Abstract
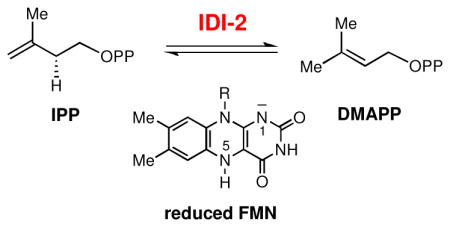
The type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase (IDI-2) catalyzes the reversible isomerization of the two ubiquitous isoprene units, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are required to initiate the biosynthesis of all isoprenoid compounds found in nature. The overall chemical transformation catalyzed by IDI-2 involves a net 1,3-proton addition/elimination reaction. Surprisingly, IDI-2 requires a reduced flavin mononucleotide (FMN) coenzyme to carry out this redox neutral isomerization. The exact function of FMN in catalysis has not yet been clearly defined. To provide mechanistic insight into the role of the reduced flavin in IDI-2 catalysis, several FMN analogues with altered electronic properties were chemoenzymatically prepared, and their effects on the kinetic properties of the IDI-2 catalyzed reaction were investigated. Linear free energy relationships (LFERs) between the electronic properties of the flavin and the steady state kinetic parameters of the IDI-2 catalyzed reaction were observed. The LFER studies are complemented with kinetic isotope effect studies and kinetic characterization of an active site mutant enzyme (Q154N). Cumulatively, the data presented in this work (and in other studies) suggest that the reduced FMN coenzyme of IDI-2 functions as an acid/base catalyst, with the N5 atom of the flavin likely playing a critical role in the deprotonation of IPP en route to DMAPP formation. Several potential chemical mechanisms involving the reduced flavin as an acid/base catalyst are presented and discussed.
Isopentenyl diphosphate:dimethylallyl diphosphate isomerases (IDI) catalyze the reversible interconversion of isopentenyl pyrophosphate (IPP, 1) and dimethylallyl pyrophosphate (DMAPP, 2) used in the biosynthesis of isoprenoids in all living organisms (see Scheme 1).1 Two structurally distinct IDIs exist in nature. The type I IDI (IDI-1) requires only a divalent metal ion for catalysis, whereas the type II enzyme (IDI-2) requires both a divalent metal ion and a reduced flavin mononucleotide (FMN) coenzyme.2 Mechanistic studies of IDI-1 indicate that it uses acid-base chemistry provided by active site amino acids to catalyze a proton addition-elimination reaction via an electron deficient carbocation-like intermediate or transition state.3–6 The mechanism of IDI-2 enzymes is less well understood and in particular, it is unclear what role the reduced flavin coenzyme plays in this isomerization reaction with no net redox change.
Scheme 1.
Potential IDI-2 chemical mechanisms
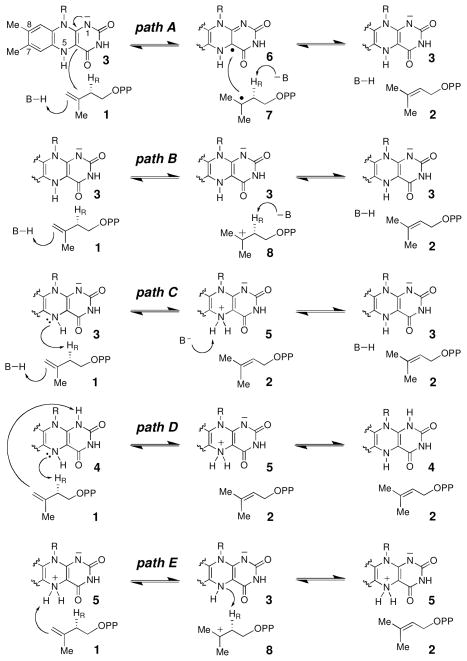
Upon binding 1 or 2, the IDI-2 bound reduced FMN is rapidly converted into a species whose UV-visible absorbance is most consistent with either an anionic reduced FMNH− (3) with slightly altered absorption properties relative to the resting state, or a protonated reduced flavin (FMNH2, either 4 or 5, Scheme 1).7,8 Following formation of the enzyme·IPP complex, several mechanisms in which the reduced flavin plays a catalytic role can be envisioned for the redox-neutral isomerization catalyzed by IDI-2 (see Scheme 1, paths A-E).7–9 The detection of trace amounts of a magnetically isolated neutral flavin semiquinone (6, Scheme 1, path A) in reaction mixtures by EPR provided initial evidence for a radical mechanism. However, no coupled substrate radical (such as 7) was observed.7,8 Subsequent attempts to detect 6 in stopped-flow studies under single turnover conditions failed,8 and several radical clock mechanistic probes also failed to reveal the existence of radical substrate intermediates.10 These results cast doubt on an isomerization mechanism involving single electron transfer. Alternatively, the anionic, reduced FMN (3) could help to electrostatically stabilize a substrate-derived carbocation (8, Scheme 1, path B), generated through acid-base chemistry mediated by active site amino acid residues. However, a recently solved crystal structure of IDI-2 in complex with reduced FMN, Mg2+, and IPP (1) showed no ionizable amino acid side chains in the vicinity of the C2 atom of the bound IPP substrate, raising questions as to the identity of the putative proton donor and acceptor in this mechanism.11 Another possibility is that the reduced FMN coenzyme participates directly in acid-base catalysis, either with the assistance of an amino acid group (Scheme 1, path C), or by serving as both an acid and a base via FMNH2 or its zwitterionic tautomer (Scheme 1, path D and E, respectively).
