Abstract
Reactivation of the fetal cardiac gene program is a characteristic feature of hypertrophied and failing hearts that correlates with impaired cardiac function and poor prognosis. However, the mechanism governing the reversible expression of fetal cardiac genes remains unresolved. Here we show that neuron-restrictive silencer factor (NRSF), a transcriptional repressor, selectively regulates expression of multiple fetal cardiac genes, including those for atrial natriuretic peptide, brain natriuretic peptide and α-skeletal actin, and plays a role in molecular pathways leading to the re-expression of those genes in ventricular myocytes. Moreover, transgenic mice expressing a dominant-negative mutant of NRSF in their hearts exhibit dilated cardiomyopathy, high susceptibility to arrhythmias and sudden death. We demonstrate that genes encoding two ion channels that carry the fetal cardiac currents If and ICa,T, which are induced in these mice and are potentially responsible for both the cardiac dysfunction and the arrhythmogenesis, are regulated by NRSF. Our results indicate NRSF to be a key transcriptional regulator of the fetal cardiac gene program and suggest an important role for NRSF in maintaining normal cardiac structure and function.
Keywords: arrhythmia/cardiomyopathy/fetal cardiac genes/transcription
Introduction
Cardiac hypertrophy is initially an adaptive response of the heart to mechanical stress, tissue injury or neurohumoral activation; however, sustained hypertrophy can lead to dilated cardiomyopathy and heart failure. Up to 50% of the deaths among heart failure patients are sudden and unexpected, and are presumably the result of lethal arrhythmias (Tomaselli and Marbán, 1999). The dysregulation of a panel of cardiac genes accounts for the biochemical, structural, functional and electrical alterations in failing hearts, but relatively little is known about the molecular pathways underlying those complex remodeling processes. One of the characteristic genetic alterations in hypertrophied and failing hearts is reactivation of a fetal cardiac gene program, i.e. upregulation of genes encoding atrial and brain natriuretic peptides (ANP and BNP, respectively), as well as fetal contractile protein isoforms such as β-myosin heavy chain (MHC) and α-skeletal actin (Chien et al., 1991). Indeed, production of ANP and BNP in cardiomyocytes is markedly augmented in hypertrophied and failing hearts and is a prognostic indicator of clinical severity (Kjær and Hesse, 2001). Accordingly, elucidation of the regulatory mechanisms governing expression of these fetal cardiac genes should enable one to understand better the processes by which cardiac hypertrophy and heart failure are established.
Altered expression of certain ion channels in diseased hearts is also indicative of the reactivation of a fetal gene program. Two ionic currents, the hyperpolarization activated non-selective cation current (If) and the T-type Ca2+ current (ICa,T), which are normally expressed in fetal ventricles but repressed in adult ventricles, are re-expressed in ventricular myocytes of hypertrophied or failing hearts, perhaps increasing the vulnerability of the hearts to ventricular arrhythmias (Nuss and Houser, 1993; Sen and Smith, 1994; Cerbai et al., 1996, 2001; Martinez et al., 1999). Moreover, increased expression of ICa,T might contribute to the progression of heart failure (Clozel et al., 1999). Thus, elucidation of the mechanisms that control the fetal cardiac gene program may also shed light on the molecular basis for arrhythmias and sudden death associated with heart failure.
Despite considerable effort, virtually the entire process by which reversible expression of fetal cardiac genes is governed remains unknown. It was shown recently that (i) a repressor element named neuron-restrictive silencer element (NRSE), also known as repressor element-1 (RE-1) (Kraner et al., 1992; Mori et al., 1992), is present in the 3′-untranslated region (UTR) of ANP; (ii) NRSE represses basal expression of ANP in ventricular myocytes by recruiting the transcriptional repressor neuron-restrictive silencer factor (NRSF), also known as RE-1 silencing transcription factor (REST) (Schoenherr and Anderson, 1995; Chong et al., 1995), which forms a complex with histone deacetylases (HDACs); and (iii) attenuation of NRSE-mediated repression is an important component of the signaling pathways by which endothelin (ET)-1 induces ANP promoter activity (Kuwahara et al., 2001). We also showed that NRSE in the 5′-flanking region of BNP represses transcription of BNP and that attenuation of NRSE-mediated repression contributes to the increase in BNP transcription induced by hypertrophic stimuli (Ogawa et al., 2002). Notably, NRSE is also reportedly present in the α-skeletal actin gene (Schoenherr et al., 1996).
The aforementioned findings suggest that the NRSE–NRSF system regulates the expression of multiple fetal cardiac genes. However, the role of the NRSE–NRSF system in the expression of endogenous fetal cardiac genes and in the regulation of cardiac function remains unknown. To address these questions, in this study, we used a recombinant adenovirus expressing a dominant-negative mutant of NRSF (dnNRSF) and transgenic mice expressing dnNRSF in their hearts. We show that the inhibition of NRSF induces endogenous expression of multiple fetal cardiac genes in ventricular myocytes and markedly reduces the response of fetal cardiac genes to hypertrophic stimuli both in vitro and in vivo, suggesting that NRSF-mediated repression contributes to the dynamic regulation of expression of fetal cardiac genes, and that persistent inhibition of cardiac NRSF repressor function leads to dilated cardiomyopathy and sudden death. We also show that the genes encoding the ion channels that carry If and ICa,T are upregulated in the ventricles of the transgenic mice, and that they are downstream targets of NRSF. Taken together, our findings indicate that NRSF is a novel and important regulator of the fetal cardiac gene program responsible for maintaining normal cardiac structure and function, and that NRSF plays a key role in mediating signaling pathways that lead to heart failure and sudden cardiac death.
Results
NRSF regulates endogenous cardiac fetal gene expression in vitro and in vivo
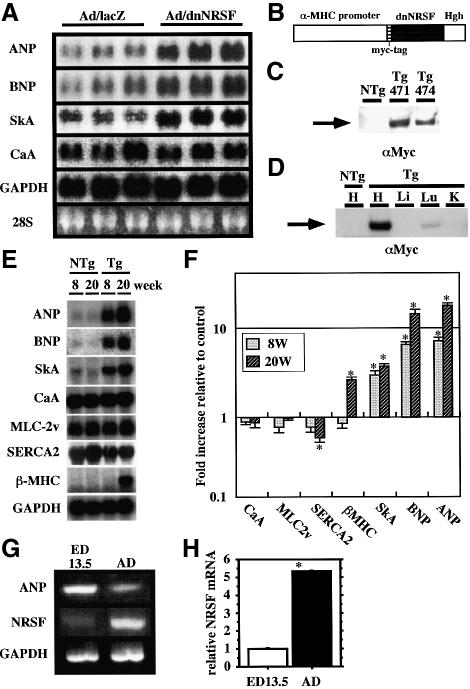
We previously used promoter–reporter constructs to show that the NRSF–NRSE system represses transcription of ANP and BNP in cultured ventricular myocytes. Moreover, the fact that NRSE is also present in the α-skeletal actin gene suggests that NRSF represses expression of multiple fetal cardiac genes (Schoenherr et al., 1996; Kuwahara et al., 2001; Ogawa et al., 2002). To confirm the function of NRSF in the regulation of endogenous fetal cardiac gene expression, we infected cultured ventricular myocytes with a recombinant adenovirus expressing dnNRSF (Ad/dnNRSF) and examined the resultant gene expression. dnNRSF contains a DNA-binding domain but lacks two identified repressor domains, and thus inhibits NRSE-mediated repression (Chen et al., 1998). We previously confirmed that this dnNRSF construct removes the repression of NRSE-containing reporter constructs exclusively in cultured ventricular myocytes (Kuwahara et al., 2001; Ogawa et al., 2002). Endogenous expression of ANP, BNP and α-skeletal actin mRNA, but not α-cardiac actin or GAPDH mRNA was markedly increased in ventricular myocytes infected with Ad/dnNRSF, as compared with cells infected with control vector (Figure 1A).
