Abstract
σE is an alternative sigma factor involved in a pathway of extracytoplasmic stress responses in Escherichia coli. Under normal growth conditions, σE activity is down-regulated by the membrane-bound anti-σE protein, RseA. Extracytoplasmic stress signals induce degradation of RseA by two successive proteolytic events: DegS-catalyzed first cleavage at a periplasmic site followed by YaeL-mediated second proteolysis at an intramembrane region. Normally, the second reaction (site-2 proteolysis) only occurs after the first cleavage (site-1 cleavage). Here, we show that YaeL variants with the periplasmic PDZ domain deleted or mutated allows unregulated cleavage of RseA and consequent σE activation. It was also found that a glutamine-rich region in the periplasmic domain of RseA was required for the avoidance of the YaeL-mediated proteolysis in the absence of site-1 cleavage. These results indicate that multiple negative elements both in the enzyme (PDZ domain) and in the substrate (glutamine-rich region) determine the strict dependence of the site-2 proteolysis on the site-1 cleavage.
Keywords: extracytoplasmic stress response/glutamine-rich regions/PDZ domain/regulated intramembrane proteolysis/site-2 protease
Introduction
Gene expression can be modulated in response to changes in extracytoplasmic environments. In this process, external signals are somehow transmitted across the membrane. Escherichia coli cells sense surface stresses such as accumulation of abnormal proteins in the periplasm and the outer membrane to induce expression of a set of genes encoding chaperones, foldases and proteases that function to cope with the stresses (Missiakas and Raina, 1998; Raivio and Silhavy, 1999, 2001). Two major stress-response pathways, Cpx and σE, are known in E.coli. The Cpx pathway utilizes the CpxA–CpxR two component phospho-relay mechanism whereas σE pathway utilizes this dedicated sigma factor specialized for the extracytoplasmic stress response.
Under normal growth conditions, a majority of σE is sequestered by its interaction with the N-terminal cytoplasmic domain of the membrane-bound anti-σE factor, RseA (De Las Peñas et al., 1997; Missiakas et al., 1997). A periplasmic protein, RseB, may also participate in the negative regulation of the σE activity through its interaction with the periplasmic domain of RseA (De Las Peñas et al., 1997; Missiakas et al., 1997). Structural studies suggested that the cytoplasmic domain of RseA and RNA polymerase competitively bind to σE (Campbell et al., 2003). RseA is a type I membrane protein having a transmembrane segment in the middle of the molecule (Missiakas et al., 1997). Exposure of the cell to an extracytoplasmic stress results in rapid degradation of RseA, liberation of σE and expression of the stress-inducible genes (Ades et al., 1999).
It has been shown that two membrane proteases, DegS and YaeL, are involved in this stress-induced degradation of RseA (Alba et al., 2002; Kanehara et al., 2002). DegS is a member of the DegP protease family (Waller and Sauer, 1996) and has an N-terminal transmembrane segment as well as a periplasmic region composed of a serine-protease domain and an evolutionarily conserved ‘PDZ’ domain generally thought to be involved in protein–protein interaction (Alba et al., 2001). In response to stress signals, DegS cleaves off a substantial portion of the periplasmic domain of RseA. The transient degradation product of RseA, referred to as RseAΔP in this paper, then receives the second proteolysis by YaeL (Alba et al., 2002; Kanehara et al., 2002). YaeL spans the membrane four times with its central periplasmic domain also containing a PDZ-like sequence (Kanehara et al., 2001). It is an E.coli homolog of S2P protease that participates in proteolytic activation of SREBP (sterol regulatory element binding protein) and ATF6 in mammalian cells (Brown et al., 2000; Weihofen and Martoglio, 2003). Cleavage of these substrates by S2P is also preceded by a cleavage by another membrane protease, S1P (Sakai et al., 1998; Ye et al., 2000), although there is no apparent sequence similarity between S1P and DegS.
Presumably, S2P cleaves the substrate protein within the transmembrane segment (Duncan et al., 1998), and so does YaeL. Indeed, the metalloprotease active site motif (HExxH) exists close to or within a transmembrane segment in both S2P and YaeL (Brown et al., 2000; Kanehara et al., 2001). Recent studies have shown that regulation of membrane protein functions through proteolysis within the membrane, or RIP (regulated intramembrane proteolysis), plays critical roles in diverse cellular processes (Brown et al., 2000; Weihofen and Martoglio, 2003). It is likely that the cytoplasmic domain of RseA is liberated as a result of YaeL-dependent proteolysis of RseAΔP. On the other hand, in vivo and in vitro studies showed that the RseA cytoplasmic domain retains an ability to bind to and inactivate σE (De Las Peñas et al., 1997; Missiakas et al., 1997). Thus, it may further be degraded by some other proteases, including ClpXP (Flynn et al., 2003).
