis. (B) Schematic of ROP13HA construct, depicting the endogenous promoter (pROP13), coding region with predicted signal peptide (SP), and C-terminal HA tag. (C) Immunofluorescence assay (IFA) with anti-HA and anti-protein phosphatase 2C-host nuclear (PP2C-hn) showing the HA fusion traffics properly to the rhoptries. (D) Western blot displaying mature ROP13 at ~38 kDa and proROP13 at ~42 kDa in wild type T. gondii and those species as well as mature ROP13HA at ~39 kDa and proROP13HA at ~44 kDa in the ROP13HA line. Anti-ROP13 antibody shows all bands while anti-HA shows only the upshifted HA bands.
3.2. ROP13 E66A mutagenesis blocks prodomain processing
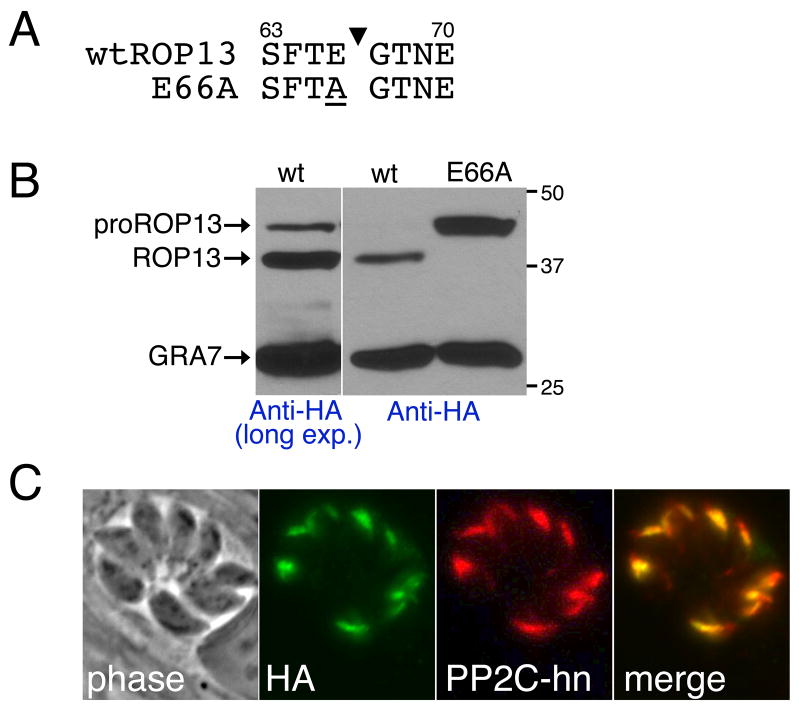
Removal of the predicted signal peptide of ROP13 results in an ~42 kDa proprotein as seen by Western blot analysis. Examination of the ROP13 sequence reveals SFTE amino acid residues at positions 63–66 (corresponding to P4-P1) that not only fit the SϕXE consensus for a rhoptry protein cleavage site, but also result in a predicted 38 kDa mature protein following cleavage. Previous mutagenesis studies of the ROP1 processing site have shown that non-conservative mutation of the residue at the P1 position (E66 for ROP13) inhibits processing. As such, E66 was mutated to an alanine, converting an acidic to a nonpolar residue, and stable parasite clones were isolated that expressed the mutant protein (Fig. 2A). Anti-HA antiserum was used to compare processing of wild type and E66A ROP13HA expressed in Toxoplasma (Fig. 2B). Processing of ROP13 was completely blocked by the E66A mutation, resulting in detection of only the 42 kDa proprotein. To ensure that defective processing is not a result of mistargeting of the mutant protein, we stained parasites expressing E66A ROP13HA and showed that the mutant protein is still trafficked to the rhoptries (Fig. 2C). These data demonstrate that mutation of glutamic acid to alanine at position 66 blocks ROP13 processing and that processing is not essential for intra-parasite trafficking. This is, to our knowledge, the first demonstration that mutation of glutamic acid to alanine is not tolerated at the P1 position of rhoptry protein processing sites. In addition, the N. caninum orthologue of ROP13 contains the similar sequence, SFAE, at residues corresponding to the SFTE site (Supplementary Fig. S1). As Toxoplasma and Neospora are so closely related, it is likely that both are cleaved at the corresponding SϕXE.
Fig. 2.