Fig. 1. NRSF regulates expression of endogenous ANP, BNP and α-skeletal actin in ventricular myocytes. (A) Northern blots showing levels of ANP, BNP, α-skeletal actin (SkA), α-cardiac actin (CaA) and GAPDH mRNA in cultured ventricular myocytes infected with Ad/lacZ or Ad/dnNRSF for 24 h. (B) A cDNA construct for the generation of dnNRSF Tg mice; Hgh, the poly(A) sequence of human growth hormone gene. (C) Western blot analysis for dnNRSF expression in ventricles from NTg and two different founders lines of dnNRSF Tg mice (Tg471 and 474). (D) Western blot analysis for dnNRSF expression in various organs from dnNRSF Tg mice: H, heart; Lu, lung; L, liver; K, kidney. (E) Representative northern blots showing levels of the indicated mRNAs in dnNRSF Tg and NTg hearts. (F) Bar graph summarizing the relative cardiac levels of the indicated mRNAs (normalized to GAPDH mRNA levels) detected in the northern blots shown in (E); bars represent means ± SEM from three independent experiments; *P < 0.05 versus NTg hearts; n = 6 each. (G) RT–PCR analysis for NRSF and ANP mRNA expression in ventricles from 13.5-day mouse embryos (ED 13.5) and 20-week-old adult mice (AD). (H) Bar graphs showing relative NRSF mRNA levels (normalized to GAPDH mRNA levels) determined by quantitative RT–PCR. Bars represent means ± SEM from three independent assays; *P < 0.05 versus ED13.5.
Because mice lacking NRSF die in utero, it is impossible to use these animals to analyze NRSF function in the post-natal heart (Chen et al., 1998). However, expression of dnNRSF in chick embryo using a retroviral vector caused ectopic expression of a specific set of neuronal genes, as did targeted deletion of NRSF in mice (Chen et al., 1998). Therefore, to investigate the role of NRSF in the post-natal heart, we produced transgenic mice that express dnNRSF under the control of the cardiac-specific α-MHC promoter (Figure 1B). Two independently derived dnNRSF transgenic (Tg) founders (Tg471 and Tg474; dnNRSF mRNAs were expressed at levels 35.5- and 27.0-fold higher than endogenous NRSF mRNAs, respectively) were obtained and investigated (Figure 1C). The data obtained from Tg471, which are essentially identical to those obtained from Tg474, are presented below. Western blot analysis confirmed that expression of dnNRSF was restricted to the heart, though weak expression was observed in the lung, where endogenous α-MHC is expressed in cardiac myocytes surrounding pulmonary veins (Figure 1D) (Subramaniam et al., 1991).
To determine the role of NRSF in the regulation of fetal cardiac gene expression in vivo, we carried out northern blot analyses of the genes for ANP, BNP, α-skeletal actin, β-MHC, α-cardiac actin, myosin light chain (MLC)-2v, sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA) 2 and GAPDH using total RNA prepared from ventricles taken from 8- and 20-week-old mice (Figure 1E and F). The expression of the genes for ANP, BNP and α-skeletal actin, which contain an NRSE, was selectively upregulated in the hearts of dnNRSF Tg mice by 8 weeks of age. In 8-week-old dnNRSF Tg and non-transgenic (NTg) mice, the expression of another fetal gene, β-MHC, which does not contain an NRSE, was not enhanced, and the heart to body weight ratios and cardiac function estimated by echocardiography and hemodynamic analysis were not different. Apparently, induction of these fetal genes in 8-week-old dnNRSF Tg hearts is a primary effect of NRSF inhibition (Figures 1E and F, and 3B and C and Table I). Taken together, these results and the data obtained from in vitro studies using cultured ventricular myocytes, which are not affected by hemodynamic alterations, clearly indicate that NRSF regulates expression of multiple fetal cardiac genes in ventricular myocytes.
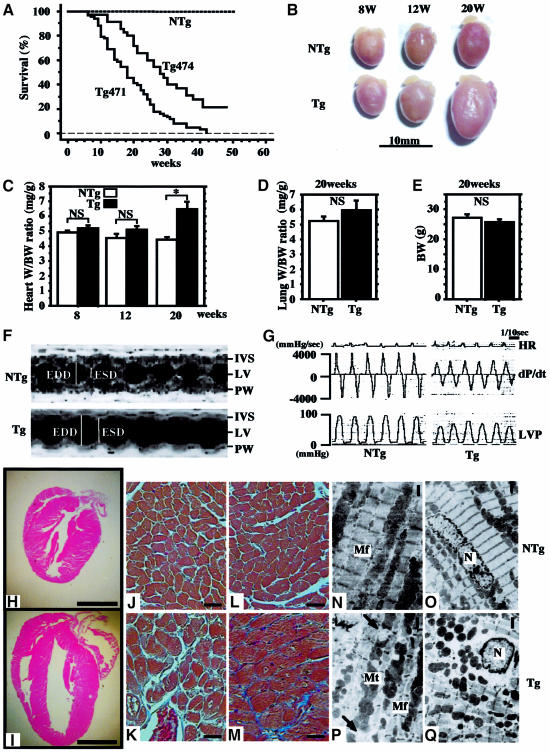
Fig. 3. Dilated cardiomyopathy in dnNRSF Tg mice. (A) Kaplan–Meier survival analysis of dnNRSF Tg471 (n = 68), Tg474 (n = 35) and NTg (n = 145) mice showing a significant difference in survival rates between dnNRSF Tg and NTg mice (log rank test; P < 0.0001). (B) Hearts of NTg and dnNRSF Tg mice at 8, 12 and 20 weeks; scale bars, 10 mm. (C) Heart to body weight ratios (mg/g), (D) lung to body weight ratios (mg/g) and (E) body weights (g) in NTg and dnNRSF Tg mice measured at the indicated times; *P < 0.05; NS, not significant. (F) Representative M-mode echocardiographic tracings from 20-week-old NTg and dnNRSF Tg mice: IVS, interventricular septum; LV; left ventricle; PW, posterior wall; ESD, end systolic dimension; EDD, end diastolic dimension. (G) Representative high fidelity left ventricular pressure tracings from 20-week-old NTg and dnNRSF Tg mice: HR, heart rate; LVP, LV pressure. (H–Q) Histological analysis of hearts from 20-week-old NTg and dnNRSF Tg mice. (H and I) Hearts from NTg (H) and dnNRSF Tg (I) mice were sectioned longitudinally and stained with HE; scale bars, 2.5 mm. (J–M) Photomicrographs of histological sections of left ventricle from NTg (J and L) and dnNRSF Tg mice (K and M) stained with Masson’s trichrome stain; collagen stains blue with trichrome stain (M); scale bars, 20 µm. (N–Q) Transmission electron micrographs of cardiac myocytes from 20-week-old NTg (N and O) and dnNRSF Tg (P and Q) mice: arrows indicate disrupted mitochondria; My, myofibrils; Mt, mitochondria; N, nucleus; scale bars, 1 µm.
