Abstract
Heparan sulfate proteoglycans cooperate with basic Fibroblast Growth Factor (bFGF/FGF2) signaling to control osteoblast growth and differentiation, as well as metabolic functions of osteoblasts. FGF2 signaling modulates the expression and activity of Runt-related transcription factor 2 (Runx2/Cbfa1), a key regulator of osteoblast proliferation and maturation. Here, we have characterized novel Runx2 target genes in osteoprogenitors under conditions that promote growth arrest while not yet permitting sustained phenotypic maturation. Runx2 enhances expression of genes related to proteoglycan-mediated signaling, including FGF receptors (e.g., FGFR2 and FGFR3) and proteoglycans (e.g., Syndecans [Sdc1, Sdc2, Sdc3], Glypicans [Gpc1], Versican [Vcan]). Runx2 increases expression of the glycosyltransferase Exostosin-1 [Ext1] and heparanase, as well as alters the relative expression of N-linked sulfotransferases (Ndst1=Ndst2>Ndst3) and enzymes mediating O-linked sulfation of heparan sulfate (Hs2st>Hs6st) or chondroitin sulfate (Cs4st>Cs6st). Runx2 cooperates with FGF2 to induce expression of Sdc4 and the sulfatase Galns, but Runx2 and FGF2 suppress Gpc6, thus suggesting intricate Runx2 and FGF2 dependent changes in proteoglycan utilization. One functional consequence of Runx2 mediated modulations in proteoglycan-related gene expression is a change in the responsiveness of bone markers to FGF2 stimulation. Runx2 and FGF2 synergistically enhance Osteopontin expression (>100 fold), while FGF2 blocks Runx2 induction of Alkaline Phosphatase. Our data suggest that Runx2 and the FGF/proteoglycan axis may form an extracellular matrix (ECM)-related regulatory feed-back loop that controls osteoblast proliferation and execution of the osteogenic program.
Keywords: Heparin, Heparan Sulfate, Proteoglycan, Glycosaminoglycan, Syndecan, Glypican, Fibroblast Growth Factor, Runx2, osteoblast
INTRODUCTION
Proteoglycans are highly glycosylated proteins that are prominently expressed in osteoblasts and functionally interact with osteogenic ligands (e.g., FGF2/bFGF, BMPs, WNTs) [Jackson et al., 2006b; De and David, 2001; Ling et al., 2009]. Glycosaminoglycans (GAGs) attached to these proteins potentiate intracellular signaling through their ability to bind distinct ligands and to support extracellular ternary complexes between proteoglycans and liganded receptors that are embedded within the cell membrane. Chemical modifications (e.g. sulfation and acetylation) of the sugar-moieties may control the specificity of the interactions and regulate signaling efficiency [Lamanna et al., 2007; Gorsi and Stringer, 2007]. For example, heparan sulfates that are attached to specific proteoglycans (e.g., syndecans, glypicans) may control Fibroblast Growth Factor (FGF) signaling through the synergistic interactions between membrane-associated FGFRs that initiate intracellular signaling through their associated tyrosine kinase activities [Jackson et al., 2006b; Marie, 2003].
FGF2 is indispensable for normal bone development and maintenance. For example, activating mutations of FGF receptors (FGFRs) cause alterations in bone development and are associated with several genetic disorders, including Pfeiffer, Apert, and Jackson-Weiss syndromes [Marie et al., 2005; Cunningham et al., 2007; Bonaventure and El, V, 2003; Bodo et al., 1999]. Patients with these syndromes characteristically exhibit increased mineralization and premature fusion of the cranial suture (craniosynostosis). FGF2 is a key signaling molecule in osteoblasts with strong anabolic functions in bone in vivo [Coffin et al., 1995; Montero et al., 2000; Yu et al., 2003; Naganawa et al., 2006] and in osteoblasts in culture [Sobue et al., 2005; Boudreaux and Towler, 1996; Molteni et al., 1999; Song et al., 2007; Ling et al., 2006; Jackson et al., 2006a; Jackson et al., 2007; Sabbieti et al., 2008; Lee et al., 2007] by stimulating bone formation, mineralization and regeneration, as well as by acting as a potent osteoblast mitogen. FGF2 knock-out mice have reduced bone mass and bone formation [Montero et al., 2000] and develop osteopenia with aging [Naganawa et al., 2006], while over-expression of the FGF2 gene in transgenic mice causes premature mineralization, flattening and shortening of the long bones [Coffin et al., 1995; Sobue et al., 2005]. Continued exposure to FGF2 during osteogenic differentiation in vitro has been shown to disrupt mineralization [Ling et al., 2006].
Runt-related transcription factor Runx2 [van Wijnen et al., 2004] and Activation Protein 1 (AP1) represent downstream gene regulatory effectors of FGF signaling in osteoblasts [Kim et al., 2003; Kim et al., 2006; Shimizu-Sasaki et al., 2001; Zhang et al., 2002; Xiao et al., 2002; Hatch et al., 2008; Okazaki et al., 1992]. Apart from mediating lineage commitment, Runx2 attenuates osteoblast proliferation [Pratap et al., 2003; Galindo et al., 2005; Galindo et al., 2007; Zaidi et al., 2007; Teplyuk et al., 2008] and performs a novel epigenetic function by associating with mitotic chromosomes [Young et al., 2007b; Young et al., 2007a; Rajgopal et al., 2007; Zaidi et al., 2003]. Consistent with functional linkage between Runx2 and FGF2, abrogation of FGF receptor signaling in mouse embryonic stem cells primes them for osteogenic stimulation and significantly induces Runx2 gene expression [Woei et al., 2007]. Runx2 has also been implicated in the etiology of osteosarcoma and cancer metastasis to bone [Thomas et al., 2004; Nathan et al., 2008; Gutierrez et al., 2008; Pratap et al., 2006] suggesting that the FGF2 dependent activity of Runx2 may regulate osteogenic commitment, bone cell growth and metastatic potential.
In this study, we have characterized genes that are modulated by re-introduction of Runx2 in Runx2 null osteoprogenitors when cells become growth arrested but have not yet differentiated into mature osteoblasts. We have previously shown that Runx2 mediates cell growth control by regulating a number of genes involved in G protein coupled receptor signaling [Teplyuk et al., 2008]. We now show that growth control by Runx2 also involves regulation by genes related to FGF signaling, a key mitogenic pathway in osteoprogenitors. We find that Runx2 up-regulates expression of several FGF receptors, proteoglycans and glycosaminoglycan modifying enzymes. FGF2 treatment strongly synergizes with Runx2 to induce the expression of the early osteogenic marker osteopontin, but FGF2 counteracts Runx2 induction of the bone-phenotypic marker Alkaline Phosphatase. Our combined results suggest that Runx2 modifies the responsiveness of osteoprogenitors to FGF stimulation during the transition from active proliferation to growth arrest.