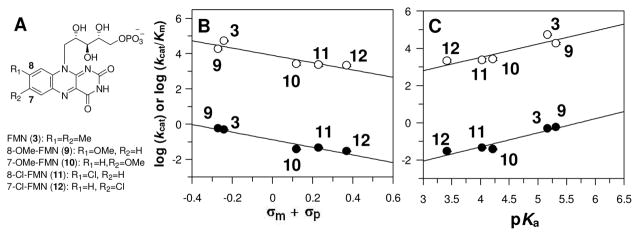
In our previous rapid-mix chemical-quench studies, it was found that a step in the kinetic mechanism prior to or concomitant with DMAPP formation limits turnover in the reaction catalyzed by IDI-2 from Staphylococcus aureus.8 In addition, a 1° substrate deuterium kinetic isotope effect was measured on kcat when (R)-[2-2H]-IPP was used as the substrate, suggesting that C2-H/D bond cleavage is at least partially rate-limiting.8 Based on these observations, we reasoned that if the reduced FMN plays a direct catalytic role in the isomerization step, then alteration of the electronic properties of the reduced flavin should affect the steady state kinetic parameters. To assess this possibility, we chemoenzymatically synthesized a series of FMN analogues (9–12, Figure 1A and Figure S1) substituted with various electron donating and electron withdrawing groups at the 7- and 8- positions (meta and para, respectively, to the N5 atom of FMN) and investigated the effects of these analogues on the kinetics of the IDI-2-catalyzed reaction. When the steady state kinetic parameters for the conversion of IPP to DMAPP were determined using IDI-2 reconstituted with the various reduced FMN analogues, a clear trend was noted. As shown in Figures 1 and S3, a linear relationship was found between the logarithm of the steady state kinetic constants (kcat and kcat/Km) versus either the sum of the Hammett inductive substituent constants12 of the 7- and 8- substituents (Figure 1B), or the estimated pKa of the N5 atom, taken to be equivalent to the pKa of the corresponding aniline derivatives (Figure 1C).13 These results, combined with our previous pre-steady state and kinetic isotope effect studies, strongly suggest that the electronic properties of the reduced FMN coenzyme of IDI-2 play a critical role in mediating the isomerization of IPP (1) to DMAPP (2). The negative slopes in the Hammett plots (ρ ~ −2.0) are consistent with a mechanism where the flavin loses electron density in the step(s) that limit steady state turnover. Similarly, the positive slopes in the Brønsted plots (β ~ 0.7) are consistent with a mechanism involving proton transfer to the N5 atom of the reduced flavin. Together, these results suggest that the flavin N5 atom may be serving as a general base catalyst in the conversion of IPP (1) to DMAPP (2).
Figure 1.
Flavin substituent effects on the steady state kinetic parameters. (A) Structures of the FMN analogues (in their oxidized forms) employed in this study. Hammett (B) and Brønsted (C) plots of log kcat/Km and log kcat (open and filled circles, respectively). The data were fitted with linear regression giving ρ values of -2.1(5) and -2.1(3) for kcat/Km and kcat, respectively, in the Hammett plots, and β values of 0.7(2) and 0.7(1) for kcat/Km and kcat, respectively, in the Brønsted plots.