Genes degS and yaeL are both essential for viability (Alba et al., 2001, 2002; Kanehara et al., 2002), but they can be disrupted in the presence of increased σE activity due to its overproduction or to the absence of RseA (Alba et al., 2002; Kanehara et al., 2002). Thus, the essential function of DegS and YaeL is to provide cells with a sufficient amount of active σE.
In the absence of DegS, the intact RseA molecule accumulates stably, indicating that YaeL cannot act against RseA without previous cleavage by DegS (Alba et al., 2001, 2002; Kanehara et al., 2002). In contrast, a C-terminally truncated RseA construct (RseA140) that mimics RseAΔP was degraded in a YaeL-dependent manner irrespective of the presence or absence of DegS (Kanehara et al., 2002). Thus, there might be some regulatory mechanism that prevents YaeL-dependent proteolysis of RseA prior to the first cleavage by DegS. This study was aimed at identifying elements required for this regulation. We investigated the role of the PDZ domain of YaeL and the periplasmic domain of RseA in regulated two-step proteolysis of RseA. Our results suggest that these enzyme and substrate domains have important roles in preventing a premature cleavage of RseA by YaeL.
Results
Loss of the PDZ domain function in YaeL results in unregulated cleavage of RseA
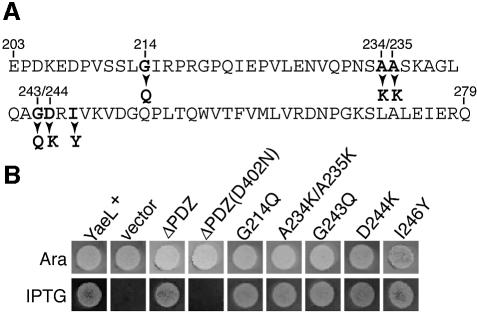
The central periplasmic domain of YaeL contains a region homologous to the PDZ domain (Kanehara et al., 2001). PDZ domains are generally considered to be involved in protein–protein interactions (Harris and Lim, 2001), and it has recently been shown that the similar domain in DegS plays an important role in sensing extracytoplasmic stresses (Walsh et al., 2003). To examine a role of the YaeL PDZ domain in the YaeL function, we constructed a YaeL derivative with its periplasmic PDZ domain deleted (YaeLΔPDZ) (Figure 1A).
Fig. 1. YaeL PDZ region and complementation abilities of mutant forms of YaeL. (A) The amino acid sequence of the YaeL PDZ homologous region and the mutational alterations constructed in this study. The entire region of this sequence was deleted in YaeLΔPDZ. (B) Complementation abilities of the PDZ mutants of YaeL. Strain KK31 [yaeL::kan/pKK6 (Para-yaeL)] was transformed with plasmids encoding the indicated forms of YaeL-His6-Myc under the lac promoter control. They were pKK11 (YaeL+-His6-Myc), pTWV228 (vector), pKK131 (YaeLΔPDZ-His6-Myc), pKK135 [YaeLΔPDZ(D402N)-His6-Myc], pKK138 [YaeL(G214Q)-His6-Myc], pKK139 [YaeL(A234K/A235K)-His6-Myc], pKK140 [YaeL(G243Q)-His6-Myc], pKK136 [YaeL(D244K)-His6-Myc] and pKK141 [YaeL(I246Y)-His6-Myc]. Cultures in l-arabinose (0.2%) were diluted 103-fold with 0.9% NaCl solution and 4 µl portions (containing ∼4 × 103 cells) were spotted on an L agar plate containing 0.2% arabinose (Ara) or 1 mM IPTG. The plates were incubated at 37°C for 14 h.
The essential role of YaeL is to provide cells with a sufficient amount of active σE through proteolytic inactivation of RseA (Alba et al., 2002; Kanehara et al., 2002). It was found that YaeLΔPDZ, expressed from a multicopy plasmid, supported the growth of the ΔyaeL strain (Figure 1B). Introduction of the D402N amino acid alteration, which had been shown to inactivate YaeL with respect to its ability to degrade RseA (Kanehara et al., 2002), into YaeLΔPDZ abolished the complementation activity. These results suggested that YaeLΔPDZ is functional and able to proteolytically inactivate RseA. To examine effects of YaeLΔPDZ on the RseA degradation, it was expressed together with HA-RseA (RseA with an N-terminally attached hemagglutinin epitope), which was detected by anti-HA immunoblotting (Figure 2). Cell fractionation experiments as described by Kanehara et al. (2001) for YaeL revealed that HA-RseA expressed from a plasmid was membrane-bound (data not shown). Additionally, we confirmed that HA-RseA had a cytosolic N-terminus and a periplasmic C-terminus (Y.Akiyama, unpublished data).