MATERIALS AND METHODS
Tissue culture and protein expression
The development of a TERT-immortalized Runx2 null mouse calvaria osteoprogenitor cell line was described previously [Bae et al., 2007]. Cells were maintained in αMEM supplemented with 10% fetal bovine serum (FBS, Atlanta), 30 mM Penicillin-Streptomycin and 100 mM L-Glutamine at 37°C and humidified 5% CO2 atmosphere. For exogenous expression of Runx2, cells were plated in 6-well plates (1 × 105 cells/well) and infected after 24 h with 100 MOI of adenovirus expressing Runx2 plus IRES-driven GFP or GFP alone control in 600 μl of medium complemented with 1% FBS as described previously [Teplyuk et al., 2008]. After 4 h, additional medium was added (400 μl of αMEM containing 1% FBS) and cells were incubated for an additional 10 h. Infection efficiency was determined by monitoring GFP positive cells. Construction of Runx2 expressing adenovirus was described previously [Teplyuk et al., 2008]. Infections were stopped by washing cells with 1X PBS and changing the media, and cells were treated 12 h later with 10 ng/ml bovine basic FGF (R&D Systems, Minneapolis, MN) for the next 24 h when indicated.
Affymetrix analysis
Initial examination of Runx2 responsiveness of proteoglycan related gene expression was based on Affymetrix data previously reported [Teplyuk et al., 2008]. In brief, we examined immortalized Runx2 null osteoprogenitors [Bae et al., 2007] that were infected with A denoviral vectors expressing a bicistronic Runx2-GFP mRNA or GFP alone. At 24 h after infection, RNA samples were harvested in triplicate and examined by Affymetrix gene expression profiling (Mouse Genome 430 2.0 Array). Statistically significant differences between probe sets were evaluated using Student’s t test (p < 0.05). Functional annotation of Affymetrix probe sets and gene ontology relationships between groups of co-regulated genes were assessed using the data base for Annotation, Visualization and Integrated Discovery (DAVID 2.0) [Dennis, Jr. et al., 2003] and information Hyperlinked Over Proteins (iHOP; http://www.ihop-net.org)[Hoffmann and Valencia, 2004].
Quantitative PCR analysis
Total RNA was extracted with Trizol reagent (Invitrogen) and purified on columns after DNaseI treatment (RNA purification kit; Zymo Research, Orange, CA). Total RNA aliquots (1 μg) were used for reverse transcription with random hexamers (Invitrogen, Carlsbad, CA). The resulting cDNA products were diluted 75 fold and 5 μl was added in a 25 μl qPCR reaction with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) for quantitation in an A BI Prism 7000 Sequence Detection System. PCR was performed using the following conditions: 1 cycle at 95°C for 10 min as the initial denaturation, 40 cycles of-step reaction at 95°C for 15 sec 2 (denaturation) and 60°C for 1 min (annealing and synthesis).
The following mouse gene primers were used in the reactions(0.5 pmol/μl each):Runx2: F 5′-CGA CAG TCC CAA CTT CCT GT-3′; R 5′-CGG TAA CCA CAG TCC CAT CT-3′; Osteocalcin: F 5′-CTG ACA AAG CCT TCA TGT CCA A-3′; R 5′-GCG CCG GAG TCT GTT CAC TA-3′; Osteopontin: F 5′-ACT CCA ATC GTC CCT ACA GTC G-3′, R 5′-TGA GGT CCT CAT CTG TGG CAT-3′; Alkaline phosphatase: F 5′-TTG TGC GAG AGA AAG AGA GAG A-3′; R 5′-GTT TCA GGG CAT TTT TCA AGG T-3′; FGF receptor 1 (FGFR1): F 5′-CCG TAT GTC CAG ATC CTG AAG A-3′; R 5′-GAT AGA GTT ACC CGC CAA GCA-3′; FGF receptor 2 (FGFR2): F 5′-GCC CTA CCT CAA GGT TCT GAA AG-3′; R 5′-GAT AGA ATT ACC CGC CAA GCA-3′; FGF receptor 3 (FGFR3): F 5′-CCC TAC GTC ACT GTA CTC AAG ACT G-3′; R 5′-GTG ACA TTG TGC AAG GAC AGA AC-3′; FGF receptor 4 (FGFR4): F 5′-CGA CGG TTT CCC CTA CGT ACA-3′; R 5′-TGC CCG CCA GAC AGG TAT AC-3′; Syndecan 1 (S dc1): F 5′-A AC CAA ATC TGG ACG GCA AA-3′; R 5′-CTA CTT ACG GGC CGC CAA A-3′; Syndecan 2 (S dc2): F 5′-TGT AGG ACC AGA CCA AGA AAA CAG-3′; R 5′-TTC TCT GGC GCC TGC TCT AG-3′; Syndecan 3 (Sdc3): F 5′-CGT AGG CCA CTG TCA TTG TCA-3′; R 5′-TGG TTA GAG GAG CCA GAT GCA-3′; Syndecan 4 (Sdc4): F 5′-CTT CCT CCA GGC GCT CTA GA-3′; R 5′-CAC GTA GTC TGA AGT GAA CCG AGT T-3′; Heparanase (Hpse): F 5′-ACC GAC GAC GTG GTA GAC TTG-3′; R 5′-GAT GGT GAT GGA CAG GAA CGA-3′; Biglycan (Bgn): F 5′-CAC CTG ACA CCA CAC TGC TAG AC-3′; R 5′-GTA GAG GTG CTG GAG GCC TTT3′; Decorin (Dcn): F 5′-ATT GAA AAC GGA GCC TTC CA-3′; R 5′-TTG AGG GAT CGC AGT TAT GTT G-3′; Versican (Vcan): F 5′-CTC ATT TCC AAA ATA GGC AGC AT-3′; R 5′-CAT GAG CTT CAC GAA AGG AAG A-3′; Perlecan (Perl): F 5′-TGC ATC CCC CGA GAC TAC CT-3′; R 5′-CTC ACA ATC AAA GTC ACC GTC ACA-3′; Glypican 1 (Glyp1): F ′-CCA GCT GCA TGG CAT CGA TG-3′; R 5′-GGC ACG AGT GTT CTG CGT ATA CAG-3′; Glypican 2 (Glyp2): F 5′-CCG TCA TCT TCA ATA GCC TGT TTT-3′; R 5′-TCC GCC AAG GTG TCA TCT AAC-3′; Glypican 4 (Glyp4): F 5′-GCT TAG CAG TTG CAA GGG ATG T-3′; R 5′-GCA GTG GGA GCA GTA GAT CAT CT-3′; Glypican 6 (Glyp 6): F 5′-GAC TGG AAG GGC CAT TCA AC-3′; R 5′-TGC ATG CTG TTT TCC TGC AT-3′; N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 1 (Ndst1): F 5′-TTT TAA CGG CCA CAA CTA TCA CA-3′; R 5′-AAG TCC GAG GTG GTG TTG GA-3′; N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 2 (Ndst2): F 5′-TCT GTG CTA GCT GAC CAG ATG AG-3′; R 5′-CAT ACC CCA GAT CCG TAG GAA TC-3′; N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 3 (Ndst3): F 5′-TCT CTA GTC CCG AAA GCC AAG A-3′; R 5′-GGA TCG CTG ATG CTG ATA CCA-3′; heparan sulfate 2-O-sulfotransferase 1 (Hs2st1): F 5′-AAG GGC CGT GGT TAG AGC TTA-3′; R 5′-AGA CAG GGC TTC TCC ATG ACA AT-3′; Heparan sulfate 6-O-sulfotransferase 1 (Hs6st1): F 5′-GTG TGC CCA CCG AGG ACT AC-3′; R 5′-TTA AAT CGT GCC CAT CAC TCT CT-3′; carbohydrate (chondroitin 4) sulfotransferase (Ch4st): F 5′-TGG AAG TGA TGA GGA TGA ACA GA-3′; R 5′-TGG GTG CAA CAT ACT TTG GAA A-3′; carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 1 (Ch6st): F 5′-CGC ACG GGT TCC TCG TTC-3′; R 5′-GAA CAC GGT GCG CTC GAT GT-3′; glucuronyl C5-epimerase (Glce): F 5′-CAA GGG CAA GCC ATC TCT ACC T-3′; R 5′-TCA TGA ACA CGG CTT TAA CTC CAT-3′; galactosamine (N-acetyl)-6-sulfate sulfatase (Galns): F 5′-CCC AGT GAC AGG GTG ATT GAT-3′; R 5′-TGT GCT TTG TAC TGG CCA AGA-3′; Exostosin 1 (Ext1): F 5′-GTG TAC CCG CAG CAG AAA GG-3′; R 5′-GTA GAA CCT GGA GCC CTC GAT-3′; Exostosin 2 (Ext2): F 5′-CAA AAT CCG AGT TCC CCT GAA-3′; R 5′-TCG ATT TCG TCG TAA GGG AAG A-3′. Data normalization of samples from cell lines was established by qPCR with primers specific for rodent GAPDH and 18S ribosomal RNA (Applied Biosystems).