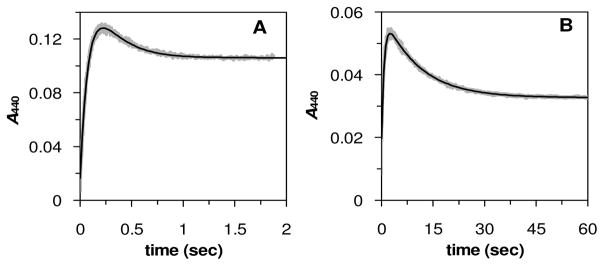
The linear relationship between the steady state kinetic constants and the electronic properties of the flavins suggest that the overall mechanism of the IDI-2 catalyzed reaction remains unchanged as the flavin substituents are altered. In support of this, the absorbance changes in the reduced, IDI-2 bound flavin analogues at 440 nm in the presence of IPP are very similar (Figure S2), suggesting that a similar flavin intermediate forms upon IPP binding. To ensure that formation of the flavin intermediate (Scheme 1, 3, 4 or 5) does not become significantly rate-limiting as the flavin structure is altered, we analyzed the effects of 7-Cl-FMN (12, the slowest of the flavin analogues employed in this study) on the pre-steady state kinetics of flavin intermediate formation and decay under single turnover conditions with IPP as the substrate. As shown in Figure 2, the pattern of pre-steady state absorption changes with FMN- and 7-Cl-FMN-reconstituted IDI-2 are very similar. Namely, the flavin intermediate accumulates rapidly relative to turnover, and then decays to an equilibrium level in a kinetically competent slow phase. As with kcat and kcat/Km, both of the observed pre-steady state rate constants (kfast and kslow) were reduced in the reaction with IDI-2•7-Cl-FMN relative to their values in the IDI-2•FMN-catalyzed reaction. However, it is important to note that the fast phase of flavin intermediate formation (kfast = 12.1 ± 0.1 s−1 and 1.30 ± 0.01 s−1) remains ~ 30 to 40 fold faster than steady state turnover (kcat = 0.47 ± 0.01 s−1 and 0.029 ± 0.001 s−1) for the FMN and 7-Cl-FMN coenzymes, respectively. Thus, there do not appear to be any major changes in the mechanism when the flavin coenzyme is altered, and the 7-Cl-FMN intermediate still accumulates rapidly in the pre-steady state, presumably due to a subsequent and partially rate determining chemistry step in the kinetic mechanism.
Figure 2.
Formation and decay of the FMN intermediate (A) and the 7-Cl-FMN intermediate (B) in the pre-steady state under single turnover conditions. The amplitudes and rates for the two kinetic phases were determined by non-linear regression as described in the supplementary information. For the FMN-catalyzed reaction, the amplitudes and rates of the fast and slow phases are: Afast = 0.179(4) AU, kfast = 12.1(1) s−1, Aslow = −0.090(4) AU, kslow = 4.3(1) s−1, and offset, C = 0.0164(1) AU. For the 7-Cl-FMN-catalyzed reaction: Afast = 0.0410(2) AU, kfast = 1.30(1) s−1, Aslow = −0.0281(2) AU, kslow = 0.096(1) s−1, and C = 0.0197(1) AU.
The reduction in both pre-steady state rate constants (kfast and kslow) with 7-Cl-FMN suggests that, in addition to the chemical step involving IPP C2-H bond cleavage, other steps in the kinetic mechanism (such as the formation of the flavin intermediate upon IPP binding) are sensitive to the electronic properties of the flavin. To assess whether these other steps are more sensitive to the flavin substituents than the C2-H bond cleavage step, we investigated the effects of the flavin analogues on the substrate deuterium kinetic isotope effect (Dkcat) using the stereospecifically deuterated analogue, (R)-[2-2H]-IPP (Figures 3 and S4). We anticipated that if other steps in the kinetic mechanism are more (or less) sensitive to the flavin substiuents than the chemical step involving C2-H/D bond cleavage, then the expression of the KIE on kcat should change. The primary Dkcat values (ranging from 1.6 – 2.8) suggest that C2-H/D bond cleavage is at least partially rate-limiting with each flavin analogue (Figure 3A). Hammett plots of the reactions with IPP and (R)-[2-2H]-IPP (Figure 3B) give ρ values within experimental error of each other. Thus, if multiple steps in the kinetic mechanism are sensitive to the flavin substituents, they appear to be effected to similar extents, such that the overall expression of the KIE on kcat is relatively unchanged. Interestingly, while the ρ values for the two reactions are within experimental error of each other, the magnitude of ρ for the (R)-[2-2H]-IPP catalyzed reaction appears to be slightly larger, to a slight increase in leading Dkcat as the flavin becomes a less efficient catalyst. This trend would suggest that the C2-H/D bond cleavage step becomes more rate limiting as the flavin becomes a poorer catalyst, but the sensitivity of our assay currently precludes this conclusion.