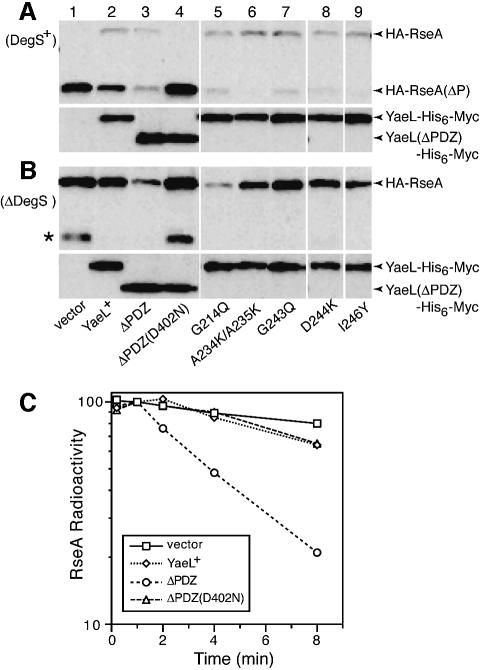
Fig. 2. Effects of YaeL PDZ mutations on degradation of RseAΔP and RseA. (A) RseA accumulation in the presence of DegS. Strain KK211 (ΔyaeL ΔrseA)/pSTD691 (HA-RseA) was further transformed with pTWV228 (vector, lane 1); pKK11 (YaeL+-His6-Myc, lane 2); pKK131 (YaeLΔPDZ-His6-Myc, lane 3); pKK135 [YaeLΔPDZ(D402N)-His6-Myc, lane 4], pKK138 [YaeL(G214Q)-His6-Myc, lane 5]; pKK139 [YaeL(A234K/A235K)-His6-Myc, lane 6]; pKK140 [YaeL(G243Q)-His6-Myc, lane 7]; pKK136 [YaeL(D244K)-His6-Myc, lane 8] and pKK141 [YaeL(I246Y)His6-Myc, lane 9]. Cells were precultured in L glucose (0.4%) medium, inoculated into M9 medium supplemented with 20 amino acids, 2 µg/ml thiamine, 0.4% glucose and 1 mM IPTG, and grown at 30°C for 3.5 h. Whole cellular proteins were subjected to SDS–PAGE and anti-HA (upper panels) and anti-Myc (lower panels) immunoblotting. (B) RseA accumulation in the absence of DegS. Strain AD1840 (ΔdegS ΔyaeL ΔrseA)/pSTD691 was further transformed with the same set of plasmids and processed as described in (A). Asterisk indicates a C-terminally cleaved product of HA-RseA that was produced by the action of some unknown periplasmic proteases and this truncated product is susceptible to active YaeL. (C) Stability of RseA in the absence of DegS. Strain AD1840/pSTD691 was further transformed with pTWV228, pKK11, pKK131 or pKK135. Cells were grown in M9 medium supplemented with 18 amino acids (other than Met and Cys), 2 µg/ml thiamine and 0.4% glucose at 30°C, and induced with 1 mM IPTG and 1 mM cAMP for 2 h. Cells were then pulse-labeled with [35S]methionine for 1.5 min followed by chase with unlabeled methionine for the indicated periods. Samples were processed for anti-HA immunoprecipitation. Radioactivities associated with HA-RseA were determined after SDS–PAGE and phosphorimaging, and are reported as values relative to the 1 min chase radioactivity for each culture set as 100%.
RseA is first cleaved by DegS on the periplasmic side and then by YaeL within the membrane (Alba et al., 2002; Kanehara et al., 2002). Thus, the ΔyaeL cells accumulated RseAΔP, a DegS degradation product of RseA, in which most of the periplasmic domain had been degraded (Figure 2A, lane 1). Co-expression of wild-type YaeL significantly lowered the level of RseAΔP (lane 2) as observed previously (Kanehara et al., 2002). YaeLΔPDZ exerted a stronger effect; it lowered the RseAΔP abundance to a barely detectable level (lane 3). On the other hand, co-expression of YaeLΔPDZ(D402N) did not affect the level of RseAΔP (lane 4). These results show that deletion of the PDZ domain from YaeL did not compromise the YaeL function to proteolyze RseAΔP. Instead, the activity appeared to be enhanced.