Western blot analysis
For western blotting, cell pellets were boiled in 100 μl of direct lysis buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% Glycerol, 12% Urea, 25 μM MG132 (Calbiochem, San Diego, CA), 100 mM DTT and 1X Complete protease inhibitors from Roche). Aliquots (5 μl) of each lysate were separated using 10% SDS-PAGE and subjected to semi-wet transfer to PVDF membranes. PBS(1X)/0.1% Tween (PBST) with 5% milk was used for 1 h to block non-specific binding. Primary and secondary antibodies were used in a 1:2,000 dilution for 1 h in PBST/1% milk. Signals were detected by chemiluminescence (Perkin Elmer Western Lightning Chemiluminescence Reagent Plus). Runx2-specific mouse monoclonal antibodies were the generous gift of Dr. Yoshiaki Ito (Cancer Research Center of Excellence, National University of Singapore). CDK2 rabbit polyclonal antibodies were purchased from Santa Cruz Biotechnology.
RESULTS
Expression of FGFR and proteoglycan genes in Runx2 null osteoprogenitors
Runx2 and FGF2 signaling both control osteoblast proliferation and maturation (Fig. 1A). Runx2 null calvarial cells represent mesenchymal precursors that are arrested in the osteoprogenitor stage [Bae et al., 2007]. Re-introduction of Runx2 into these cells by adenoviral infection (Fig. 1B) decreases cell proliferation and increases Alkaline Phosphatase expression marking the onset of osteogenic differentiation (Fig. 1C), consistent with previous observations [Teplyuk et al., 2008; Bae et al., 2007]. To understand how Runx2 controls proliferation of osteoprogenitors, we have previously performed Affymetrix gene expression profiling and established that Runx2 controls genes for distinct G protein coupled receptor signaling pathways [Teplyuk et al., 2008] and enzymes controlling sterol metabolism [Bae et al., 2007], as well as enhances expression of classical Runx2 targets such as osteocalcin, bone sialoprotein and MMP13 (Fig. 1D). During the course of these studies we observed that a subset of Runx2 responsive genes encode proteoglycans and related proteins (Fig. 1E).
Figure 1. Runx2 and FGF2 signaling control both proliferation and maturation of osteoblast.
A. The schematic depicts the dual role of Runx2 and FGF2 signaling in control of osteoblast proliferation and differentiation. Runx2 may act opposite to FGF2 signaling during the G1 phase of the cell cycle in osteoprogenitors. FGF2 has potent mitogenic effects and stimulates the proliferative expansion of osteoprogenitor cells, while Runx2 suppresses cell cycle progression and may drive cells into quiescence. Runx2 and FGF2 signaling work synergistically on the early steps of osteoblast differentiation, both inducing expression of the early osteoblastic markers (e.g. Osteopontin). B. The experimental system used in this study is a complementation assay in which Runx2 protein is exogenously expressed in osteoprogenitor cells with a homozygous Runx2 null background. Runx2 protein levels were detected by western blot analysis 36 h after infection with recombinant adenovirus expressing Runx2 and GFP (lane ‘Runx2’), after infection with the same vector lacking Runx2 but containing GFP (abbreviated EV; lane ‘Control EV’) or in mock-infected cells (lane ‘No DNA’). CDK2 protein levels were used as internal endogenous control. C. Re-introduction of Runx2 protein expression in Runx2 null cells decreases proliferation as monitored by cell counting (left bar graph) and increased osteogenic differentiation as assessed by Alkaline phosphatase (AP) staining (right bar graph) after cells were grown for 8 days in osteogenic media (50 μg/ml Ascorbic acid, 10 mM β-Glycerophosphate) and treated with 200 ng/ml BMP2 for 24 h. AP staining was quantified by densitometry using ImageJ software. Error bars represent standard error of mean (SE) between three different quantifications. D. Runx2 regulates distinct expression programs. Runx2 regulation of ECM proteins, G protein signaling and sterol metabolism in osteoprogenitor cells were identified previously by Affymetrix gene expression profiling [Teplyuk et al., 2008]. This study focuses on Runx2 regulation of proteoglycan related gene. E. Membrane and extracellular matrix related proteoglycans were identified as a component of Runx2 responsive programs by hierarchical clustering, as well as the DAVID 2.0 database (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov) [Dennis, Jr. et al., 2003] and information Hyperlinked Over Proteins (iHOP, http://www.ihop-net.org) [Hoffmann and Valencia, 2004].
Because proteoglycans support cell signaling in osteoblasts, we further examined the Runx2 dependent regulation of this set of genes, which includes FGF receptors, membrane-anchored proteoglycans and enzymes that modify glycosaminoglycans. We first assessed the basal level of this set of genes in Runx2 null cells that represent calvarial derived osteoprogenitors (Fig. 2). These cells robustly express several proteoglycans (e.g., Biglycan, Perlecan, Versican, Glypican-1 and -4, as well as Syndecan-2, -3 and -4), Exostosin-1 and FGF Receptor-1. Heparin related modifying enzymes, as well as FGF receptors-2, -3 and -4, are expressed at significantly lower but detectable levels. Thus, as expected, structural proteins that interface with the extra-cellular matrix are generally expressed at higher levels than the enzymes that control their modifications.
Figure 2. Relative basal expression levels of genes encoding proteoglycans, FGF receptors and proteoglycan modifying enzymes in osteoprogenitor cells.