Mutagenesis of rhoptry protein 13 (ROP13) amino acid E66 to A blocks processing. (A) Amino acid sequence (P4-P1, P1’-P4’) and location of the ROP13 processing site (black arrowhead) and E66A processing mutation (underlined). (B) Western blot showing both pro- and mature ROP13-HA upon long exposure and only mature ROP13HA after shorter exposure of wild type Toxoplasma gondii lysate. The E66A mutation blocks processing of HA-tagged ROP13, leading to detection of only proROP13. GRA7, a dense granule protein, was used as a loading control. (C) Immunofluorescence assay (IFA) of E66A mutants with anti-HA and anti- protein phosphatase 2C-host nuclear (PP2C-hn) showing the E66A mutation does not disrupt ROP13 rhoptry localization.
3.3. Targeted disruption of rop13
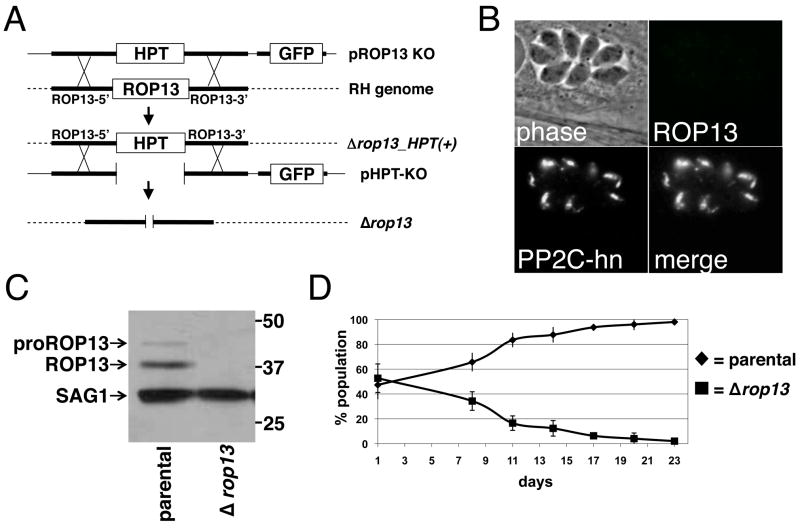
To directly assess ROP13 function, we disrupted rop13 by homologous recombination in RHΔhpt strain parasites (Fig. 3A). The rop13 knockout plasmid pROP13KO contains the selectable marker HPT, surrounded by ~4.4 kb of the rop13 5’ and 2.5 kb of the 3’ flanking regions. The construct also contains a downstream GFP marker useful for selecting against heterologous recombinants. The rop13 knockout plasmid was linearized and transfected into RHΔhpt strain parasites, selected for HPT with MPA and xanthine, and stably selected parasites were cloned by limiting dilution. GFP-negative clones were then tested for ROP13 immunoreactivity and showed no staining by IFA (Fig. 3B). Western blot analysis confirmed the successful generation of a ROP13 knockout strain (Fig. 3C). These data reveal that ROP13 is not essential for growth in rich media in vitro.
Fig. 3.
Targeted disruption of rhoptry protein 13 (ROP13). (A) Schematic showing the ROP13 knockout construct used to generate Δrop13_HPT(+) parasites and pHPT-KO construct used to generate Δrop13 parasites. (B) Immunofluorescence assay (IFA) showing lack of ROP13 staining but presence of protein phosphatase 2C-host nuclear (PP2C-hn), another rhoptry marker, in Δrop13 parasites. (C) Δrop13 Western blot showing detection of pro- and mature ROP13 in wild type T. gondii but absence of both in Δrop13 parasite lysate. SAG1, a surface antigen, is used as a loading control. (D) Results of a competitive growth rate assay comparing parental RH and Δrop13 parasites. Error bars indicate mean +/− 1 S.D. HPT, selectable marker hypoxanthine-xanthine-guanine phosphoribosyl transferase.
To exclude possible polar effects of the HPT marker and to enable complementation using HPT, we removed the HPT gene by transfecting Δrop13_HPT(+) parasites with the pHPT-KO plasmid, which contains the rop13 flanks but lacks the HPT marker. The transfected parasites were selected against HPT with 6-thioxanthine and cloned by limiting dilution. A resulting Δrop13 line was tested for growth under positive selection for HPT with MPA and xanthine. No growth was observed, indicating removal of hpt.