Table I. Echocardiographic and hemodynamic analysis at 8 or 20 weeks of age.
| 8 weeks |
20 weeks |
|||
|---|---|---|---|---|
| NTg | Tg | NTg | Tg | |
| Echocardiographic data | n = 6 | n = 6 | n = 5 | n = 5 |
| LVDd (mm) | 4.02 ± 0.11 | 4.00 ± 0.29 | 4.04 ± 0.23 | 5.32 ± 0.22a |
| LVDs (mm) | 2.76 ± 0.14 | 2.87 ± 0.20 | 2.58 ± 0.15 | 4.66 ± 0.15a |
| IVST (mm) | 0.66 ± 0.038 | 0.65 ± 0.028 | 0.58 ± 0.08 | 0.60 ± 0.06 |
| PWT (MM) | 0.67 ± 0.042 | 0.69 ± 0.034 | 0.60 ± 0.07 | 0.54 ± 0.05 |
| FS (%) | 32.8 ± 1.88 | 27.9 ± 1.54 | 36.2 ± 2.08 | 12.0 ± 0.84a |
| EF (%) | 69.7 ± 2.56 | 62.67 ± 2.20 | 71.8 ± 2.75 | 30.4 ± 1.94 |
| Hemodynamic data | n = 4 | n = 5 | n = 6 | n = 5 |
| dP/dtmax (mmHg/s) | 4865 ± 201 | 4757 ± 325 | 4090 ± 188 | 2328 ± 264a |
| dP/dtmin (mmHg/s) | –4935 ± 218 | –4756 ± 237 | –4150 ± 193 | –2264 ± 292a |
| HR (/min) | 498 ± 26.9 | 533 ± 29.4 | 503 ± 42.8 | 487 ± 43.6 |
| LVSP (mmHg) | 98.9 ± 4.53 | 95.1 ± 3.6 | 88.4 ± 3.54 | 68.3 ± 5.34a |
| LVEDP (mmHg) | 2.25 ± 0.56 | 2.36 ± 0.69 | 3.83 ± 1.04 | 7.68 ± 2.30 |
Values are means ± SEM. LVDd, left ventricular end diastolic dimension; LVDs, left ventricular end systolic dimension; IVST, interventricular septal thickness; PWT, posterior wall thickness; FS, fractional shortening; EF, ejection fraction; dP/dt, first derivative of pressure; HR, heart rate; LVSP, left ventricular systolic pressure; LVEDP, left ventricular end diastolic pressure.
aP < 0.05 dnNRSF Tg versus NTg mice.
Several earlier studies reported that expression of NRSF mRNA is barely detectable in fetal hearts, though its expression is detected in neonatal ventricular myocytes and adult hearts (Schoenherr and Anderson, 1995; Chong et al., 1995; Palm et al., 1998; Grimes et al., 2000; Kuwahara et al., 2001). We therefore used RT–PCR to examine expression of NRSF and ANP in fetal ventricles obtained from 13.5-day mouse embryos and adult ventricles from 20-week-old mice. Consistent with those earlier reports, expression of NRSF mRNA was significantly lower in the embryonic than adult ventricles. Conversely, expression of ANP mRNA was significantly lower in adult ventricles than embryonic ventricles (Figure 1G). These results, which were confirmed by quantitative real-time RT–PCR (Figure 1H), suggest that NRSF plays a role in the developmental regulation of fetal cardiac gene expression and further support the notion that NRSF represses expression of fetal cardiac genes.
NRSF plays an important role in the re-expression of fetal cardiac genes induced by hypertrophic stimuli
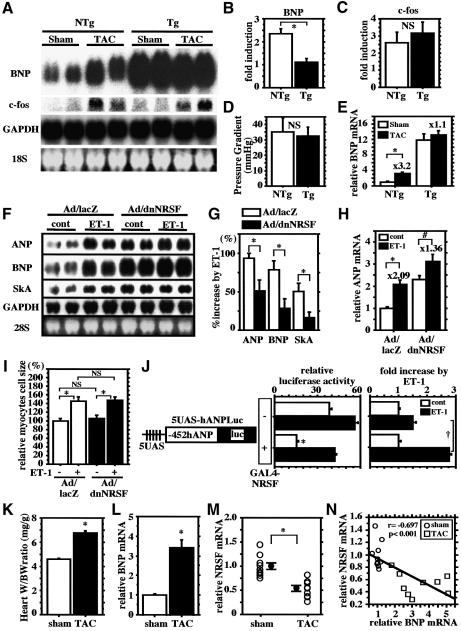
To examine further the role of NRSF in the re-expression of endogenous fetal cardiac genes in vivo, we subjected the hearts of dnNRSF Tg mice and their NTg littermates to acute pressure overload created by transverse aortic constriction (TAC). As previously reported (Harada et al., 1998), acute pressure overload induced expression of c-fos and BNP in NTg hearts within 30 min after the onset of TAC (Figure 2A–C). Northern blot analysis showed that basal levels of BNP expression were higher in 8-week-old dnNRSF Tg than NTg hearts, but that induction of BNP expression by TAC was less pronounced in dnNRSF Tg hearts (Figure 2A and B). In contrast, the inducibility of c-fos expression and peak-to-peak systolic pressure gradients across the stenosis were similar in dnNRSF Tg and NTg hearts (Figure 2A, C and D). These results were confirmed by quantitative RT–PCR analysis for BNP mRNA expression (Figure 2E) and indicate that BNP expression is constitutively elevated in hearts expressing dnNRSF, though its inducibility is diminished.
Fig. 2. NRSF regulates the inducible response of cardiac fetal gene expression to hypertrophic stimuli. (A) Northern blots showing levels of BNP, c-fos and GAPDH mRNA in sham- or TAC-operated NTg and dnNRSF Tg hearts. (B and C) Bar graphs summarizing the fold increases in BNP (B) and c-fos (C) mRNA detected in the northern blots shown in (A); n = 4 each. (D and E) Pressure gradients across the stenosis after TAC (D) and quantitative real-time RT–PCR analyses for BNP mRNA expression (E); n = 4 each. (F) Representative northern blots showing levels of ANP, BNP, SkA and GAPDH mRNA in cultured ventricular myocytes infected with Ad/lacZ or Ad/dnNRSF and subsequently treated for 24 h with or without 10 nM ET-1. (G) Bar graphs summarizing the percentage increases in the levels of ANP, BNP and SkA mRNA elicited by 10 nM ET-1 detected in the northern blots shown in (F); *P < 0.05 versus Ad/lacZ; n = 5 each. (H) Bar graph summarizing the results of a quantitative real-time RT–PCR analysis of ANP mRNA expression; *P < 0.01, P < 0.05; n = 4 each. The numbers above each bar indicate the fold increase induced by ET-1. For (B), (C), (E), (G) and (H), relative mRNA levels (normalized to GAPDH mRNA levels) are presented as means ± SEM. (I) Surface areas of 40 randomly selected cells in each group normalized to control myocytes (=100%) (mean ± SEM); *P < 0.05; NS, not significant. (J) The promoter activities of 5UAS-hANPLuc in ventricular myocytes treated or not with 10 nM ET-1 for 48 h in the absence or presence of a GAL4-NRSF fusion protein. The activities of the construct transfected into myocytes treated with 10 nM ET-1 are expressed as the fold increase over the value for myocytes treated without ET-1 in the right panel. Bars represent means ± SEM of the relative luciferase activity from at least three separate assays carried out in triplicate; *P < 0.05 versus GAL4-NRSF(–) ET-1(–); †P < 0.05. (K) Heart weight to body weight ratios in sham- and TAC-operated mice. Bars represent means ± SEM. (L and M) Quantitative RT–PCR analysis of BNP (L) and NRSF (M) mRNA levels. Relative mRNA levels (normalized to GAPDH mRNA levels) are presented as means ± SEM; levels in sham-operated mice are assigned a value of 1.0; *P < 0.05 versus sham-operated mice. (N) Inverse correlation between expression of ventricular BNP and NRSF mRNAs.