We determined basal mRNA expression by qPCR analysis in Runx2 null osteoprogenitor cells to assess the relative expression of selected genes in the absence of Runx2 and to determine which genes are characteristic for immature cells within the early osteogenic lineage. The mRNA levels of different genes were plotted as a percentage of GAPDH mRNA levels. Genes were arbitrarily divided based on their expression level into robustly expressed (A) and weakly expressed (B) genes, with the dividing point at 1% of GAPDH expression. Error bars represented Standard error of mean (SE) between three different populations of cells.
Runx2 regulates genes encoding proteoglycans and glycosaminoglycan modifying enzymes in osteoprogenitors
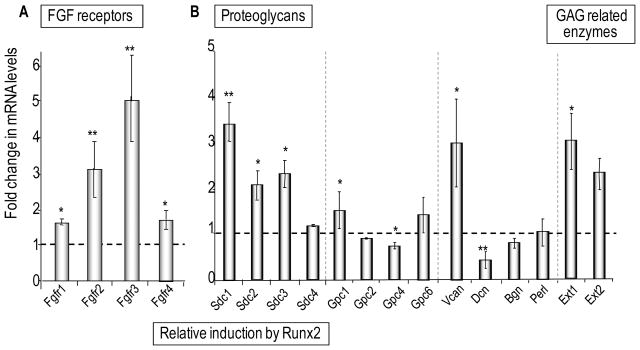
To validate our preliminary findings using Affymetrix profiling that proteoglycan related genes are responsive to elevation of Runx2 levels, we performed quantitative RT-PCR analysis. We analyzed triplicate RNA samples from Runx2 null cells infected by an adenoviral vector expressing exogenous wild type Runx2 protein or a corresponding empty vector. Upon exogenous expression of Runx2, all four FGF receptors (FGFR-1, -2, -3 and -4) are up regulated (Fig. 3A). FGFR-3 mRNA levels increase by approximately five-fold, whereas FGFR1 mRNA levels increase by only a modest amount (~1.6–.7 fold), yet FGFR-1 remains the most prominently expressed FGF receptor following the reintroduction of Runx2 into Runx2 null cells. Similarly, expression of Exostosin-1 and Syndecan-1, -2 and -3, as well as other plasma membrane chondroitin and heparan sulfate proteoglycans (Versican and Glypican-1) is increased, while expression of Glypican-4 and the dermatan sulfate proteoglycan Decorin is modestly diminished in the presence of Runx2 (Fig. 3B).
Figure 3. Responsiveness of FGF receptors and proteoglycans gene expression by Runx2 in osteoprogenitors.
Expression levels of FGF receptors (A) or proteoglycans (B) were determined by qPCR analysis in Runx2 null osteoprogenitors infected with vectors that either do or do not express Runx2 protein. Relative mRNA levels were plotted as fold change of exogenous Runx2 expressing cells over the GFP expressing control and normalized to 18S ribosomal RNA level. Statistical significance of differences was determined by Student’s T-test. Values with P < 0.05 are indicated by asterisks and values with P < 0.01 have two asterisks. Error bars represented Standard error of mean (SE) between three independent experiments. Panel B (right part) also contains data for two glycosaminoglycan (GAG) modifying enzymes that did not fit the scale of the graphs in Figure 4A.
We note that the magnitude of the fold-increase for FGF receptors, proteoglycans and proteoglycan related enzymes (see Figs. 3 and 4) is in several cases inversely related to their basal expression in the absence of Runx2 (see Fig. 2). Highly expressed genes (e.g., FGFR1, Glypican 1) exhibit modest quantitative changes upon forced expression of Runx2, while moderately expressed genes (e.g., FGFR3, Syndecan 1) tend to be stimulated to a greater degree. This finding is not unexpected as transcription of genes that are already prominently expressed in Runx2 null osteoprogenitors is at least in part controlled independently of Runx2.
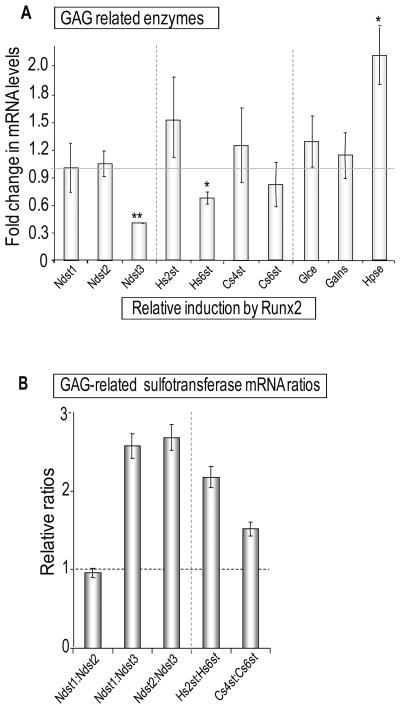
Figure 4. Regulation of glycosaminoglycan modifying enzymes gene expression by Runx2 in osteoprogenitors.
Fold changes in mRNA expression of genes encoding glycosaminoglycan (GAG) modifying enzymes including different heparan sulfate and chondroitin sulfate sulfotransferases species upon expression of Runx2 (A), or the same data as in Panel B plotted in a manner that emphasizes relative changes in the expression ratios of enzymes involved in N- versus O-linked modifications of GAGs (B). Similar to Fig. 3, relative mRNA levels were normalized to 18S ribosomal RNA levels and plotted as fold change upon exogenous Runx2 expression in Runx2 null cells. Error bars represent SE between three independent experiments. Statistical significance of differences was determined by Student’s T-test. Values with P < 0.05 are indicated by asterisks and values with P < 0.01 have two asterisks. Error bars represented Standard error of mean (SE) between three independent experiments. Data on expression of two glycosyltransferases (Ext 1 and Ext2) that did not fit the scale of the graph in Fig. 4A were included in Fig. 3B.
The signaling function of proteoglycans can be modified by changes in the sulfation pattern of their glycosaminoglycan side-chains. Therefore, we assessed whether Runx2 regulates enzymes that can modify the sulfation status of sugar-moieties. The expression of the heparin-related enzymes heparanase and heparan sulfate 2-O sulfotransferase (Hs2st) is increased, while expression of at least two sulfo-transferases (i.e., heparan glucosaminyl sulfotransferase 3 [Ndst3] and heparan sulfate 6-O sulfotransferase [Hs6st]) is repressed by Runx2 (Fig. 4A). Consequently, Runx2 alters the mRNA expression ratios of N-linked sulfotransferases (Ndst1 and Ndst2 versus Ndst3) and enzymes mediating O-linked sulfation of heparan sulfate (Hs2st versus Hs6st) or chondroitin sulfate (Cs4st versus Cs6st) (Fig. 4B). A number of other genes encoding proteoglycans and enzymes related to heparan sulfate synthesis and metabolism did not reveal statistically significant differences in expression upon Runx2 introduction in osteoprogenitor cells (Figs. 3 and 4). We conclude that Runx2 coordinately and selectively regulates the levels of several distinct proteoglycans, FGF receptors and enzymes that modify sugar moieties.