3.4. ROP13 deletion results in a mild growth phenotype
To determine the fitness of the parasite without ROP13, we performed a competitive growth rate assay. Δrop13 parasites were mixed in a 1:1 ratio with the parental line and allowed to infect and grow in HFFs. When the host cells lysed, IFA was performed to count how many of each type of parasite remained. Parental parasites outcompeted the Δrop13 parasites (Fig. 3D), indicating that deletion at the rop13 locus is disadvantageous to Toxoplasma growth in vitro. We also assessed whether deletion of rop13 affected virulence of the parasite in mice and found that all mice died with the same kinetics following infection with either wild type or knockout strains (data not shown).
Several attempts to complement the disrupted gene were performed, with or without the selectable marker HPT, and using either the HA-tagged or untagged version of rop13. Although in each case the complemented protein localized properly to the rhoptries, we saw variable levels of expression between independent clones. The growth phenotype was not complemented in any of these clones, indicating that either exact levels or timing of expression of ROP13 are necessary for complementation or the observed growth defect was the result of an off-target effect.
3.5. ROP13 is a soluble protein
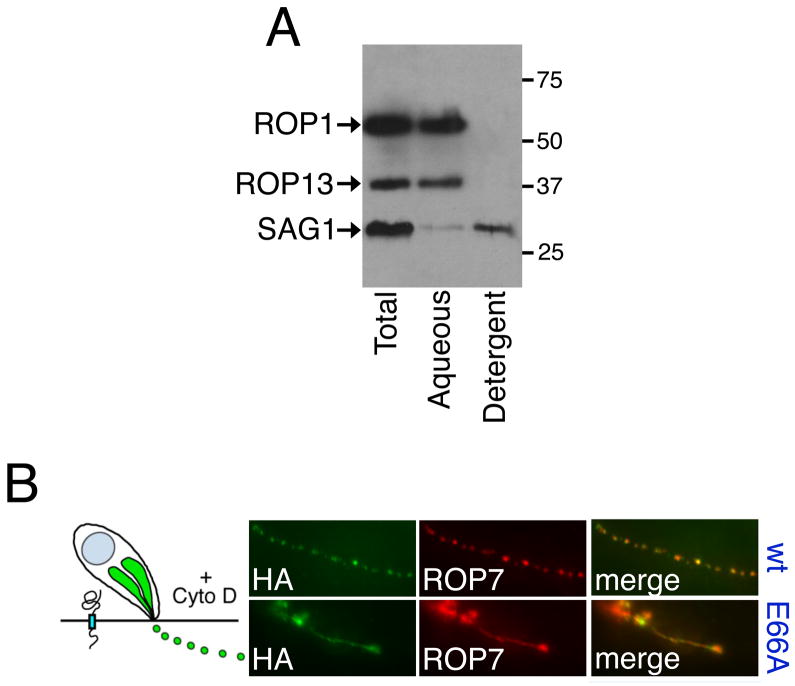
Most known ROPs are either soluble or PVM-associated. Determination of membrane association and integration has been a particularly difficult issue with the ROP2 family proteins. ROP2 was initially believed to be an integral membrane protein because it contains a hydrophobic domain and fractionates with membranes following carbonate extraction (Beckers et al., 1994). However, it was later determined that this family is not composed of integral membrane proteins, but that the putative transmembrane domain was instead merely the hydrophobic core of a kinase domain (Labesse et al., 2009). Membrane association was later shown to be conferred by alpha helices in an N-terminal region of the protein (Labesse et al., 2009; Reese and Boothroyd, 2009). To address this issue with ROP13, a non-ROP2 family member, we first bioinformatically assessed whether ROP13 is soluble or membrane-associated. TMpred predicts one transmembrane domain for the protein, although TMHMM does not predict that this region is a transmembrane domain (Hofmann and Stoffel, 1993; Krogh et al., 2001). We therefore performed a TX-114 extraction to determine ROP13’s solubility. As assessed by Western blot, ROP13 clearly fractionates with ROP1 (Fig. 4A), a known soluble protein (Rome et al., 2008), and not with the glycosylphosphatidylinositol (GPI)-anchored SAG1 (Burg et al., 1988), indicating that ROP13 is soluble and not membrane-associated.
Fig. 4.
Rhoptry protein 13 (ROP13) is a soluble effector protein ejected into the host cell. (A) TX-114-extracted RH lysate (separated by SDS-PAGE and Western blotted) was probed with anti-ROP1 (a known soluble protein), anti-SAG1 (a known glycosylphosphatidylinositol (GPI)-anchored membrane protein), and anti-ROP13. (B) Immunofluorescence assay (IFA) showing ROP13 and E66A mutant in evacuoles as detected by hemagglutinin (HA) antibody and colocalized with anti-ROP7.