Expression of wild-type YaeL only insignificantly affected the accumulation level of intact RseA in the ΔdegS ΔyaeL strain (Figure 2B, lanes 1 and 2), confirming that the YaeL-dependent proteolysis of RseA requires its prior cleavage by DegS (Alba et al., 2002; Kanehara et al., 2002). Strikingly, the expression of YaeLΔPDZ markedly reduced the abundance of RseA (lane 3). YaeLΔPDZ(D402N) was not effective in this respect (lane 4), suggesting that the observed decrease in RseA upon YaeLΔPDZ expression was due to its proteolytic function. Pulse–chase experiments showed that RseA molecules newly synthesized in the absence of DegS were degraded rapidly in the presence of YaeLΔPDZ, but not in the presence of the wild-type YaeL protein (Figure 2C). These results indicate that YaeLΔPDZ, unlike the wild-type YaeL, can degrade the intact RseA protein.
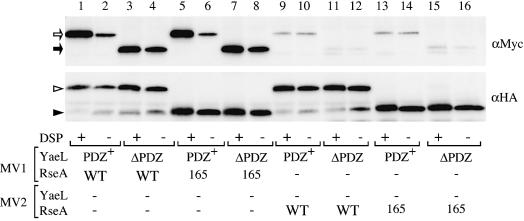
Consistent with YaeL and YaeLΔPDZ being a protease directly degrading RseA, we observed physical interactions between the presumed enzyme and substrate. YaeL and YaeLΔPDZ derivatives, each having a protease motif mutation and a C-terminally attached His6-Myc, were expressed together with HA-RseA. Then membranes were either treated with a cleavable crosslinker, dithio-bis(succinimidyl propinate) (DSP), or mock-treated, solubilized with N-dodecyl-β-d-maltoside and subjected to anti-HA immunoprecipitation. As shown in Figure 3 (lanes 1–4), both YaeL(H22F)-His6-Myc and YaeLΔPDZ(D402N)-His6-Myc were pulled down with HA-RseA. The crosslinking was not essential but it increased the efficiency of co-isolation significantly. In these experiments, we detected in vivo association between these proteins, because very little co-isolation was observed when they were present in different vesicles before detergent solubilization (Figure 3, lanes 9–12). The absence of the PDZ domain did not compromise the protein association. Rather, immunoprecipitation without crosslinking suggested that YaeLΔPDZ has an enhanced ability of RseA-binding (Figure 3, lane 4). These results indicate that YaeL as well as its ΔPDZ derivative interact directly with RseA.
Fig. 3. Crosslinking and co-immunoprecipitation of RseA and YaeL. For lanes 1–8, membrane vesicles carrying indicated combinations of YaeL(H22F)-His6-Myc (shown as PDZ+), YaeLΔPDZ(D402N)-His6-Myc (shown as ΔPDZ), HA-RseA (shown as WT) and HA-RseA165 (shown as 165), were treated with or without DSP. For lanes 9–16, YaeL and RseA were present in separate membrane vesicles (MV1 and MV2), which were mixed and DSP-treated or mock-treated. Membrane proteins were solubilized with 1% N-dodecyl-β-d-maltoside and subjected to immunoprecipitation using immobilized mouse-monoclonal anti-HA antibodies. Proteins recovered were solubilized in SDS, reduced with β-mercaptoethanol to cleave the crosslinkages, and analyzed by SDS–PAGE/immunoblotting using anti-Myc (upper panel) or anti-HA (lower panel) rabbit antibodies. Open arrow, closed arrow, open arrowhead and closed arrowhead indicate YaeL(H22F)-His6-Myc, YaeLΔPDZ(D402N)-His6-Myc, HA-RseA and HA-RseA165, respectively. Direct SDS–PAGE and immunoblotting (without the HA immunoprecipitaiton) demonstrated that all the samples used above contained similar amounts of the YaeL derivatives (not shown).
To delineate the significance of the PDZ sequence further, we constructed YaeL variants with a single or multiple amino acid substitution(s), G214Q, A234K/A235K, G243Q, D244K and I246Y, in the PDZ domain (Figure 1A). The residues selected for mutagenesis are well-conserved among PDZ domains from various species (data not shown). All the mutant forms of the yaeL gene complemented the growth defect of the yaeL disruption strain (Figure 1B). Expression of any of these mutant proteins in the ΔyaeL strain greatly lowered the accumulation level of RseAΔP (Figure 2A, lanes 5–9). As observed with YaeLΔPDZ, these missense mutant proteins [other than YaeL(G243Q)] lowered the accumulation of intact RseA in the absence of DegS (Figure 2B, lanes 5–9); among them YaeL(G214Q) showed the strongest effect, comparable to that of YaeLΔPDZ. Taken together, it was suggested strongly that the PDZ domain of YaeL exerts a specific negative effect on the YaeL activity, preventing an uncontrolled cleavage of intact RseA without imposition of proper stress signals that activate DegS.