Runx2 opposes FGF2 dependent down-regulation of proteoglycan related genes
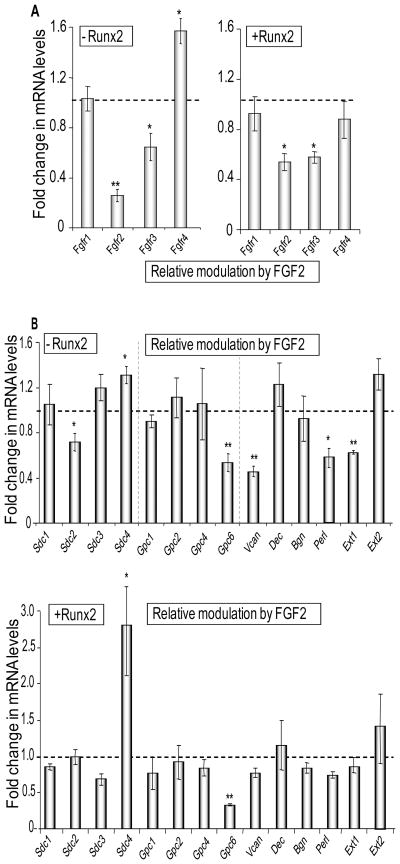
The observation that Runx2 induces multiple components of the FGF receptor-proteoglycan axis and other genes related to plasma membrane signal transduction suggests that Runx2 may regulate the sensitivity of osteoblast precursors to FGF signaling. Runx2 activity itself is controlled by FGF2 and we have previously shown that osteoblast-specific heparan sulfate upregulates Runx2 expression [Jackson et al., 2007]. Therefore, we examined how expression of proteoglycan related genes in osteoprogenitors responds to FGF2 signaling in the presence or absence Runx2. The results show that FGF2 selectively suppresses expression of FGFR2 and FGFR3, independent of Runx2, and has no effect on FGFR1 expression (Fig. 5A). FGF2 slightly increases FGFR4 levels in the absence of Runx2. The selective down-regulation of FGFR2 and FGFR3 by FGF2 in Runx2 null cells is consistent with negative feed-back regulation of FGF signaling that occurs independently of Runx2.
Figure 5. FGF2 responsiveness of genes encoding FGF receptors and Proteoglycans in the absence or presence of Runx2.
Cells infected with an Adenoviral vector expressing Runx2 or the corresponding empty vector were treated 24 h after infection with 10 ng/ml of bovine FGF2 in DMSO or a corresponding amount of DMSO control for the next 24 h. Relative mRNA expression levels of genes encoding FGF receptors (A) and Proteoglycans (B) were detected by qPCR analysis. The mRNA levels were normalized to 18S ribosomal RNA and plotted as fold change after FGF2 treatment versus DMSO control in the absence (left panel in A, upper panel in B) or in the presence (right panel in A and lower panel in B) of Runx2 expression. Statistical significance of differences was determined by Student’s T-test and values with P < 0.05 are indicated by asterisks, and values with P < 0.01 have two asterisks. Error bars represented Standard error of mean (SE) between three independent experiments.
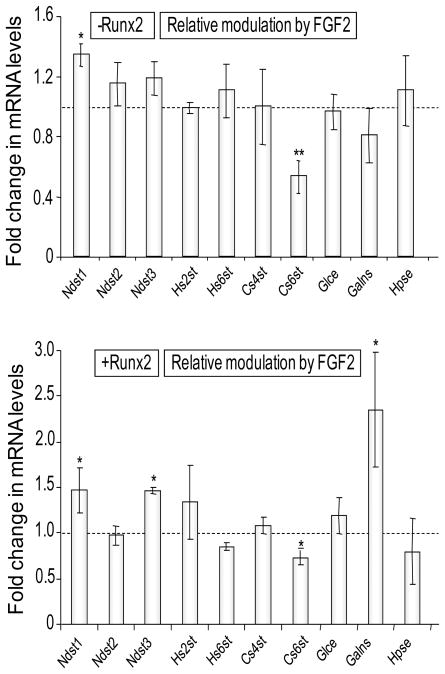
FGF2 signaling also reduces Syndecan-2, Glypican-6, Versican, Exostosin-1 and Perlecan in Runx2 null cells (Fig. 5B). However, the presence of Runx2 prevents the FGF2 dependent suppression of these same genes with the exception of Glypican-6. Hence, Runx2 opposes the FGF2 dependent down-regulation of four out of nine highly expressed proteoglycans (see Fig. 1) that are produced by Runx2 null osteoprogenitors. Similarly, Runx2 attenuates the decrease of Cs6st upon FGF2 administration, and renders two genes (the sulfotransferase Ndst3 and the sulfatase Galns) modestly responsive to FGF2 signaling, but has no other effect on glycosaminoglycan modifying enzymes (Fig. 6). Apart from opposing effects between Runx2 and FGF2, the data also show that Runx2 and FGF2 cooperate to stimulate expression of Sdc4 and the sulfatase Galns (Figs. 5 and 6). Hence, our results suggest that Runx2 alters the sensitivity of osteoprogenitors to FGF signaling through selective modulations in the expression of proteoglycan related genes.
Figure 6. FGF2 responsiveness of genes encoding glycosaminoglycan modifying enzymes in the absence or presence of Runx2.
Similar to Figure 5, cells infected with an Adenoviral vector expressing Runx2 or the corresponding empty vector were treated 24 h after infection with 10 ng/ml of bovine FGF2 in DMSO or a corresponding amount of DMSO control for the next 24 h. Relative mRNA expression levels of genes encoding glycosaminoglycan modifying enzymes were detected by qPCR analysis. The mRNA levels were normalized to 18S ribosomal RNA and plotted as fold change after FGF2 treatment versus DMSO control in the absence (upper panel) or in the presence (lower panel) of Runx2 expression. Statistical significance of differences was determined by Student T-test and values with P < 0.05 are indicated by asterisks, and values with P < 0.01 have two asterisks. Error bars represented Standard error of mean (SE) between three independent experiments.
Runx2 potentiates the FGF2 dependent induction of osteopontin and FGF2 opposes Runx2 dependent upregulation of osteocalcin and alkaline phosphatase
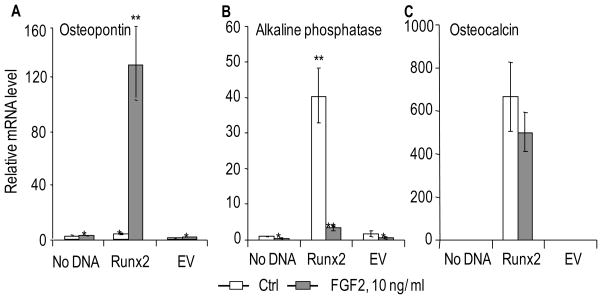
Because Runx2 modulates the expression of proteoglycan genes that are linked to FGF2 signaling, we investigated whether these modulations have functional consequences for responsiveness of immortalized Runx2 null osteoprogenitors to FGF2. We evaluated whether Runx2 and FGF2 cooperate in the expression of osteoblast phenotypic markers that reflect progression along the osteogenic lineage. Runx2 null calvarial osteoprogenitors were infected with adenoviral vectors expressing Runx2 followed by treatment with a low dose (10 ng/ml) of FGF2 under low serum conditions. We note that osteoprogenitor cells cultured in low fetal bovine serum conditions (0.1–% FBS) (see Fig. 7) exhibit more pronounced FGF2 effects than when treatment is performed in high (10%) FBS (data not shown). The RNA expression of osteogenic markers was evaluated after 24 h of FGF2 treatment by qPCR analysis (this treatment period spans approximately one cell cycle). Runx2 and FGF2 can each robustly stimulate expression of the early osteogenic marker osteopontin (i.e., Runx2 alone increases the expression of osteopontin by ~15 to 20 fold, while and treatment by FGF2 alone increases the expression 3 to 4 fold) (Fig. 7A and data not shown). The combined treatment by both Runx2 and FGF2 dramatically increases the expression of osteopontin (>100 fold) (Fig. 7A). Hence, Runx2 and FGF2 synergistically enhance osteopontin gene expression.