Figure 3.
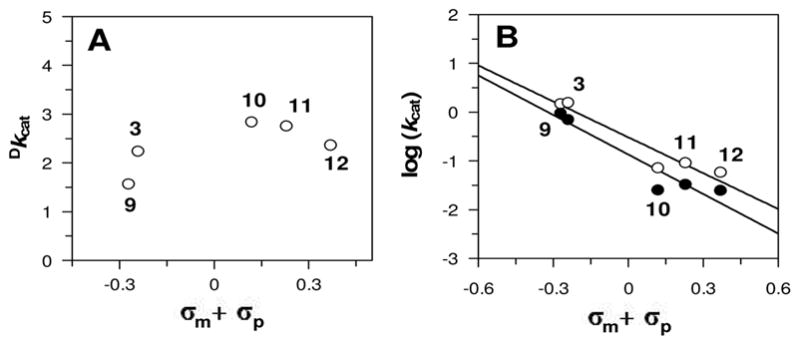
Flavin substituent effects on Dkcat. Reactions with IPP and (R)-[2-2H]-IPP (open and filled circles, respectively, in panel B) were carried out under pseudo-first order conditions as described in the supporting information. (A) Calculated Dkcat from the data in panel (B). (B) Hammett plots: ρ = −2.5(4) and −2.7(5) for IPP and (R)-[2-2H]-IPP, respectively.
Additional support for a direct catalytic role for the FMN coenzyme in the IDI-2-catalyzed reaction can be gleaned from previous studies. For example, the reduced FMN can be covalently modified at the N5 atom by various electrophilic IPP and DMAPP analogues,10,14,15 suggesting that the N5 atom of the reduced flavin carries sufficient electron density to serve as a nucleophile with these electrophilic inhibitors. In addition, the nearly complete inactivity of IDI-2 reconstituted with reduced 5-deazaFMN16,17 suggests that the N5 atom of reduced FMN is critical for isomerization chemistry and that the role of the flavin in catalysis likely extends beyond the simple electrostatic stabilization of a substrate carbocation intermediate (Scheme 1, path B). Interestingly, the N5 atom of the reduced FMN appears to be positioned appropriately for a role as an acid-base catalyst, as it is only 3.2 Å away from C2 of IPP (1) in the crystal structure.11 The substrate in this crystal structure is bound in a mode that orients the pro-R C2 proton of IPP towards the N5 atom of FMN, consistent with the known stereospecificity of C2-H abstraction.18 Furthermore, a hydrogen bond formed between the carbonyl group of a conserved M66 residue and the N5 atom of FMN (separated by 2.9 Å),11 may help to modulate the pKa of the N5 atom for an acid/base catalyst function during turnover. In conjunction with the linear free energy relationships (LFER) studies presented here, these results are all consistent with an IDI-2 chemical mechanism involving general base catalysis by the N5 atom of the reduced flavin coenzyme (Scheme 1, C–E).
Although the N5 atom of FMN is a reasonable candidate for one of the acid/base groups required for turnover, the proton source for the isomerization of IPP eludes us. If the mechanism of path C in Scheme 1 is operative, then the proton required to complete the isomerization of IPP may be supplied by a proton relay composed of H10-E222-Q154 (S. aureus numbering), that connects the enzyme surface to the active site. Interestingly, the absolutely conserved Q154 residue is in close proximity to the bound IPP substrate in the crystal structure and is oriented on the opposite face of the IPP molecule relative to the FMN N5 atom.11 As such, it appears to be positioned appropriately for an anti 1,3-proton addition-elimination reaction. Kinetic studies of the Q154N mutant enzyme indicate a substantial reduction in kcat (170-fold) relative to the wild type enzyme with the Km for IPP (1) being relatively unchanged, suggesting that this residue may indeed play a crucial role in acid-base catalysis (Figure S5).
Alternatively, the flavin could carry out both the protonation and deprotonation steps, precluding the need for amino acid-derived acid/base functional groups (Scheme 1, paths D and E). If the 1,5-dihydro-FMNH2 tautomer (4) is utilized as the active species in the isomerization reaction (Scheme 1, path D), the N1 atom may serve as the proton source, with N5 serving as the base in the forward direction. In this mechanism, the trends observed in the LFER studies could be explained if the N5 atom “senses” the inductive effects of the substituents more strongly than the N1 atom. If the zwitterionic FMNH2 tautomer (5) is employed (Scheme 1, path E), the substrate protonation step may be faster than the deprotonation step in this obligate stepwise mechanism, leading to the observed LFER trends. In this mechanism, however, it is unclear whether a primary deuterium Dkcat would be measured, as deprotonation of the 3° carbocation (8) is expected to be rapid and, thus, would probably not limit the rate of steady state turnover. In these latter two mechanisms, the Q154 residue may function to keep the substrate and flavin oriented properly to ensure optimal proton transfer rates.