PDZ domain of YaeL is required for the down-regulation of σE activity
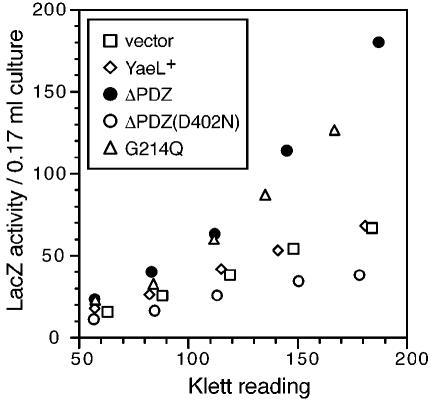
To substantiate that the PDZ mutant forms of YaeL possess unregulated protease activity, wild-type YaeL, YaeLΔPDZ, YaeLΔPDZ(D402N) or YaeL(G214Q) was expressed in a yaeL+ strain that carried a rpoHP3-lacZ reporter for the σE-directed transcription (Figure 4). YaeLΔPDZ and YaeL(G214Q) indeed elevated the LacZ activities (up to 2.5-fold), while wild-type YaeL and YaeLΔPDZ(D402N) did not. The other PDZ missense mutants other than YaeL(G243Q) also significantly elevated the LacZ activities (data not shown). Thus, the PDZ-defective YaeL variants can dominantly activate the σE pathway gene expression. Presumably, this was brought about by active and DegS-independent proteolysis of RseA. The role of the intact PDZ domain is to prevent this unregulated activation of σE.
Fig. 4. Activation of σE upon expression of YaeL PDZ variants. Plasmids pKK11 (YaeL+-His6-Myc), pTWV228 (vector), pKK131(YaeLΔPDZ-His6-Myc), pKK135 [YaeLΔPDZ(D402N)-His6-Myc] and pKK138 [YaeL(G214Q)-His6-Myc] were introduced into TR71 [yaeL+ λ(rpoHP3-lacZ)]. Cells were grown in L medium containing 5 mM cAMP at 30°C, during which samples were withdrawn and assayed for β-galactosidase (LacZ) activity. Results were plotted against the turbidity of culture at each point.
Periplasmic domain of RseA is required for the regulated cleavage by YaeL
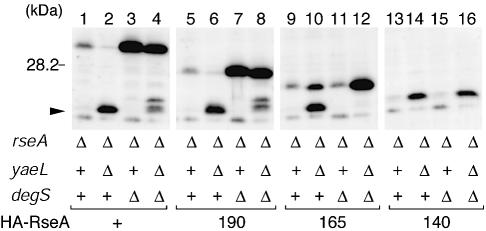
The above results show that the PDZ domain of YaeL is crucial for the regulated two-step proteolysis of RseA. Since the regulation is also lost in RseA140 lacking most part of the periplasmic domain (C-terminal 74 residues) and mimicking RseAΔP (Kanehara et al., 2002), the periplasmic region of RseA must have some role in the regulation. To address the role played by the periplasmic domain of RseA, we first constructed two variants of RseA, RseA190 and RseA165, by deleting C-terminal 26 and 51 residues, respectively (Figure 5). These mutant forms of RseA as well as wild-type RseA were expressed from a plasmid in ΔrseA strains additionally carrying either the ΔdegS and/or the ΔyaeL mutation (Figure 6). RseA190 (lanes 5–8) and the full-length RseA (lanes 1–4) were similar in that they were stable in the ΔdegS strain but converted to the RseAΔP species in the degS+ ΔyaeL strain. In contrast, RseA165 (lanes 9–12) and RseA140 (lanes 13–16) were similar in that they only accumulated in the ΔyaeL strain, irrespective of the degS state.
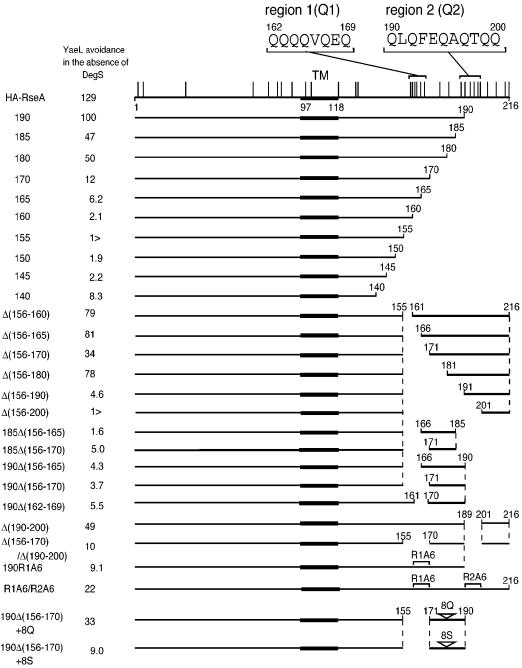
Fig. 5. Schematic representations of mutant forms of RseA with C-terminal or internal deletions, amino acid substitutions and/or insertions. The horizontal lines indicate the portions in RseA carried on individual derivatives (the residue numbers in the authentic RseA protein are shown). The thick parts represent the transmembrane segment (TM). Vertical lines on the top line indicate the positions of glutamine residues. Amino acid sequences of region 1 (Q1) and region 2 (Q2) are shown. YaeL avoidance in the absence of DegS is expressed as the accumulation ratio (%) of each RseA derivative in AD1839 (ΔdegS yaeL+ ΔrseA) cells and in AD1840 (ΔdegS ΔyaeL ΔrseA) cells (see Figures 6 and 8).