3.6. Epitope-tagged ROP13 is injected into the host cytoplasm
Soluble ROPs are suspected to be secreted into the host cytoplasm where they are presumed to act as effectors, altering the host response to the invading parasite. However, such small quantities of these proteins are injected into the host that it can be very difficult to detect them. The use of cytochalasin D to arrest invasion of Toxoplasma into the host cell allows detection of small amounts of ROPs injected into the host cytoplasm in unique structures known as evacuoles (Håkansson et al., 2001). We could not detect ROP13 in evacuoles using our polyclonal antibody (Gilbert et al., 2007), however when we performed evacuole experiments on ROP13HA-expressing parasites, the tagged protein localized to evacuoles along with known evacuole marker ROP7 (Fig. 4B upper) (El Hajj et al., 2006; Rome et al., 2008). The E66A processing mutant line was also found to be present in evacuoles (Fig. 4B), indicating that processing is not essential for injection of ROP13 from the rhoptries into the host cell. To address whether ROP13 injected into the host cell remains in the cytosol or localizes to the PVM, we performed IFA to detect ROP13HA from early-invaded parasites (30 min or 1 h) in host cells concurrently infected with ROP13 knockout parasites. HA-tagged ROP13 could not be detected on or in knockout parasite vacuoles, suggesting that the protein’s final destination is in the host cytosol.
3.7. Exogenously-expressed mature ROP13 is cytosolic
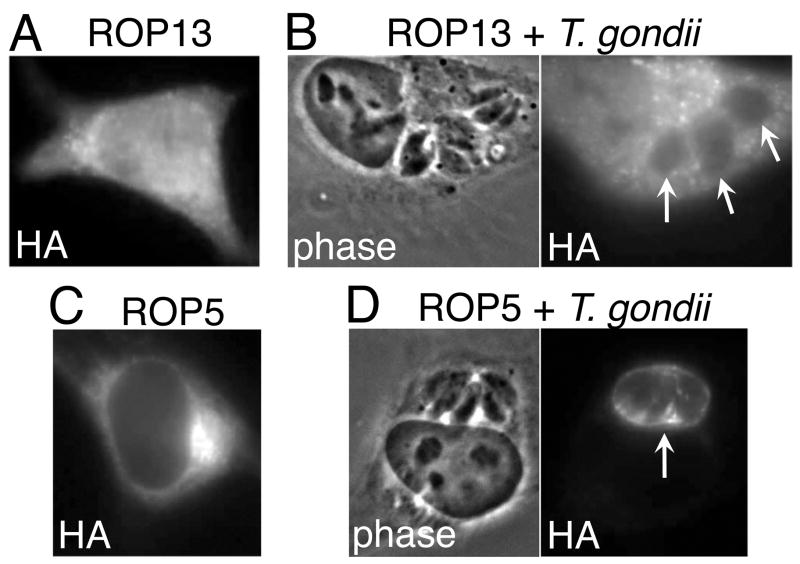
ROPs have been shown to be secreted into the host cytoplasm and then targeted to specific compartments (Reese and Boothroyd, 2009). The evacuole experiment above indicates that ROP13 is able to access the host cytoplasm, but the parasite-injected protein is likely in too low abundance to determine its destination within the host cell through IFA detection of endogenous levels of the protein. To further assess host localization of ROP13, we engineered a construct that contained mature ROP13 with C-terminal HA and PTP tags for expression in mammalian cells. The construct was then transiently transfected into HeLa cells and IFA performed. ROP13 could be detected in very few (≪1%) HeLa cells, although >50% of the cells expressed a control protein (PP2C-hn) in parallel experiments. Similar results were seen with various cell types (BHK-21, HEK 293 and CHO-K1), promoters (MoMuLV, CMV), and tags (plus or minus the PTP tandem affinity tag), and with or without the prodomain. This indicates that ROP13 over-expression was not inhibited due to cell, promoter, tag or processing-specific effects and instead suggests that this protein may be toxic to the host cell. In those cells in which ROP13 could be detected, it was diffusely cytoplasmic (Fig. 5A). Cytosolic localization of exogenously-expressed protein has been seen for the rhoptry effector and ROP2 family member ROP18, although upon infection of these host cells with Toxoplasma, ROP18 localizes to the PVM (El Hajj et al., 2007). To determine whether this occurs with ROP13, we infected the ROP13-transfected HeLa cells with Toxoplasma and performed IFAs. In the cells that expressed detectable levels of ROP13 and were parasite-infected, ROP13 did not relocalize to the PVM, instead remaining in the HeLa cytoplasm (Fig. 5B). ROP5 is a protein known to be injected into the host cell and subsequently to relocate to the PVM (Reese and Boothroyd, 2009). We engineered HA-tagged ROP5 as a control to show cytoplasmic localization of transiently expressed protein in HeLa cells and relocalization of that protein to the PVM of parasites infecting these host cells (Fig. 5C and D). These results suggest that ROP13 functions in the host cytoplasm, where over-expression may be toxic to the host cell.