Consistent with that finding, expression of dnNRSF markedly reduced the fold increases in endogenous ANP, BNP and α-skeletal actin expression induced by ET-1 in cultured ventricular myocytes (Figure 2F–H), whereas it had no significant effect on ET-1-induced increases in cell size (Figure 2I). In addition, when we incubated ventricular myocytes co-transfected with an ANP promoter–reporter construct containing GAL4-binding sites (5UAS-hANPLuc) and a plasmid encoding a GAL4 DNA-binding domain–NRSF fusion protein (GAL4-NRSF) with ET-1, we found that in the presence of GAL4-NRSF, the response of 5UAS-hANPLuc to ET-1 was significantly increased, though the basal control activity of 5UAS-hANPLuc was significantly reduced (Figure 2J). This means that NRSF-mediated repression is necessary for full induction of fetal cardiac gene expression during cardiac muscle cell hypertrophy, which suggests that NRSF contributes not only to the determination of basal expression of fetal cardiac genes but also to the increase in transcriptional efficiency of fetal cardiac genes induced by hypertrophic stimuli.
We next examined cardiac NRSF expression in a mouse model of chronic pressure overload-induced left ventricular hypertrophy created by TAC. In mice subjected to TAC for 2 weeks, heart to body weight ratios and ventricular BNP expression were significantly higher than in sham-operated mice (Figure 2K and L), whereas NRSF mRNA expression was significantly lower in TAC hearts than in control sham-operated hearts (Figure 2M), i.e. ventricular expression of NRSF mRNA showed a significant inverse correlation with ventricular expression of BNP mRNA (Figure 2N). Collectively, these lines of data indicate an involvement of NRSF in molecular pathways leading to the upregulation of fetal gene expression during cardiac hypertrophy and support the idea that release of NRSF-mediated repression contributes to the reinduction of fetal cardiac genes.
Inhibition of NRSF in ventricular myocytes leads to increased mortality and dilated cardiomyopathy in vivo
Persistent inhibition of NRSF had a marked effect on cardiac structure and function. dnNRSF Tg mice started to die by 8 weeks of age, and by 40 weeks 70–95% had died. In contrast, 100% of their NTg littermates remained alive (Figure 3A). Gross examination revealed the hearts of 20-week-old dnNRSF Tg mice to be much larger than those of NTg mice, despite the fact that at 8 weeks of age there is no significant difference in heart size (Figure 3B). The heart to body weight ratios in 8- and 12-week-old dnNRSF Tg and NTg mice were also not significantly different, but by 20 weeks the ratio in dnNRSF Tg mice was 1.48-fold higher than in NTg mice (Figure 3C). Consistent with these data, at 20 weeks of age, β-MHC expression was upregulated and SERCA2 expression was downregulated in dnNRSF Tg hearts as compared with NTg hearts, whereas their expression was unchanged in both mouse types at 8 weeks of age (Figure 1E and F). The manner in which dnNRSF Tg mice died was characterized as sudden death; they were unexpectedly found dead within 24 h of displaying apparently normal behavior and levels of activity. Consistent with this diagnosis, body weights and lung to body weight ratios were not different between NTg and dnNRSF Tg mice, even at 20 weeks of age (Figure 3D and E), and there were no apparent signs of lung or liver congestion (data not shown).
To understand better the changes in the hearts of dnNRSF Tg mice, we carried out a series of detailed echocardiographic, hemodynamic and histological analyses. Two-dimensionally targeted M-mode echocardiography, performed at 8 and 20 weeks, revealed a progressive decline in fractional shortening (FS) and ejection fraction (EF) in 20-week-old dnNRSF Tg mice (Table I and Figure 3F), though FS and EF did not yet differ significantly in 8-week-old mice (Table I). Left ventricular end diastolic dimension (LVDd) and end systolic dimension (LVDs) were also not altered at 8 weeks of age, but by 20 weeks LVDd and LVDs were 30 and 80%, respectively, higher in dnNRSF Tg than NTg mice (Table I). Wall thickness was not significantly affected, indicating progressive LV dysfunction without LV wall hypertrophy in dnNRSF Tg mice.
The results of a hemodynamic analysis using a high-fidelity micromanometer catheter were consistent with the echocardiographic findings and indicated that inhibition of cardiac NRSF is sufficient to cause progressive LV dysfunction (Figure 3G). At 20 weeks of age, dnNRSF Tg mice showed diminished systolic ventricular pressure and significant depression of both the maximal first derivative of the LV pressure (dP/dtmax) and the maximal negative derivative of LV pressure (dP/dtmin), conditions that were not seen at 8 weeks of age (Table I). Histological examination of cross-sections confirmed LV enlargement without significant increases in wall thickness in 20-week-old dnNRSF Tg hearts (Figure 3H and I). In addition, a marked heterogeneity in the size of ventricular myocytes was noted, with some appearing hypertrophied and others appearing atrophied (Figure 3J–M). No significant inflammatory cell infiltrates were observed in dnNRSF Tg hearts, though Masson’s trichrome staining showed the presence of interstitial fibrosis (Figures 3M). Electron microscopic examination of the ultrastructure of ventricular myocytes revealed that, in 20-week-old dnNRSF Tg mice, myofibrils were sparse and misarranged, Z-bands were discontinuous, and mitochondria were deformed or completely disrupted (compare Figure 3N and O with P and Q; arrows indicate disrupted mitochondria). All of these features of dnNRSF Tg mice are characteristic of dilated cardiomyopathy in humans, suggesting that NRSF plays a critical role in the maintenance of normal cardiac structure and function.
Sudden death and ventricular arrhythmias in dnNRSF transgenic mice
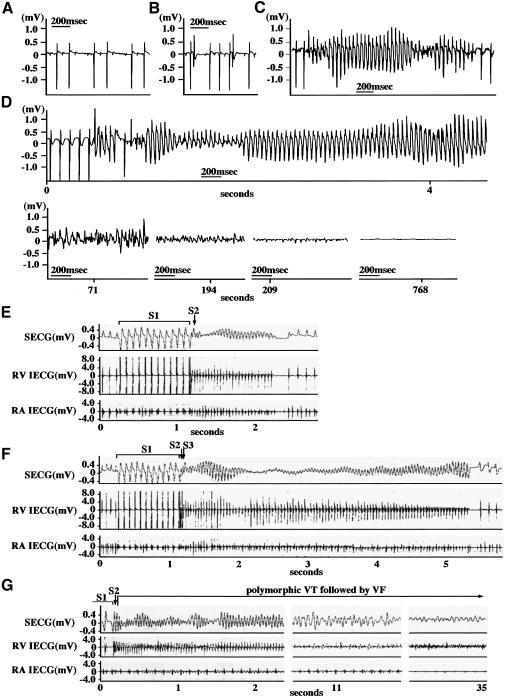
Given that dnNRSF Tg mice appeared to die of sudden death, we hypothesized that life-threatening arrhythmias must have occurred. To test this hypothesis, we continuously monitored electrocardiographic data from the mice using implantable radio telemetry. We observed no significant difference in heart rate, QRS time or QT interval between NTg and dnNRSF Tg mice (data not shown). On the other hand, the basal electrocardiograph (ECG) patterns showed the PQ interval to be longer in dnNRSF Tg than NTg mice (dnNRSF Tg, 56.0 ± 3.0 versus NTg, 35.5 ± 0.3 ms; n = 4 each, P < 0.001). More ominously, second degree AV block (Figure 4A) and ventricular ectopies, including runs of ventricular tachycardia (VT) (Figure 4B and C), were observed in dnNRSF Tg mice. No such rhythm disturbances were observed in NTg mice. During the monitoring period, three dnNRSF Tg mice died suddenly. In each case, VT and ventricular fibrillation (VF) followed by asystole were recorded at the time of death (Figure 4D). Thus, dnNRSF Tg mice exhibit a variety of arrhythmias that are often observed in the human cardiomyopathic heart, culminating in sudden arrhythmic death.