Fig. 6. YaeL-avoidance is preserved in RseA190 but lost in RseA165. Plasmids pKK55 (HA-RseA, lanes 1–4), pSTD670 (HA-RseA190, lanes 5–8), pSTD671 (HA-RseA165, lanes 9–12) and pKK58 (HA-RseA140, lanes 13–16) were introduced into a ΔrseA strain (AD1811, lanes 1, 5, 9 and 13) and its derivatives carrying ΔyaeL (KK211, lanes 2, 6, 10 and 14), ΔdegS (AD1839, lanes 3, 7, 11 and 15) and the both mutations (AD1840, lanes 4, 8, 12 and 16). Plasmid-bearing cells were grown in L-broth containing 1 mM IPTG and 1 mM cAMP at 30°C for 3.5 h. Proteins were analyzed by SDS–PAGE and anti-HA immunoblotting. Arrowhead indicates RseAΔP.
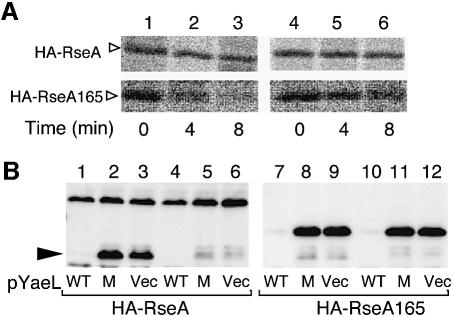
In other words, YaeL can degrade RseA165 lacking C-terminal 51 residues without prior action of DegS. This was demonstrated by pulse–chase experiments (Figure 7A). RseA165 was rapidly degraded in the ΔdegS yaeL+ strain, in sharp contrast to full-length RseA, which was stable in the same strain (lanes 1–3). To confirm that this degradation was YaeL-dependent, we expressed YaeL or its protease motif variant (FEXXH) together with RseA165 in either the degS+ ΔyaeL or the ΔdegS ΔyaeL strain (Figure 7B). Accumulation of RseA165 was abolished by co-expression of YaeL (lanes 7 and 10) but not by YaeL(FEXXH) (lanes 8 and 11). YaeL only slightly lowered the accumulation level of wild-type RseA in these strains (lanes 1 and 4), while it reduced the amount of RseAΔP to an almost undetectable level (lane 1). These results show that RseA165 is subject to YaeL-dependent proteolysis in the absence of DegS. Interestingly, RseA165 was converted to RseAΔP in the degS+ΔyaeL strain (Figure 6, lane 10), indicating that it is still susceptible to DegS. Thus, RseA165 acquired the sensitivity to YaeL, while retaining the sensitivity to DegS. It is suggested then that the periplasmic RseA segment of residues 166–190 contains some element required for the YaeL-avoidance. As expected, HA-RseA165 retained the ability to interact either with YaeL(H22F)-His6-Myc (Figure 3, lanes 5 and 6) or with YaeLΔPDZ(D402N)-His6-Myc (Figure 3, lanes 7 and 8). Our results so far indicate that direct interaction between the YaeL PDZ domain and the RseA periplasmic region, if any, is not strong enough to give a positive signal in two-hybrid analyses (data not shown).
Fig. 7. YaeL-dependent and DegS-independent degradation of RseA165. (A) Cells of AD1839 (ΔdegS ΔrseA, lanes 1–3) and AD1840 (ΔdegS ΔyaeL ΔrseA, lanes 4–6), each carrying pKK55 (HA-RseA+, upper panels) or pSTD671 (HA-RseA165, lower panels) were grown in M9 medium supplemented with 18 amino acids (other than Met and Cys), 2 µg/ml thiamine and 0.4% glucose at 30°C, and induced with 1 mM IPTG and 1 mM cAMP. Cells were then pulse-labeled with [35S]methionine for 1 min followed by chase with unlabeled methionine for the indicated periods. Samples were processed for anti-HA immunoprecipitation, SDS–PAGE and phosphorimaging. (B) Strains KK211 (ΔyaeL ΔrseA)/pKK55 (HA-RseA) (lanes 1–3), AD1840 (ΔdegS ΔyaeL ΔrseA)/pKK55 (lanes 4–6), KK211/pSTD671 (HA-RseA165) (lanes 7–9) and AD1840/pSTD671 (lanes 10–12) were transformed further with one of the following plasmids: pSTD630 (YaeL+-His6-Myc; WT) for lanes 1, 4, 7 and 10; pSTD631 [YaeL(H22F)-His6-Myc; M] for lanes 2, 5, 8 and 11 and pMPM-T3 (vector; V) for lanes 3, 6, 9 and 12. Plasmid-bearing cells were grown at 30°C in L-broth containing 1 mM IPTG and 1 mM cAMP. Total cellular proteins were subjected to SDS–PAGE and anti-HA immunoblotting.