Figure 7. FGF2 signaling synergizes with Runx2 for the induction of Osteopontin, but antagonizes the induction of Alkaline Phosphatase in osteoprogenitor cells.
Similar to Figure 5, cells infected with an Adenoviral vector expressing Runx2 or the corresponding empty vector were treated 24 h after infection with 10 ng/ml of bovine FGF2 in DMSO or a corresponding amount of DMSO control for the next 24 h. Expression of the osteoblastic markers Osteopontin (A), Alkaline phosphatase (B) and Osteocalcin (C) was determined by qPCR analysis. The mRNA levels for the three genes were normalized using 18S ribosomal RNA and plotted relative to the value of Osteocalcin mRNA in the control sample (no infection, DMSO treated) which was arbitrarily set as 1. Statistical significance of differences was determined by Student’s T-test and values with P < 0.05 are indicated by asterisks, and values with P < 0.01 have two asterisks. Error bars represented Standard error of mean (SE) between three independent experiments.
We evaluated the expression of other osteogenic markers including Alkaline Phosphatase and Osteocalcin, which are both expressed at very low levels in osteoprogenitors. Consistent with previous data using similar samples [Teplyuk et al., 2008; Bae et al., 2007], expression of both Alkaline Phosphatase and Osteocalcin is significantly induced by Runx2 (Figs. 7B and 7C). The expression of bone sialoprotein (BSP) gene was not detectable in our experiments with proliferating osteoprogenitor cells (data not shown). Strikingly, FGF2 treatment almost completely abrogates the Runx2 mediated induction of Alkaline Phosphatase gene expression (Fig. 7B), but only marginally decreases the induction of osteocalcin (Fig. 7C). The synergistic (Fig. 7A) and antagonistic (Fig. 7B) actions of FGF2 and Runx2 clearly indicate cross-talk between Runx2 and FGF signaling (Fig. 8).
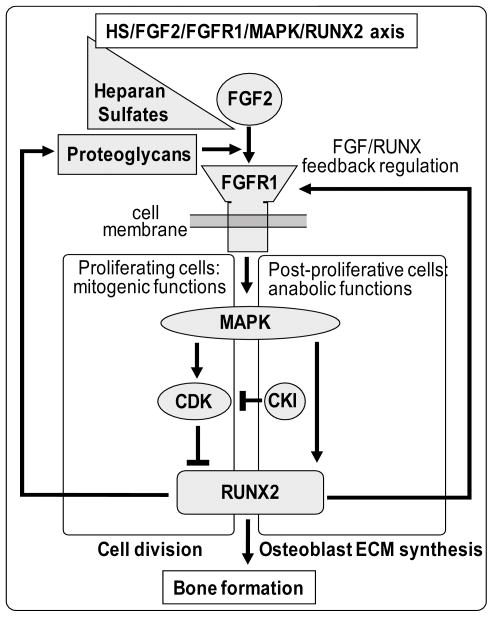
Figure 8. Cross-talk between the Runx2 and FGF2 signaling axes during osteoblastic lineage progression.
The model depicts several well-known aspects of the FGF signaling cascade including the synergy between FGF signaling and proteoglycans, as well as downstream effects on FGF signaling on MAPK and CDK related pathways. We propose that Runx2 may participate in two distinct feed-back loops. In actively dividing cells, FGF2 is mitogenic and activates MAPKs and CDKs. This activation may both promote and attenuate Runx2 activity to generate short-term changes in proteoglycan expression that transiently modulate responsiveness to FGF2. In post-proliferative cells, FGF2 functions anabolically and CDK effects on Runx2 are blocked by CDK inhibitors (CKIs). Consequently, the FGF/MAPK/Runx2 pathway may generate a long-term sustained response in which Runx2 modulates a program of proteoglycan expression to promote osteoblast maturation. The ideas presented in the model are consistent with references presented in the main text, but remain to be experimentally tested.
DISCUSSION
In this study, we have characterized novel Runx2 target genes in Runx2 null osteoprogenitors under conditions that promote growth arrest while not yet permitting sustained phenotypic maturation. Runx2 null cells in which Runx2 is re-introduced will only differentiate when cultured in the presence of osteogenic media and/or BMP2 [Bae et al., 2007]. Our studies have been aimed at identifying sets of co-regulated genes and pathways, including genes that are both directly and indirectly controlled by Runx2, during Runx2 induced inhibition of osteoblast proliferation [Teplyuk et al., 2008]. We focused here on proteoglycan genes, because proteoglycans are key regulators of cell signaling pathways that promote osteoblast proliferation and differentiation [Jackson et al., 2006b; De and David, 2001; Ling et al., 2009]. For example, proteoglycans support FGF2 signaling, and FGF2 signaling modulates the activity of Runx2 and osteoblast-specific gene expression [Kim et al., 2003; Kim et al., 2006; Shimizu-Sasaki et al., 2001; Zhang et al., 2002; Xiao et al., 2002; Hatch et al., 2008; Okazaki et al., 1992].
We first validated initial Affymetrix gene expression profiling studies indicating that Runx2 selectively regulates expression of proteoglycan genes. Our data reveal that Adenoviral expression of Runx2 in Runx2 null cells prominently increases mRNA levels of FGFR2 and FGFR3, Syndecan-1, -2 and -3, as well as Glypican-1, Versican and Exostosin-1. While FGFR1 is most prominently expressed in immature proliferating osteoblastic precursors, expression of both FGFR2 and FGFR3 expression is enhanced during osteoblast maturation [Jackson et al., 2006b; Marie, 2003; Jackson et al., 2006a]. Thus, Runx2 modulates the repertoire of FGF receptors, as well as proteoglycans that support FGF signaling. We also find that Runx2 controls glycosaminoglycan modifying enzymes. Glycosaminoglycans are highly complex sugar moieties that are covalently modified by sulfation. The distribution and concentration of sulfation on sugar chains will modulate their ability to bind selective ligands (e.g., FGF2, BMPs or WNTs). Runx2 alters the expression ratio of Hs2st versus Hs6st, as well as Cs4st versus Cs6st, suggesting that Runx2 may indirectly alter O-linked sulfation patterns of heparan sulfates or chondroitin sulfates. Because the oxygen at position 2 (2-O) of iduronate in heparan sulfate contributes to FGF2 binding, the Runx2 dependent increase in the ratio of 2-O/6-0 sulfotransferases may modulate the affinity of FGF2 for specific proteoglycans.