Fig. 4. Spontaneous and inducible arrhythmias in dnNRSF Tg mice. (A–D) ECGs from dnNRSF Tg mice obtained with implantable radio telemetry: second degree AV block (A); isolated ventricular ectopies (B); non-sustained ventricular tachycardia (VT) (C). Sustained VT and ventricular fibrillation (VF) followed by asystole recorded at the time of sudden death (D). (E–G) ECG obtained from dnNRSF Tg mice during electrophysiological studies: polymorphic VT (E and F); sustained polymorphic VT followed by VF (G).
To evaluate further the vulnerability of dnNRSF Tg hearts to ventricular arrhythmias, we performed an intracardiac electrophysiological analysis using an octapolar EP catheter (Gehrmann and Berul, 1999). To avoid the secondary effects of structural remodeling on the electrophysiological properties, we used 12-week-old dnNRSF Tg and NTg mice whose heart to body weight ratios (Figure 3C) and right ventricular effective refractory periods (dnNRSF Tg, 22.9 ± 1.3 ms versus NTg, 26.0 ± 1.8 ms; n = 6 each) did not differ significantly. When programmed electrical stimulation was applied, VT was induced in six out of 10 dnNRSF Tg mice versus none out of 10 NTg mice (Figures 4E–G). Clearly, dnNRSF Tg hearts were more vulnerable to ventricular arrhythmias than NTg hearts.
Increased expression of fetal-type ion channel genes in dnNRSF transgenic ventricle
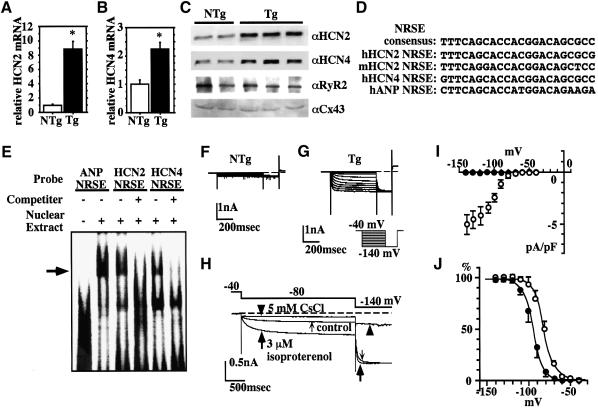
The fact that dnNRSF Tg mice exhibit increased vulnerability to arrhythmias and mortality even before development of severe structural abnormalities and cardiac dysfunction suggests that primary alterations in ion channel gene expression are involved. In addition, altered ion channel expression may also participate in the progression of cardiomyopathy in dnNRSF Tg mice. We therefore used murine genome U74A arrays (Affymetrix) to quantify simultaneously expression of ∼6000 genes and ∼6000 expressed sequence tags (ESTs) in 6-week-old NTg and dnNRSF Tg ventricles, and found that expression of HCN2/HAC1 is significantly increased in 6-week-old dnNRSF Tg hearts. HCN2 encodes a channel that carries a hyperpolarization-activated current (If) observed in fetal but not normal mature ventricular myocytes, but is reinduced in ventricular myocytes of hypertrophied or failing hearts and may contribute to the increased arrhythmogenicity (Cerbai et al., 1996, 2001). We therefore used quantitative RT–PCR to examine the expression of HCN2 and HCN4 mRNA, which are the predominant isoforms expressed in the ventricle (Santoro and Tibbs, 2000). We found that expression of both is significantly upregulated in the hearts of 8-week-old dnNRSF Tg mice (Figure 5A and B). Moreover, western blot analysis confirmed that expression of HCN2 and HCN4 proteins was also upregulated in dnNRSF Tg mice, though ryanodine receptor 2 and connexin 43 were not (Figure 5C). More importantly, sequences highly similar to the 22 bp consensus NRSE (>95% identity) were detected within the first intron of both HCN2 and HCN4 (Figure 5D), and electrophoretic mobility shift assays (EMSAs) clearly showed that NRSF binds to NRSE in both genes (Figure 5E). As our previous experiments showed that NRSE can repress multiple promoters in ventricular myocytes (Kuwahara et al., 2001), and others have reported that NRSF represses transcription irrespective of the location or orientation of its binding site within a gene (Thiel et al., 1998), our present findings indicate both HCN2 and HCN4 to be downstream targets of NRSF-regulated pathways.
Fig. 5. Increased expression of If in dnNRSF Tg hearts. (A and B) Relative levels of HCN2 (A) and HCN4 (B) mRNA in the hearts of 8-week-old NTg and dnNRSF Tg mice; *P < 0.05 versus NTg. (C) Western blot analysis of HCN2, HCN4, ryanodine receptor (RYR) 2 and connexin (Cx) 43 in the hearts of 8-week-old NTg and dnNRSF Tg mice. (D) Sequences similar to NRSE in mouse (m) and human (h) HCN genes. (E) EMSA was carried out using radiolabeled oligonucleotides from NRSE in ANP, HCN2 or HCN4 as probes. Unlabeled oligonucleotide from NRSE ANP was used as a cold competitor. (F and G) Representative membrane currents elicited by hyperpolarization after complete inhibition of IK1 in isolated ventricular myocytes from NTg (F) and dnNRSF Tg (G) mice; the pulse protocol is shown in (G). (H) Pharmacological properties of If in ventricular myocytes from dnNRSF Tg mice. Isoproterenol (3 µM) increased the amplitude of If during the first conditioning pulse (large arrows), as compared with control (small arrows). The initial current jump at the onset of the second pulse (–140 mV) was concomitantly increased. If was completely inhibited by 5 mM Cs+ (arrow heads). (I) Current–voltage relationship of If. The amplitude of the Cs+-sensitive component was measured at the end of conditioning pulses in ventricular myocytes from NTg (filled circles; n = 8) and dnNRSF Tg (open circles; n = 6) mice. (J) If activation curves measured in the absence (filled circles; n = 6) or presence (open circles; n = 6) of 3 µM isoproterenol. In (I) and (J), symbols represent means ± SEM.
In order to obtain functional evidence of the re-expression of HCN2 and HCN4, we carried out patch–clamp studies using ventricular myocytes isolated from 8- to 14-week-old dnNRSF Tg mice. After the inward-rectifier K+ current (IK1) was completely inhibited with 0.5 mM Ba2+, no time-dependent current was elicited by hyperpolarization in NTg mice (Figure 5F). In all myocytes from dnNRSF Tg mice, in contrast, hyperpolarizations more negative than –70 mV activated time-dependent inward currents (Figure 5G). That 5 mM Cs+ completely blocked these currents confirmed them to be If (Figure 5H). The current–voltage relationship of If was obtained by subtracting the Cs+-sensitive component; no Cs+-sensitive current was recorded in myocytes from NTg mice (Figure 5I). As in pacemaker cells, If in ventricular myocytes from dnNRSF Tg mice were augmented by β-adrenergic stimulation (3 µM isoproterenol; Figure 5H) due to a rightward shift in the voltage-dependent activation curve (the potential of half-maximal activation was shifted from –93.3 ± 1.3 to –79.7 ± 1.0 mV; Figure 5J).