Fig. 5.
Exogenous rhoptry protein 13 (ROP13) is detected in the cytoplasm of the host cell and not the parasitophorous vacuole membrane (PVM) of Toxoplasma gondii. (A) Immunofluorescence assay (IFA) showing ROP13- hemagglutinin (HA) cytoplasmic localization (as detected by anti-HA antibody) in transiently transfected HeLa cells. (B) IFA showing ROP13HA cytoplasmic localization (as detected by anti-HA antibody) in transiently transfected HeLa cells infected with RH parasites. The arrows show parasite-containing vacuoles, devoid of tagged protein. (C) IFA showing ROP5 cytoplasmic localization (as detected by anti-HA antibody) in transiently transfected HeLa cells. (D) IFA showing ROP5HA PVM localization (as detected by anti-HA antibody) in transiently transfected HeLa cells infected with RH parasites. The arrow shows a parasite-containing vacuole with tagged protein.
4. Discussion
The Toxoplasma rhoptries have emerged as the key organelles that hijack host cell functions by injecting proteins into the host cell at the onset of invasion. Our previous proteomic analysis of the rhoptries showed that the organelle is dominated by a large family of kinases and pseudokinases but also contains many proteins of unknown function (Bradley et al., 2005). To date, most of the work in this area has focused on rhoptry kinases and pseudokinases of the ROP2 family. Members of the family have been shown to interact with host STAT signaling pathways (ROP16), serve as strong determinants of parasite virulence (ROP18), and potentially modulate host organelle association (ROP2) (Sinai and Joiner, 2001; Saeij et al., 2006, 2007; Taylor et al., 2006). Much less is known about the other, more unique effectors, although many kinase and non-kinase rhoptry proteins alike are processed in the parasite prior to secretion.
Many rhoptry proteins are initially synthesized as pre-pro-proteins, with an N-terminal signal sequence targeting them to the secretory pathway, followed by a prodomain that is processed en route to the rhoptries (Soldati et al., 1998). The prodomain is hypothesized to be useful for either targeting to the rhoptries (as opposed to other secretory organelles) or maintaining a certain tertiary structure, thereby keeping the protein in zymogen form, although it is possible that it performs both functions. For ROP1 and ROP4, the prodomain is sufficient for targeting a reporter protein to the organelle (Bradley and Boothroyd, 2001; Striepen et al., 2001; Bradley et al., 2004). Similar to what has been shown for ROP1, we show here that processing of the ROP13 prodomain is not necessary for either targeting to the rhoptries or for secretion during invasion (Bradley et al., 2002). While the prodomain of ROP13 is similar in length to the ROP1 prodomain and those predicted for the ROP2 family proteins, common sequences are not obvious, suggesting that secondary structure is important for function. Whether the prodomain region also plays a role in modulating activity of rhoptry proteins will be simple to assess using the processing mutants described here once ROP13’s precise function is determined.
Rhoptry protein processing is likely to be performed by TgSUB2, a T. gondii subtilisin-like protease that recognizes the consensus sequence SϕXE at the P4-P1 positions and cleaves after the glutamic acid residue. Many rhoptry protein cleavage events have been hypothesized based on size shifts from pro- to mature forms and sequence analysis for candidate cleavage sites, but experimental evidence has only been provided for ROP1 and TgSUB2 (Bradley et al., 2002; Miller et al., 2003). Mutagenesis studies of ROP1 and TgSUB2 have shown that cleavage can be completely blocked by mutagenesis of the glutamic acid to a methionine or arginine. Mutation of the P1 position of ROP1 to aspartic acid only partially disrupts processing and is not found in known or predicted cleavage sites to date. Here, we show that ROP13’s SFTE does not tolerate a non-conservative mutation to alanine at the P1 position, adding both a third protein to the short list of ROPs experimentally shown to be processed at the consensus SϕXE and a new mutagenesis option for complete disruption of processing. If TgSUB2 is verified to be the protease responsible for cleavage, the recognition site is unusual for subtilisin proteases (which generally cleave after basic or aspartic acid residues) and indicates that parasite-specific inhibitors of the enzyme can be designed for therapeutic intervention (Bergeron et al., 2000; Withers-Martinez et al., 2004). Additional identification of rhoptry protein cleavage sites and further mutagenesis studies will provide a clearer picture of this unusual cleavage event.