Glutamine-rich segments are involved in the YaeL-avoidance
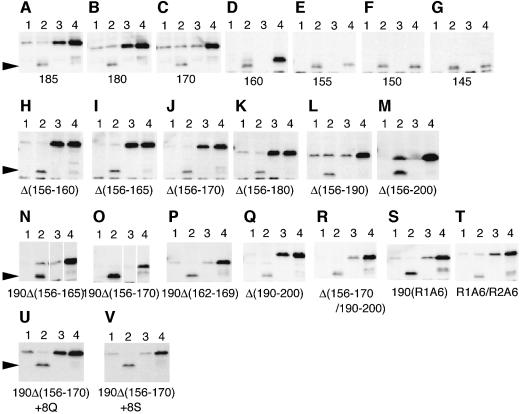
While the mode of YaeL–RseA interaction detected in our pull-down assay should have been related to the ability of the enzyme to degrade the substrate, these two proteins should also interact for the purpose of the negative regulation, in which the periplasmic region of RseA is involved. To further identify RseA element(s) that are required for the regulation, a series of C-terminal deletion mutants of RseA were constructed (Figure 5). They were expressed in four sets of strains: degS+ yaeL+, degS+ ΔyaeL, ΔdegS yaeL+ and ΔdegS ΔyaeL (Figure 8). YaeL-avoidance in the absence of DegS was assessed by comparing accumulation levels in the ΔdegS yaeL+ strain (lanes 3) and in the ΔdegS ΔyaeL strain (lanes 4). Lower accumulation in the former strain indicates the occurrence of DegS-independent and YaeL-dependent degradation. As shown in Figure 8, YaeL-avoidance was retained in RseA185 (Figure 8A) and RseA180 (Figure 8B) but was lost in RseA170 and the smaller derivatives (Figure 8C–G). Cleavage by DegS was observed for RseA155 (Figure 8E) and the larger derivatives (Figure 8A–D), consistent with the recent observation that DegS cleaves RseA between Val148 and Ser149 in vitro (Walsh et al., 2003). Thus, N-terminal 180 residues of RseA is sufficient for the YaeL-avoidance.
Fig. 8. YaeL-avoidance assays for various mutant forms of RseA. Cells carrying a plasmid encoding a HA-RseA derivative as indicated were analyzed for the RseA accumulation as described in the legend to Figure 6. The host strains used were AD1811 (ΔrseA, lanes 1); KK211 (ΔyaeL ΔrseA, lanes 2); AD1839 (ΔdegS ΔrseA, lanes 3) and AD1840 (ΔdegS ΔyaeL ΔrseA, lanes 4).
However, our internal deletion analysis indicated that multiple regions can contribute to the YaeL-avoidance in intact RseA. An internal deletion of the residues 156 to 180 did not abolish the YaeL-avoidance (Figure 8H–K), while larger internal deletions, Δ(156–190) (Figure 8L) and Δ(156–200) (Figure 8M), abolished it. One possible explanation for the above results would be that RseA has two or more regions that can confer the YaeL-avoidance independently. One of such regions may be around residue 170 and the other around residue 190. Consistent with this notion, degradation of RseA190 (Figure 8N and O) and RseA185 (data not shown) became DegS-independent when an internal deletion, Δ(156–165) or Δ(156–170), was introduced into them.
It is remarkable that the periplasmic domain of RseA has a high content of glutamine residues (22%) as compared with its cytoplasmic domain (8.3%) or with the overall E.coli proteins (4.4%). In particular, two regions, residue 162–169 designated here as region Q1 and residue 190–200 designated as region Q2 can be noted for their high glutamine content (6/8 and 6/11, respectively, see Figure 5). These regions Q1 and Q2 roughly overlap with the YaeL-avoidance elements suggested from our deletion mapping. Although deletion of either Q1 or Q2 alone only weakly affected the stability of RseA in the absence of DegS (Figure 8J and Q), deletion of both Q1 and Q2 in RseA (Figure 8R) as well as deletion of Q1 in RseA190 (Figure 8P) resulted in the YaeL-dependent destabilization. We then mutagenized Q1 and Q2 to replace their glutamine residues by alanines. The alanine substituted version of Q1 in RseA190 (Figure 8S) and those of Q1 and Q2 in RseA (Figure 8T) had greatly reduced ability to confer the YaeL-avoidance character. These results show that the presence of either Q1 or Q2 is required for the effective negative regulation of YaeL-dependent proteolysis in the absence of DegS.