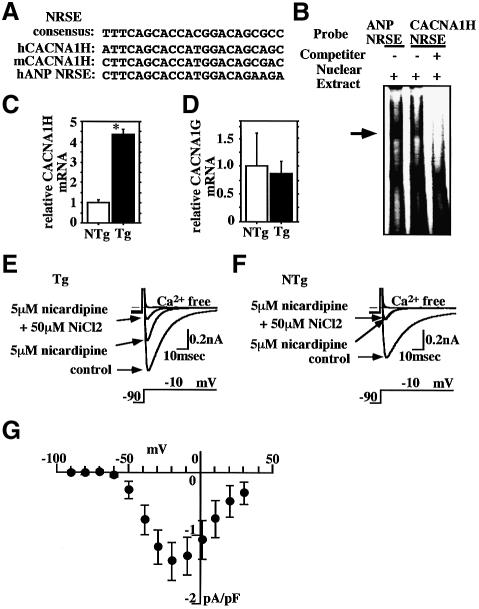
The T-type Ca2+ current (ICa,T) is also expressed in fetal ventricular myocytes, repressed in mature ventricular myocytes, and reactivated in ventricular myocytes in hypertrophied or failing hearts (Nuss and Houser, 1993; Sen and Smith, 1994; Martinez et al., 1999). Several experiments using a T-type Ca2+ channel blocker suggest its involvement in the progression of heart failure (Clozel et al., 1999). Of the two T-type Ca2+ channel genes expressed in heart, CACNA1H and CACNA1G, the former reportedly predominates in fetal ventricle, but is markedly repressed in normal adult heart and reinduced under pathological conditions (Perez-Reyes, 2002; Niwa et al., 2003). Notably, we identified NRSE-like sequences in the first intron of CACNA1H (Figure 6A). The NRSE-like sequences in mouse and human CACNA1H are 93 and 97%, respectively, identical to the consensus NRSE sequences, and EMSA carried out using NRSE as a probe clearly indicated that NRSF binds to these sequences (Figure 6B). In addition, quantitative RT–PCR showed increased expression of CACNA1H, but not CACNA1G, in the ventricles of 8-week-old dnNRSF Tg mice (Figure 6C and D). Finally, the Ni2+ sensitivity of CACNA1H-related currents has been shown to be much higher than that of CACNA1G-related currents (Lee et al., 1999). Consistent with that finding, highly Ni2+-sensitive ICa,T were recorded from ventricular myocytes from dnNRSF Tg mice (Figure 6E) but not NTg mice (Figure 6F), demonstrating the functional expression of CACNA1H (Figure 6G). Taken together, these results indicate that NRSF regulates expression of genes encoding the channels that carry the fetal ionic currents If and ICa,T.
Fig. 6. Increased expression of ICa,T in dnNRSF Tg hearts. (A) Sequences similar to NRSE in mouse (m) and human (h) CACNA1H. (B) EMSA was carried out using a radiolabeled oligonucleotide from NRSE in ANP or CACNA1H as a probe. Unlabeled oligonucleotide from NRSE in ANP was used as a cold competitor. (C and D) Relative levels of CACNA1H (C) and 1G (D) mRNA in the hearts of 8-week-old NTg and dnNRSF Tg mice; *P < 0.05 versus NTg. (E and F) Representative T-type and L-type Ca2+ currents in ventricular myocytes from dnNRSF Tg (E) and NTg (F) hearts; the pulse protocol is shown at the bottom. Na+ current was completely inhibited by 50 µM tetrodotoxin throughout the experiments. Inhibition of the L-type Ca2+ current with 5 µM nicardipine revealed the presence of the T-type Ca2+ current. (G) The peak I–V relationship of the Ni2+-sensitive T-type Ca2+ current measured in the presence of 5 µM nicardipine in ventricular myocytes obtained from dnNRSF Tg mice (n = 5). The threshold for the activation was between –60 and –50 mV; symbols represent means ± SEM.
Discussion
NRSF is an important regulator of the fetal cardiac gene program
Reactivation of a fetal gene program is a common feature of cardiac hypertrophy and heart failure; despite much study, however, the mechanism governing such gene reprogramming remains unclear. Our findings that NRSE is selectively present in multiple fetal cardiac genes and that the inhibition of NRSE-mediated repression increases their expression in ventricular myocytes, both in vitro and in vivo, are indicative of the significance of the NRSE–NRSF system to the regulation of the fetal cardiac gene program. Indeed, we showed there to be a reciprocal relationship between the levels of NRSF and ANP mRNA expression in fetal and adult ventricles, which strongly suggests that NRSF represses fetal cardiac gene expression in terminally differentiated ventricular myocytes. We also demonstrated that inhibition of NRSF diminishes the capacity of hypertrophic stimuli to increase the transcriptional efficiency of fetal cardiac genes. Conversely, the presence of full-length NRSF significantly reduces the basal ANP promoter activity and increases the responsiveness of its activity to ET-1. Thus, NRSF appears to be involved not only in the repression of basal expression of fetal cardiac genes but also in the reinduction of fetal cardiac gene expression during cardiac hypertrophy. That ventricular NRSF expression is diminished in a mouse model of load-induced cardiac hypertrophy, in sharp contrast to BNP expression, is consistent with that view.
NRSF contributes to the maintenance of normal cardiac structure and function
The present findings suggesting a decrease in NRSF repressor activity to be a component of the hypertrophic signal leading to the upregulation of fetal gene expression prompted us to examine the consequences of persistent inhibition of NRSF on the function of post-natal hearts. It is notable that heart-specific inactivation of NRSF was sufficient to cause dilated cardiomyopathy, which suggests that NRSF plays a critical role in the structural and functional alterations that occur during the development of heart failure. It is not completely clear at present which of the primary genetic alterations induced by NRSF inhibition initiate the subsequent remodeling process that leads to dilated cardiomyopathy, though there is a possibility that constitutive expression of α-skeletal actin in adult heart contributes to the progression to LV dysfunction. In normal, mature ventricular myocytes, cardiac α-actin comprises ∼80% of the total actin protein, with α-skeletal actin comprising the remaining 20%. Induction of α-skeletal actin expression in dnNRSF Tg hearts could change the normal ratio of the two actin isoforms, thereby impairing cardiac function; indeed, actin dysfunction is known to lead to heart failure (Olson et al., 1998). Also, the fact that constitutive expression of β-MHC, another fetal cardiac isoform of a contractile protein, is sufficient to reduce systolic function further supports this view (Tardiff et al., 2000). It is unlikely that increased production of ANP and BNP causes the dilated cardiomyopathy and sudden death seen in dnNRSF Tg mice: BNP transgenic mice exhibit no pathological phenotypes in the heart, though BNP knockout mice do show interstitial fibrosis (Ogawa et al., 1994; Tamura et al., 2000). In addition, dnNRSF Tg mice lacking the guanylyl cyclase-A gene, encoding the receptor for ANP and BNP, still showed increased mortality and severe cardiomyopathic phenotype (unpublished observation).
We recently found that the class II HDACs, HDAC4 and 5, which reportedly act as signal-responsive repressors in molecular pathways governing cardiac hypertrophy and heart failure (Zhang et al., 2002), associate with NRSF and modulate its repressor function in ventricular myocytes (unpublished observation). Thus further investigation will be required before complete characterization of the mechanisms involved in the development of dilated cardiomyopathy, the modulation of NRSF function, and the role of NRSF in pathogenesis of heart failure is achieved.