In summary, LFER studies using several flavin analogues have provided evidence that the reduced flavin coenzyme of S. aureus IDI-2 plays a direct role in the isomerization of IPP and DMAPP. Our data are most consistent with a mechanism (Scheme 1C) where the N5 atom of the flavin serves as a general acid-base catalyst, perhaps in conjunction with Q154, to effect the isomerization of the IPP double bond using proton addition-elimination chemistry. These studies can now be added to the growing body of experimental evidence7–11,14,15,17 suggesting that the flavin coenzyme of IDI-2 serves a novel function as an acid-base catalyst. Studies aimed at elucidating additional features of the IDI-2 catalyzed reaction are currently in progress.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Steve Sorey for his assistance in collecting the KIE data, Drs. Steven O. Mansoorabadi and Mark Ruszczycky for their helpful discussions, and the NIH (GM40541) and Welch Foundation (F-1511) for funding.
Footnotes
Supporting Information Available: Chemoenzymatic synthesis and absorbance spectra of IDI-2 bound flavin analogues, kinetic data for the LFER, KIE and Q154A mutant enzyme studies, and descriptions of all assays and data fitting are available free of charge.
References
- 1.Ramos-Valdiva AC, van der Heijden R, Verpoorte R. Nat Prod Rep. 1997;14:591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 2.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. Proc Natl Acad Sci USA. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reardon JE, Abeles RH. Biochemistry. 1986;25:5609–5616. doi: 10.1021/bi00367a040. [DOI] [PubMed] [Google Scholar]
- 4.Muehlbacher M, Poulter CD. Biochemistry. 1988;27:7315–7328. doi: 10.1021/bi00419a021. [DOI] [PubMed] [Google Scholar]
- 5.Wouters J, Oudjama Y, Berkley SJ, Tricot C, Stalon V, Droogmans L, Poulter CD. J Biol Chem. 2003;278:11903–11908. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Poulter CD. J Am Chem Soc. 2006;128:11545–11550. doi: 10.1021/ja063073c. [DOI] [PubMed] [Google Scholar]
- 7.Rothman S, Helm TR, Poulter CD. Biochemistry. 2007;46:5437–5445. doi: 10.1021/bi0616347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibodeaux CJ, Mansoorabadi SO, Kittleman W, Chang W-c, Liu H-w. Biochemistry. 2008;47:2547–2558. doi: 10.1021/bi701467g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansoorabadi SO, Thibodeaux CJ, Liu H-w. J Org Chem. 2007;72:6329–6342. doi: 10.1021/jo0703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston JB, Walker JR, Rothman SC, Poulter CD. J Am Chem Soc. 2007;129:7740–7741. doi: 10.1021/ja072501r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unno H, Yamashita S, Ikeda Y, Sekiguchi S, Yoshida N, Yoshimura T, Kusunoki M, Nakayama T, Nishino T, Hemmi H. J Biol Chem. 2009;284:9160–9167. doi: 10.1074/jbc.M808438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansch C, Leo A, Taft RW. Chem Rev. 1991;91:165–195. [Google Scholar]
- 13.Perrin DD, editor. Dissociation constants of organic bases in aqueous solution. Butterworths; London: 1965. [Google Scholar]
- 14.Hoshino T, Tamegai H, Kakinuma K, Eguchi T. Bioorg Med Chem. 2006;14:6555–6559. doi: 10.1016/j.bmc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Rothman SC, Johnston JB, Lee S, Walker JR, Poulter CD. J Am Chem Soc. 2008;130:4906–4913. doi: 10.1021/ja7108954. [DOI] [PubMed] [Google Scholar]
- 16.Hemmi H, Ikeda Y, Yamashita S, Nakayama T, Nishino T. Biochem Biophys Res Commun. 2004;322:905–910. doi: 10.1016/j.bbrc.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Kittleman W, Thibodeaux CJ, Zhang H, Liu Y-n, Liu H-w. Biochemistry. 2007;46:8401–8413. doi: 10.1021/bi700286a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao C-l, Kittleman W, Zhang H, Seto H, Liu H-w. Org Lett. 2005;7:5677–5680. doi: 10.1021/ol0524050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.