Since it is an effector protein, ROP13 is not merely of interest due to its processing, but also for how it interacts with the host cell. As such, we disrupted the ROP13 gene in an attempt to determine the protein’s function. As with several other ROP knockouts in the laboratory-adapted type I RH strain, no gross defect was seen in vitro (Kim et al., 1993; Gilbert et al., 2007; Lodoen et al., 2009). The subtle growth defect that was observed by competition assay could not be complemented, which could be due to a requirement for exactly the correct level or timing of expression of ROP13 or a polar effect on neighboring genes. In addition, the lack of a clear phenotype in vivo is not surprising, given the extreme virulence of the RH strain of Toxoplasma and the fact that similar results have been seen for other ROP effector proteins in this highly virulent strain (Kim et al., 1993; Gilbert et al., 2007; Lodoen et al., 2009). The wide host range of the parasite and its ability to infect any nucleated cell also suggests that any given effector may be important for a certain class of host or a specific cell type and therefore dispensable in rich growth conditions in the human fibroblasts examined here (Boothroyd, 2009).
As effectors, ROPs are secreted during invasion of Toxoplasma into the host cell, allowing them to interact with host cell components, either at the PVM surface (e.g. ROP2, ROP5, ROP18), in the nucleus (PP2C-hn and ROP16) or in the cytoplasm (toxofilin) (El Hajj et al., 2006; Gilbert et al., 2007; Saeij et al., 2007; Ravindran and Boothroyd, 2008; Labesse et al., 2009; Lodoen et al., 2009). We have shown that ROP13 belongs to the last category, being a soluble protein that can be detected in the host cytoplasm. Due to the relatively low amounts injected, it is difficult to detect ROPs in the host cytoplasm. Thus, few ROPs have as of yet been detected there, where they are likely to act like bacterial effector proteins and co-opt host signaling pathways for optimizing intracellular survival. Those that have been detected have primarily been seen in evacuoles, membranous whorls injected into the host cell from cytochalasin D-arrested parasites. Prior to this study, evacuoles were suggested to be involved in formation of the PV, as ROP1 can be transferred via evacuole from cytochalasin D-arrested parasites to ROP1 PVs already formed in the host cell (Håkansson et al., 2001). Further support of the vacuole-formation theory was provided by detection of ROP18 transiently expressed in mammalian cells interacting with the nascent PVM after infection with Toxoplasma (El Hajj et al., 2007). Here we show that ROP13 can access the host cell, as it can be seen in evacuoles (the lack of detection with our polyclonal antibody is likely due to its relatively poor sensitivity). ROP13, however, cannot be detected on the PVM in transiently-transfected mammalian cells, suggesting that its target is located in the host cytosol, and not all ROPs in evacuoles are directed to the PV.
Little is known about the specific pathways modulated by Toxoplasma effector proteins once they have entered the host cell. The only role known is that of ROP16, which is involved in activating the host STAT3/6 signaling pathway, but even its direct target remains obscure (Saeij et al., 2007). In this paper, we have shown that despite some bioinformatic data to the contrary, ROP13 is soluble and can be injected into the host cell cytoplasm. We engineered ROP13 with a PTP tag for identification of its target upon expression in mammalian cells, but its apparent toxicity indicates that a tightly regulated approach will be necessary to identify its target within the host cell. Toxicity upon over-expression of bacterial effector proteins is relatively common and likely reflects the need for effectors to be potent modulators of host functions as they are injected in low abundance into the host cell (Galán, 2009). Recently, crystal structure studies or expression in yeast in combination with yeast deletion strains have been utilized to determine the function of bacterial effector proteins (Janjusevic et al., 2006; Kramer et al., 2007). These approaches may similarly be effective to elucidate the precise function of ROP13 and other rhoptry effector proteins in Toxoplasma infections.
Supplementary Material
Alignment of the primary amino acid sequences of the rhoptry protein 13 (ROP13) orthologues of Neospora caninum (NcROP13) and Toxoplasma gondii (TgROP13). Identical residues are highlighted in yellow, while similar residues are highlighted in green. The P4-P1 SϕXE residues are boxed in blue.