Our results also demonstrate that NRSF regulates ventricular expression of genes encoding fetal-type ion channels carrying If and ICa,T. In adult hearts, expression of If and ICa,T is restricted to pacemaker cells and his-Purkinje fibers. Although it is still controversial, If and ICa,T appear to participate in automaticity of the pacemaker cells (Santoro and Tibbs, 2000; Shorofsky and Balke, 2001). Re-expression of these channels in the hypertrophied or failing ventricle may therefore increase the automaticity of ventricular myocytes, giving rise to arrhythmias. Alternatively, the arrhythmogenesis in dnNRSF Tg mice may be due to Ca2+ overload and delayed after-depolarization of ventricular myocytes. The channel carrying If is highly permeable to both Na+ and K+, and continuous activation of If at the resting membrane potential, particularly during adrenergic stimulation, may cause Na+ to accumulate within myocytes. The resultant decrease of driving force for the Na+–Ca2+ exchanger could in turn lead to Ca2+ overload. Likewise, overexpression of ICa,T could also contribute to Ca2+ overload, thereby leading to arrhythmogenesis.
In similar fashion, alterations in If and ICa,T may contribute to the development of cardiac dysfunction in dnNRSF Tg mice. Recent reports indicate that intracellular Na+ is elevated in hypertrophied and failing hearts, and that the increased intracellular Na+ may play an important role for the initiation and progression of heart failure (Pieske and Houser, 2003; Pogwizd et al., 2003). An increase in Na+ influx carried by If can directly affect intracellular Ca2+ handling via the Na+–Ca2+ exchanger and lead to cardiac dysfunction, while increased ICa,T expression reportedly increases diastolic intracellular Ca2+, profoundly affecting cardiac function (Clozel et al., 1999). Taken together, these findings suggest that dysregulation of the expression of the fetal ionic channels carrying If and ICa,T may contribute, at least in part, to the progression of dilated cardiomyopathy, as well as to the increased vulnerability to arrhythmias in dnNRSF Tg mice.
In summary, we have shown NRSF to be a crucial transcriptional regulator contributing to the maintenance of the normal mature phenotype of cardiac myocytes by repressing expression of several fetal cardiac genes, including those encoding natriuretic peptides (ANP and BNP), a fetal isoform of a contractile protein (α-skeletal actin) and fetal ion channels (HCN2, HCN4 and CACNA1H). NRSF is also involved in the reactivation of the fetal gene program during cardiac muscle cell hypertrophy, and persistent inhibition of NRSF function causes dilated cardiomyopathy and leads to the increased vulnerability to arrhythmias and sudden death. NRSF thus appears to play a key role in the progression of cardiac dysfunction and lethal arrhythmias associated with heart failure. Normalization of dysregulated cardiac gene expression through restoration or enhancement of NRSF repressor function may represent a new therapeutic strategy for preventing the progression of heart failure and sudden death.
Materials and methods
Plasmid constructs
Five GAL4-binding sites were inserted upstream of the ANP promoter (–452 to +97), which had been inserted into the BglII–HindIII sites of PGV-B2 (TOYO INK Inc., Tokyo, Japan), yielding a plasmid designated 5UAS-ANPLuc. Plasmids encoding mouse full-length NRSF and dnNRSF were kindly provided by D.J.Anderson. A plasmid encoding GAL4-NRSF was constructed within the pBIND vector (Promega, Madison, WI) as previously described (Kuwahara et al., 2001).
Ventricular myocyte culture and transfection
Preparation, transfection, adenoviral infection and culture of rat neonatal ventricular myocytes were carried out as previously described (Kuwahara et al., 2001).
Generation of α-MHC-dnNRSF transgenic mice
A dnNRSF cDNA (along with a 5′ myc epitope tag) was cloned into the SalI–HindIII site of a pBluescript II KS (+) plasmid containing the α-MHC promoter (a generous gift from J.Robbins). The α-MHC-dnNRSF transgenic construct was then released from the vector backbone by digestion with NotI and purified for injection into the pronucleus of fertilized oocytes derived from C57BL/6J mice, after which the surviving embryos were transferred to the oviducts of pseudopregnant MCH(ICR) mice. Transgenic founders were identified by either Southern blot analysis or PCR. dnNRSF expression was confirmed by western blot analysis using anti-myc tag antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Determination of expression levels of the dnNRSF gene is described in the Supplementary data available at The EMBO Journal Online. Pressure overload was produced by TAC as described previously (Harada et al., 1998). The care of the animals and all experiments were conducted in accordance with the institutional guidelines of Kyoto University Graduate School of Medicine.
Intracardiac electrophysiology
Mice were intubated and anesthetized with 0.5–1.5% isoflurane, and surface ECGs (limb leads) were placed. With a 1.7 French octapolar catheter (CIBer mouse EP, NuMe, Hopkinton, NY) inserted via a jugular vein, the standard EPS protocol was performed as described previously (Gehrmann and Berul, 1999). Rapid ventricular pacing using the extrastimulation (S1S2) technique was performed with 1–2 extra stimuli to determine the ventricular refractory period and to attempt induction of ventricular arrhythmias. The stimulation was performed at twice the ventricular diastolic capture threshold.
Analysis of electrocardiographs by telemetry
To monitor ambulatory ECGs, radio frequency transmitters (TA 10ETA-F20; Data Science, St Paul, MN) were implanted as previously described (Lee et al., 1998).
Electrophysiological recording from isolated ventricular myocytes
Myocytes were isolated from the left ventricle as previously described (Powell et al., 1980). Whole-cell patch–clamp was then carried out using K+-rich pipet solution containing 145 mM KCl, 5.0 mM MgATP, 5 mM Na2 creatine phosphate, 5 mM EGTA, 0.1 mM Na2GTP, 5 mM HEPES 5 (pH 7.2 with KOH) and physiological bathing solution (phosphate-buffered saline; PBS) containing 140 mM NaCl, 5.4 mM KCl; 0.5 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES and 5 mM glucose (pH 7.4 with NaOH). If was recorded in PBS containing 0.5 mM BaCl2. Alternatively, when recording ICa,T, the pipet solution contained: 100 mM CsCl, 50 mM N-methyl-d-glutamine, 10 mM tetraethyl ammonium chloride (TEA), 5 mM MgATP, 10 mM BAPTA and 5 mM HEPES (pH 7.2 with HCl). After formation of the giga-seal in PBS, the myocytes were superfused with TEA–Cs bathing solution containing 100 mM NaCl, 40 mM TEA, 5.4 mM CsCl, 0.5 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, 5 mM glucose and 0.05 mM tetrodotoxin (pH 7.4 with NaOH). All recordings were made at 32–34°C.
Statistical analysis
Data are presented as means ± SEM. Unpaired t-test was used for comparison between two groups, and ANOVA with post hoc Fisher’s tests was used for comparison among groups. Values of P < 0.05 were considered significant. An inverse correlation between cardiac BNP and NRSF mRNA levels was examined by linear regression analysis.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Ms Okamura for her secretarial work. This work was supported in part by research grants from the Japanese Ministry of Education, Science and Culture, the Japanese Ministry of Health and Welfare, the Japanese Society for the Promotion of Science ‘Research for the Future’ program (JSPS-RFTF96I00204 and JSPS-RFTF98L00801), JSPS Research Fellowships for Young Scientists, Japanese Heart Foundation Research Grant, Japanese Heart Foundation/Pfizer Pharmaceuticals Inc. Grant on Cardiovascular Disease Research, Kanae Foundation for Life and Socio-Medical Science, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research (OPSR) of Japan, and the Takeda Science Foundation.