We directly tested whether Runx2 alters the FGF2 responsiveness of immortalized Runx2 null osteoprogenitors. The experiments revealed that Runx2 and FGF2 synergistically enhance expression of Osteopontin, as well as Syndecan-4 and the heparanase Galns. In addition, Runx2 prevents or reduces FGF2 dependent downregulation of Syndecan-2, Exostosin-1, Versican and Perlecan. Thus, our results together are consistent with the concept that Runx2 promotes modifications in the composition of the extracellular matrix to modify responsiveness of osteoprogenitors to FGF signaling.
Runx2 activity and/or expression is controlled by many osteogenic and mitogenic signaling pathways. For example, BMP2 promotes the osteogenic program by inducing Smad1 and Smad5 that together with Smad4 can interact with Runx2 to control BMP2 dependent targets [Javed et al., 2008]. However, the Runx2 co-factors Smad4 and Smad5 themselves are also Runx2 targets [Young et al., 2007a; Galindo et al., 2006], thus generating a BMP2-Smad-Runx2 feed-forward loop. Similarly, Runx2 regulates the RNA helicase Ddx5, and Ddx5 interacts with Runx2 to control osteoblast differentiation [Jensen et al., 2008]. Runx2 control is regulated by cAMP signaling, hence Runx2 enhancement of G protein coupled receptor signaling through Gsα proteins is expected to generate a self-sustained molecular circuit [Teplyuk et al., 2008]. Our current results indicate that Runx2 coordinately controls proteoglycan components of the extracellular matrix and selectively synergizes with FGF2 to enhance Syndecan-4 and osteopontin gene expression. We conclude that many target genes controlled by Runx2 represent components of reinforcing molecular pathways that mediate cell growth and differentiation of osteoblasts.
Acknowledgments
We thank Margaretha van der Deen, Anurag Gupta, David Leong and Victor Nurcombe for stimulating discussions, for biological samples and/or technical advice. We also thank Judy Rask for assistance with preparation of the manuscript. We thank Jim Neil (University of Glasgow, Scotland), as well as Yoshiaki Ito and Dominique Voon (Cancer Research Centre of Excellence, National University of Singapore) for sharing unpublished data on Runx target genes.
Contract Grant Sponsor: NIH grants R01 AR49069 and R01 AR039588. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- FGF-2
Fibroblast Growth Factor
- bFGF
basic FGF
- FGFR
FGF receptor
- Runx2
Runt-related transcription factor
- Cbfa1
Core Binding Factor Alpha 1
- BMP
bone morphogenetic protein
- Wnt
Wingless/Integration protein
- Sdc
syndecan
- Gpc
glypican
- Vcan
versican/chondroitin sulfate proteoglycan 2
- Hspg2
heparin sulfate proteoglycan 2/perlecan
- Ext1
Exostosin-1
- Ndst1
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
- Hs2st1
heparan sulfate 2-O-sulfotransferase 1
- Hs6st1
heparan sulfate 6-O-sulfotransferase 1
- Chst11/Cs4st
carbohydrate (chondroitin 4) sulfotransferase 11 / Chondroitin 4-O-sulfotransferase 1
- Chst3/Cs6st
carbohydrate (chondroitin 6) sulfotransferase 3 / Chondroitin 6-O-sulfotransferase 1
- Galns
galactosamine (N-acetyl)-6-sulfate sulfatase
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- RT-PCR
reverse transcriptase polymerase chain reaction
Footnotes
DISCLOSURE STATEMENT SUMMARY: The authors have nothing to disclose.
References
- Bae JS, Gutierrez S, Narla R, Pratap J, Devados R, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J Cell Biochem. 2007;100:434–449. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]
- Bodo M, Baroni T, Carinci F, Becchetti E, Bellucci C, Conte C, Pezzetti F, Evangelisti R, Tognon M, Carinci P. A regulatory role of fibroblast growth factor in the expression of decorin, biglycan, betaglycan and syndecan in osteoblasts from patients with Crouzon’s syndrome. Eur J Cell Biol. 1999;78:323–330. doi: 10.1016/S0171-9335(99)80066-1. [DOI] [PubMed] [Google Scholar]
- Bonaventure J, El GV. Molecular and cellular bases of syndromic craniosynostoses. Expert Rev Mol Med. 2003;5:1–17. doi: 10.1017/S1462399403005751. [DOI] [PubMed] [Google Scholar]
- Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271:7508–7515. doi: 10.1074/jbc.271.13.7508. [DOI] [PubMed] [Google Scholar]
- Coffin JD, Florkiewicz RZ, Neumann J, Mort-Hopkins T, Dorn GW, Lightfoot P, German R, Howles PN, Kier A, O’Toole BA. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6:1861–1873. doi: 10.1091/mbc.6.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ML, Seto ML, Ratisoontorn C, Heike CL, Hing AV. Syndromic craniosynostosis: from history to hydrogen bonds. Orthod Craniofac Res. 2007;10:67–81. doi: 10.1111/j.1601-6343.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- De CB, David G. Developmental roles of the glypicans. Semin Cell Dev Biol. 2001;12:117–125. doi: 10.1006/scdb.2000.0240. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Galindo M, Kahler RA, Teplyuk NM, Stein JL, Lian JB, Stein GS, Westendorf JJ, van Wijnen AJ. Cell cycle related modulations in Runx2 protein levels are independent of lymphocyte enhancer-binding factor 1 (Lef1) in proliferating osteoblasts. J Mol Histol. 2007;38:501–506. doi: 10.1007/s10735-007-9143-0. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M, Teplyuk N, Yang X, Young DW, Javed A, Lian JB, Stein JL, Stein GS, van Wijnen AJ. Post-mitotic expression of transcription factor RUNX2 defines a novel regulatory transition during the G1 phase of proliferating pre-osteoblasts. J Bone Miner Res. 2006;21:S384. (Abstract) [Google Scholar]
- Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development. Trends Cell Biol. 2007;17:173–177. doi: 10.1016/j.tcb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, Thomas DM, Hinds PW. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci U S A. 2008;105:18402–18407. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch NE, Li Y, Franceschi RT. FGF2 stimulation of the pyrophosphate generating enzyme, PC-1, in pre-osteoblast cells is mediated by RUNX2. J Bone Miner Res. 2008 doi: 10.1359/JBMR.081213. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Kumarasuriyar A, Nurcombe V, Cool SM. Long-term loading inhibits ERK1/2 phosphorylation and increases FGFR3 expression in MC3T3-E1 osteoblast cells. J Cell Physiol. 2006a;209:894–904. doi: 10.1002/jcp.20779. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Murali S, van Wijnen AJ, Stein GS, Nurcombe V, Cool SM. Heparan sulfate regulates the anabolic activity of MC3T3-E1 preosteoblast cells by induction of Runx2. J Cell Physiol. 2007;210:38–50. doi: 10.1002/jcp.20813. [DOI] [PubMed] [Google Scholar]