Acknowledgments
We thank Joe Schwartzman for ROP1 antibody, John Boothroyd for SAG1 antibody, and the members of the Bradley laboratory for helpful discussions. This work was supported by the Microbial Pathogenesis Training Grant T32-AI07323 to J.M.T. and B.E.H. and NIH Grant 1R01AI064616 to P.J.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: supplementary data associated with this article
References
- Beckers CJ, Dubremetz JF, Mercereau-Puijalon O, Joiner KA. The Toxoplasma gondii rhoptry protein ROP 2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host cell cytoplasm. J Cell Biol. 1994;127:947–961. doi: 10.1083/jcb.127.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers CJM, Wakefield T, Joiner KA. The expression of Toxoplasma proteins in Neospora caninum and the identification of a gene encoding a novel rhoptry protein. Molecular and Biochemical Parasitology. 1997;89:209–223. doi: 10.1016/s0166-6851(97)00120-5. [DOI] [PubMed] [Google Scholar]
- Bergeron F, Leduc R, Day R. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J Mol Endocrinol. 2000;24:1–22. doi: 10.1677/jme.0.0240001. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii Rhoptry Neck Protein Complex at the Host Cell Membrane to Form the Moving Junction during Invasion. PLoS Pathog. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd JC, Dubremetz JF. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Micro. 2008;6:79–88. doi: 10.1038/nrmicro1800. [DOI] [PubMed] [Google Scholar]
- Boothroyd JC. Expansion of host range as a driving force in the evolution of Toxoplasma. Mem Inst Oswaldo Cruz. 2009;104:179–184. doi: 10.1590/s0074-02762009000200009. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Boothroyd JC. The pro region of Toxoplasma ROP1 is a rhoptry-targeting signal. International Journal for Parasitology. 2001;31:1177–1186. doi: 10.1016/s0020-7519(01)00242-9. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Hsieh CL, Boothroyd JC. Unprocessed Toxoplasma ROP1 is effectively targeted and secreted into the nascent parasitophorous vacuole. Molecular and Biochemical Parasitology. 2002;125:189–193. doi: 10.1016/s0166-6851(02)00162-7. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Li N, Boothroyd JC. A GFP-based motif-trap reveals a novel mechanism of targeting for the Toxoplasma ROP4 protein. Molecular and Biochemical Parasitology. 2004;137:111–120. doi: 10.1016/j.molbiopara.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. Proteomic Analysis of Rhoptry Organelles Reveals Many Novel Constituents for Host-Parasite Interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Sibley LD. Rhoptries: an arsenal of secreted virulence factors. Current Opinion in Microbiology. 2007;10:582–587. doi: 10.1016/j.mib.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg JL, Perelman D, Kasper LH, Ware PL, Boothroyd JC. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- Donald RGK, Carter D, Ullman B, Roos DS. Insertional Tagging, Cloning, and Expression of the Toxoplasma gondii Hypoxanthine-Xanthine-Guanine Phosphoribosyltransferase Gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- El Hajj H, Lebrun M, Fourmaux MN, Vial H, Dubremetz JF. Characterization, biosynthesis and fate of ROP7, a ROP2 related rhoptry protein of Toxoplasma gondii. Molecular and Biochemical Parasitology. 2006;146:98–100. doi: 10.1016/j.molbiopara.2005.10.011. [DOI] [PubMed] [Google Scholar]
- El Hajj H, Lebrun M, Arold ST, Vial H, Labesse G, Dubremetz JF. ROP18 Is a Rhoptry Kinase Controlling the Intracellular Proliferation of Toxoplasma gondii. PLoS Pathogens. 2007;3:e14. doi: 10.1371/journal.ppat.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE. Common Themes in the Design and Function of Bacterial Effectors. Cell Host & Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Ravindran S, Turetzky JM, Boothroyd JC, Bradley PJ. Toxoplasma gondii Targets a Protein Phosphatase 2C to the Nuclei of Infected Host Cells. Eukaryotic Cell. 2007;6:73–83. doi: 10.1128/EC.00309-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson S, Charron AJ, Sibley LD. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO. 2001;20:3132–3144. doi: 10.1093/emboj/20.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMBASE - A database of membrane spanning protein segments. Biological Chemistry. 1993:374. [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A Bacterial Inhibitor of Host Programmed Cell Death Defenses Is an E3 Ubiquitin Ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- Karasov AO, Boothroyd JC, Arrizabalaga G. Identification and disruption of a rhoptry-localized homologue of sodium hydrogen exchangers in Toxoplasma gondii. International journal for parasitology. 2005;35:285–291. doi: 10.1016/j.ijpara.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Kim K, Soldati D, Boothroyd J. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science. 1993;262:911–914. doi: 10.1126/science.8235614. [DOI] [PubMed] [Google Scholar]