References
- Cerbai E., Barbieri,M. and Mugelli,A. (1996) Occurrence and properties of the hyperpolarization-activated current If in ventricular myocytes from normotensive and hypertensive rats during aging. Circulation, 94, 1674–1681. [DOI] [PubMed] [Google Scholar]
- Cerbai E. et al. (2001) The properties of the pacemaker current If in human ventricular myocytes are modulated by cardiac disease. J. Mol. Cell. Cardiol., 33, 441–448. [DOI] [PubMed] [Google Scholar]
- Chen Z.F., Paquette. A.J. and Anderson,D.J. (1998) NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nature Genet., 20, 136–142. [DOI] [PubMed] [Google Scholar]
- Chien K.R., Knowlton,K.U., Zhu,H. and Chien,S. (1991) Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J., 5, 3037–3046. [DOI] [PubMed] [Google Scholar]
- Chong J.A. et al. (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell, 80, 949–957. [DOI] [PubMed] [Google Scholar]
- Clozel J.P., Ertel,E.A. and Ertel,S.I. (1999) Voltage-gated T-type Ca2+ channels and heart failure. Proc. Assoc. Am. Physicians, 111, 429–437. [DOI] [PubMed] [Google Scholar]
- Gehrmann J. and Berul,C.I. (1999) Cardiac electrophysiology in genetically engineered mice. J. Cardiovasc. Electrophysiol., 11, 354–368. [DOI] [PubMed] [Google Scholar]
- Grimes J.A. et al. (2000) The co-repressor mSin3A is a functional component of the REST–CoREST repressor complex. J. Biol. Chem., 275, 9461–9467. [DOI] [PubMed] [Google Scholar]
- Harada K., Komuro,I., Zou,Y., Kudoh,S., Kijima,K., Matsubara,H., Sugaya,T., Murakami,K. and Yazaki,Y. (1998) Acute pressure overload could induce hypertrophic response in the heart of angiotensin II type1a knockout mice. Circ. Res., 82, 779–785. [DOI] [PubMed] [Google Scholar]
- Kjær A. and Hesse,B. (2001) Heart failure and neuroendocrine activation: diagnostic, prognostic and therapeutic perspectives. Clin. Physiol., 21, 661–672. [DOI] [PubMed] [Google Scholar]
- Kraner S.D., Chong,J.A., Tsay,H.J. and Mandel,G. (1992) Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron, 9, 37–44. [DOI] [PubMed] [Google Scholar]
- Kuwahara K. et al. (2001) The neuron-restrictive silencer element–neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol. Cell. Biol., 21, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Gomora,J.C., Cribbs,L.L. and Perez-Reyes,E. (1999) Nickel block of three cloned T-type calcium channels: low concentrations selectively block α1H. Biophys. J., 77, 3034–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. et al. (1998) Conditional lineage ablation to model human diseases. Proc. Natl Acad. Sci. USA, 95, 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M.L., Heredia M.P. and Delgado,C. (1999) Expression of T-type Ca2+ channels in ventricular cells from hypertrophied rat hearts. J. Mol. Cell. Cardiol., 31, 1617–1625. [DOI] [PubMed] [Google Scholar]
- Mori N., Schoenherr,C.J., Vandenbergh,D.J. and Anderson,D.J. (1992) A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron, 9, 45–54. [DOI] [PubMed] [Google Scholar]
- Niwa N., Hojo,M., Takemura,H., Horiba,M., Lee,J., Yasui,K. and Kodama,I. (2003) Recapturation of early embryonic T-type Ca2+ channel genes (α1h) in mouse remodeled hearts after myocardial infarction. Circ. J., 67, 503. [Google Scholar]
- Nuss H.B. and Houser,S.R. (1993) T-type Ca2+ current is expressed in hypertrophied adult feline left ventricular myocytes. Circ. Res., 73, 777–782. [DOI] [PubMed] [Google Scholar]
- Ogawa E. et al. (2002) Fibronectin stimulates BNP gene transcription by inhibiting neuron-restrictive silencer element-dependent repression. Cardiovasc. Res., 53, 451–459. [DOI] [PubMed] [Google Scholar]
- Ogawa Y. et al. (1994) Molecular cloning of the complementary DNA and gene that encode mouse brain natriuretic peptide and generation of transgenic mice that overexpress the brain natriuretic peptide gene. J. Clin. Invest., 93, 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson T.M., Michels,V.V., Thibodeau,S.N., Tai,Y.S. and Keating,M.T. (1998) Actin mutation in dilated cardiomyopathy, a heritable form of heart failure. Science, 280, 750–752. [DOI] [PubMed] [Google Scholar]
- Palm K., Belluardo,N., Metsis,M. and Timmusk,T. (1998) Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci., 18, 1280–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Reyes E. (2002) Molecular physiology of low-voltage-activated T-type calcium channels. Physiol. Rev., 83, 117–161. [DOI] [PubMed] [Google Scholar]
- Pieske B. and Houser,S.R. (2003) [Na+]I handling in the failing human heart. Cardiovasc. Res., 57, 874–886. [DOI] [PubMed] [Google Scholar]
- Pogwizd S.M., Sipido,K.R., Verdonck,F. and Bers,D.M. (2003) Intracellular Na in animal model of hypertrophy and heart failure: contractile function and arrhythmogenesis. Cardiovasc. Res., 57, 887–896. [DOI] [PubMed] [Google Scholar]
- Powell T., Terrar,D.A. and Twist,V.W. (1980) Electrophysiological properties of individual cells isolated from adult rat ventricular myocardium. J. Physiol., 302, 131–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro B. and Tibbs,G. (2000) The HCN gene family: molecular basis of the hyperpolarization-activated pacemaker channels. Ann. NY Acad. Sci., 868, 741–764. [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J. and Anderson,D.J. (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science, 267, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J., Paquette,A.J. and Anderson,D.J. (1996) Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl Acad. Sci. USA, 93, 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen L. and Smith,T.W. (1994) T-type Ca2+ channels are abnormal in genetically determined cardiomyopathic hamster hearts. Circ. Res., 75, 149–155. [DOI] [PubMed] [Google Scholar]
- Shorofsky S.R. and Balke,W. (2001) Calcium currents and arrhythmias: insights from molecular biology. Am. J. Med., 110, 127–140. [DOI] [PubMed] [Google Scholar]
- Subramaniam A., Jones,W.K., Gulick,J., Wert,S., Neumann,J. and Robbins,J. (1991) Tissue-specific regulation of the α-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem., 266, 24613–24620. [PubMed] [Google Scholar]
- Tamura N. et al. (2000) Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc. Natl Acad. Sci. USA, 97, 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff J.C., Hewett,T.E., Factor,S.M., Vilkstrom,K.L., Robbins,J. and Leinwand,L.A. (2000) Expression of the β (slow)-isoform of MHC in the adult mouse heart causes dominant-negative functional effects. Am. J. Physiol., 278, H412–H419. [DOI] [PubMed] [Google Scholar]
- Thiel G., Lietz,M. and Cramer,M. (1998) Biological activity and modular structure of RE-1 silencing transcription factor (REST), a repressor of neuronal genes. J. Biol. Chem., 273, 26891–26899. [DOI] [PubMed] [Google Scholar]
- Tomaselli G.F. and Marbán,E. (1999) Electrophysiological remodeling in hypertrophy and heart failure. Cardiovasc. Res., 42, 270–283. [DOI] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey,T.A., Chang,S., Antos,C.L., Hill,J.A. and Olson,E.N. (2002) Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell, 110, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]