- Jackson RA, Nurcombe V, Cool SM. Coordinated fibroblast growth factor and heparan sulfate regulation of osteogenesis. Gene. 2006b;379:79–91. doi: 10.1016/j.gene.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ED, Niu L, Caretti G, Nicol SM, Teplyuk N, Stein GS, Sartorelli V, van Wijnen AJ, Fuller-Pace FV, Westendorf JJ. p68 (Ddx5) interacts with Runx2 and regulates osteoblast differentiation. J Cell Biochem. 2008;103:1438–1451. doi: 10.1002/jcb.21526. [DOI] [PubMed] [Google Scholar]
- Kim BG, Kim HJ, Park HJ, Kim YJ, Yoon WJ, Lee SJ, Ryoo HM, Cho JY. Runx2 phosphorylation induced by fibroblast growth factor-2/protein kinase C pathways. Proteomics. 2006;6:1166–1174. doi: 10.1002/pmic.200500289. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Bae SC, Choi JY, Kim HJ, Ryoo HM. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J Biol Chem. 2003;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome--the enigma of encoding and decoding heparan sulfate sulfation. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- Lee JY, Choo JE, Choi YS, Lee KY, Min DS, Pi SH, Seol YJ, Lee SJ, Jo IH, Chung CP, Park YJ. Characterization of the surface immobilized synthetic heparin binding domain derived from human fibroblast growth factor-2 and its effect on osteoblast differentiation. J Biomed Mater Res A. 2007;83:970–979. doi: 10.1002/jbm.a.31351. [DOI] [PubMed] [Google Scholar]
- Ling L, Murali S, Dombrowski C, Haupt LM, Stein GS, van Wijnen AJ, Nurcombe V, Cool SM. Sulfated glycosaminoglycans mediate the effects of FGF2 on the osteogenic potential of rat calvarial osteoprogenitor cells. J Cell Physiol. 2006;209:811–825. doi: 10.1002/jcp.20760. [DOI] [PubMed] [Google Scholar]
- Ling L, Nurcombe V, Cool SM. Wnt signaling controls the fate of mesenchymal stem cells. Gene. 2009 doi: 10.1016/j.gene.2008.12.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Marie PJ. Fibroblast growth factor signaling controlling osteoblast differentiation. Gene. 2003;316:23–32. doi: 10.1016/s0378-1119(03)00748-0. [DOI] [PubMed] [Google Scholar]
- Marie PJ, Coffin JD, Hurley MM. FGF and FGFR signaling in chondrodysplasias and craniosynostosis. J Cell Biochem. 2005;96:888–896. doi: 10.1002/jcb.20582. [DOI] [PubMed] [Google Scholar]
- Molteni A, Modrowski D, Hott M, Marie PJ. Differential expression of fibroblast growth factor receptor-1, -2, and -3 and syndecan-1, -2, and -4 in neonatal rat mandibular condyle and calvaria during osteogenic differentiation in vitro. Bone. 1999;24:337–347. doi: 10.1016/s8756-3282(98)00191-4. [DOI] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganawa T, Xiao L, Abogunde E, Sobue T, Kalajzic I, Sabbieti M, Agas D, Hurley MM. In vivo and in vitro comparison of the effects of FGF-2 null and haplo-insufficiency on bone formation in mice. Biochem Biophys Res Commun. 2006;339:490–498. doi: 10.1016/j.bbrc.2005.10.215. [DOI] [PubMed] [Google Scholar]
- Nathan SS, Pereira BP, Zhou YF, Gupta A, Dombrowski C, Soong R, Pho RW, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Elevated expression of Runx2 as a key parameter in the etiology of osteosarcoma. Mol Biol Rep. 2008;36:153–158. doi: 10.1007/s11033-008-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R, Ikeda K, Sakamoto A, Nakano T, Morimoto K, Kikuchi T, Urakawa K, Ogata E, Matsumoto T. Transcriptional activation of c-fos and c-jun protooncogenes by serum growth factors in osteoblast-like MC3T3-E1 cells. J Bone Miner Res. 1992;7:1149–1155. doi: 10.1002/jbmr.5650071006. [DOI] [PubMed] [Google Scholar]
- Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, Robinson JA, Choi J-Y, Komori T, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Rajgopal A, Young DW, Mujeeb KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J Cell Biochem. 2007;100:1509–1517. doi: 10.1002/jcb.21137. [DOI] [PubMed] [Google Scholar]
- Sabbieti MG, Agas D, Materazzi S, Capacchietti M, Materazzi G, Hurley MM, Menghi G, Marchetti L. Prostaglandin F2alpha involves heparan sulphate sugar chains and FGFRs to modulate osteoblast growth and differentiation. J Cell Physiol. 2008;217:48–59. doi: 10.1002/jcp.21471. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sasaki E, Yamazaki M, Furuyama S, Sugiya H, Sodek J, Ogata Y. Identification of a novel response element in the rat bone sialoprotein (BSP) gene promoter that mediates constitutive and fibroblast growth factor 2-induced expression of BSP. J Biol Chem. 2001;276:5459–5466. doi: 10.1074/jbc.M008971200. [DOI] [PubMed] [Google Scholar]
- Sobue T, Naganawa T, Xiao L, Okada Y, Tanaka Y, Ito M, Okimoto N, Nakamura T, Coffin JD, Hurley MM. Over-expression of fibroblast growth factor-2 causes defective bone mineralization and osteopenia in transgenic mice. J Cell Biochem. 2005;95:83–94. doi: 10.1002/jcb.20389. [DOI] [PubMed] [Google Scholar]
- Song SJ, Cool SM, Nurcombe V. Regulated expression of syndecan-4 in rat calvaria osteoblasts induced by fibroblast growth factor-2. J Cell Biochem. 2007;100:402–411. doi: 10.1002/jcb.21068. [DOI] [PubMed] [Google Scholar]
- Teplyuk NM, Galindo M, Teplyuk VI, Pratap J, Young DW, Lapointe D, Javed A, Stein JL, Lian JB, Stein GS, van Wijnen AJ. Runx2 regulates G-protein coupled signaling pathways to control growth of osteoblast progenitors. J Biol Chem. 2008;283:27585–27597. doi: 10.1074/jbc.M802453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, Diyagama D, Grim JE, Clurman BE, Bowtell DD, Lee JS, Gutierrez GM, Piscopo DM, Carty SA, Hinds PW. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, Stein GS, Gergen JP, Groner Y, Hiebert SW, Ito Y, Liu P, Neil JC, Ohki M, Speck N. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23:4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- Woei NK, Speicher T, Dombrowski C, Helledie T, Haupt LM, Nurcombe V, Cool SM. Osteogenic differentiation of murine embryonic stem cells is mediated by fibroblast growth factor receptors. Stem Cells Dev. 2007;16:305–318. doi: 10.1089/scd.2006.0044. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277:36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SH, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc Natl Acad Sci USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sobue T, Hurley MM. FGF-2 increases colony formation, PTH receptor, and IGF-1 mRNA in mouse marrow stromal cells. Biochem Biophys Res Commun. 2002;290:526–531. doi: 10.1006/bbrc.2001.6217. [DOI] [PubMed] [Google Scholar]