- Kim K, Weiss LM. Toxoplasma: the next 100 years. Microbes and Infection. 2008;10:978–984. doi: 10.1016/j.micinf.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RW, Slagowski NL, Eze NA, Giddings KS, Morrison MF, Siggers KA, Starnbach MN, Lesser CF. Yeast Functional Genomic Screens Lead to Identification of a Role for a Bacterial Effector in Innate Immunity Regulation. PLoS Pathog. 2007;3:e21. doi: 10.1371/journal.ppat.0030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a Hidden Markov Model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Labesse G, Gelin M, Bessin Y, Lebrun M, Papoin J, Cerdan R, Arold ST, Dubremetz JF. ROP2 from Toxoplasma gondii: A Virulence Factor with a Protein-Kinase Fold and No Enzymatic Activity. Structure. 2009;17:139–146. doi: 10.1016/j.str.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Lodoen MB, Gerke C, Boothroyd JC. A highly sensitive FRET-based approach reveals secretion of the actin-binding protein toxofilin during Toxoplasma gondii infection. Cellular Microbiology. 2009 doi: 10.1111/j.1462-5822.2009.01378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Thathy V, Ajioka JW, Blackman MJ, Kim K. TgSUB2 is a Toxoplasma gondii rhoptry organelle processing proteinase. Molecular Microbiology. 2003;49:883–894. doi: 10.1046/j.1365-2958.2003.03604.x. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. Toxoplasmosis. The Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Ravindran S, Boothroyd JC. Secretion of Proteins into Host Cells by Apicomplexan Parasites. Traffic. 2008;9:647–656. doi: 10.1111/j.1600-0854.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- Reese ML, Boothroyd JC. A Helical Membrane-Binding Domain Targets the Toxoplasma ROP2 Family to the Parasitophorous Vacuole. Traffic. 2009;10:1458–1470. doi: 10.1111/j.1600-0854.2009.00958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. Intervacuolar Transport and Unique Topology of GRA14, a Novel Dense Granule Protein in Toxoplasma gondii. Infect Immun. 2008;76:4865–4875. doi: 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. Polymorphic Secreted Kinases Are Key Virulence Factors in Toxoplasmosis. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij JPJ, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski B, Nguyen T, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryotic Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber F, Dubremetz JF, Boothroyd JC. Analysis of Toxoplasma gondii stably transfected with a transmembrane variant of its major surface protein, SAG1. Journal of cell science. 1998;111 ( Pt 1):23–29. doi: 10.1242/jcs.111.1.23. [DOI] [PubMed] [Google Scholar]
- Sinai AP, Joiner KA. The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J Cell Biol. 2001;154:95–108. doi: 10.1083/jcb.200101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D, Lassen A, Dubremetz JF, Boothroyd JC. Processing of Toxoplasma ROP1 protein in nascent rhoptries. Molecular and Biochemical Parasitology. 1998;96:37–48. doi: 10.1016/s0166-6851(98)00090-5. [DOI] [PubMed] [Google Scholar]
- Straub KW, Cheng SJ, Sohn CS, Bradley PJ. Novel components of the Apicomplexan moving junction reveal conserved and coccidia-restricted elements. Cellular Microbiology. 2009;11:590–603. doi: 10.1111/j.1462-5822.2008.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Soldati D, Garcia-Reguet N, Dubremetz JF, Roos DS. Targeting of soluble proteins to the rhoptries and micronemes in Toxoplasma gondii. Molecular and Biochemical Parasitology. 2001;113:45–53. doi: 10.1016/s0166-6851(00)00379-0. [DOI] [PubMed] [Google Scholar]
- Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. A Secreted Serine-Threonine Kinase Determines Virulence in the Eukaryotic Pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. International Journal for Parasitology. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Martinez C, Jean L, Blackman M. Subtilisin-like proteases of the malaria parasite. Mol Microbiol. 2004;53:55–63. doi: 10.1111/j.1365-2958.2004.04144.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the primary amino acid sequences of the rhoptry protein 13 (ROP13) orthologues of Neospora caninum (NcROP13) and Toxoplasma gondii (TgROP13). Identical residues are highlighted in yellow, while similar residues are highlighted in green. The P4-P1 SϕXE residues are boxed in blue.