Finally, we inserted eight consecutive glutamine residues into the C-terminal part of RseA190Δ(156–170) (Figure 8U). The polyglutamine insertion was found to stabilize the protein significantly in the ΔdegS yaeL+ background, while it was still degraded by the coordinated actions of DegS and YaeL. As a control, we inserted eight consecutive serine residues into the same position (Figure 8V). The polyserine insertion did not affect the stability of RseA190Δ(156–170). We propose that polyglutamine in a C-terminal region can confer the YaeL-avoidance character to RseA. Its exact position may not be crucial.
Discussion
Transmembrane signal transduction from the periplasmic side to the cytosolic side should accompany the extracytoplasmic stress responses that occur in E.coli. In the σE pathway, two-step proteolysis of RseA is the key mechanism of this signaling. While the site-1 cleavage of RseA is catalyzed by DegS, the second proteolysis is dependent on YaeL with intact metalloprotease sequence motifs. In vivo studies suggest strongly that the latter reaction is catalyzed by YaeL (Alba et al., 2002; Kanehara et al., 2002). In support of this notion, we showed that YaeL and RseA interact with each other (Figure 3). Although biochemical verification is still required, these two proteins should represent an enzyme (YaeL) and a substrate (RseA).
While YaeL-dependent cleavage of RseA is essential for activation of σE, YaeL never acts to cleave intact RseA in the absence of DegS. On the other hand, the degradation intermediate of RseA, RseAΔP, can never be detected under any growth conditions, unless YaeL has been impaired genetically (Alba et al., 2002; Kanehara et al., 2002). Thus, the rate-limiting step of RseA degradation should be the site-1 cleavage reaction by DegS. Once this happens, YaeL will rapidly introduce the site-2 proteolysis to the DegS reaction product (RseAΔP), resulting in the elimination of the cellular anti-σE activity. It follows then that the primary point of regulation of RseA degradation by extracytoplasmic stresses lies in the DegS-catalyzed site-1 cleavage. Clearly, this cascade-like regulatory system relies on a mechanism that prevents the otherwise very efficient YaeL proteolytic action against RseA from operating prematurely.
We have shown here that the PDZ domain of YaeL and the glutamine-rich regions of RseA, both localized in the periplasmic space, are required for the inhibition of the YaeL proteolysis of RseA. It is remarkable that both the enzyme and the substrate contain elements for this negative regulation of proteolysis. A question here is whether YaeL is an RseA-specific protease or it acts against some other protein substrates as well. Multiple enzyme and substrate elements for the negative regulation might have evolved as a means to preserve the enzymatic activity of YaeL toward other substrates while specifically blocking its action against RseA.
The use of overproduced RseA in our experiments might have complicated the results through a decreased σE activity that leads to decreased abundance of σE-controlled proteases and protein folding factors in the periplasm/outer membrane. However, this concern is ungrounded since we obtained essentially the same results using an RseA derivative in which the RseA cytoplasmic domain had been replaced with an unrelated protein, maltose-binding protein (MBP). This RseA derivative, MBP–RseA, which lacks the anti-σE activity, is degraded by YaeLΔPDZ, but not by YaeLΔPDZ(D402N) or YaeL+ in the absence of a prior periplasmic cleavage, whereas it is sensitized to YaeL by the RseA165 and RseA140 C-terminal deletions (K.Kanehara and Y.Akiyama, unpublished data). Thus, the YaeL-sensitivity we assigned for each construction of RseA can be regarded to be independent of the feedback consequences of its overproduction/degradation.
It is known in some cases that the PDZ module in a protein is not essential for the enzymatic activity of the protein (Li et al., 2002; Walsh et al., 2003). Our results suggest that the YaeL variant lacking the PDZ domain is still functional as a protease. In fact, the mutant protein seemed to have gained the ability to degrade the intact RseA molecule without any previous site-1 cleavage. Thus, the role of the PDZ domain is to negatively regulate the YaeL action against the intact RseA substrate. We identified several PDZ amino acid substitutions that compromised this regulatory function. One of such residues, G214, corresponds to G324 in the PDZ3 domain of PSD90, a residue that is important for the formation of the carboxylate-binding loop of a critical role in the target protein recognition (Doyle et al., 1996). The G214Q YaeL variant most strongly activated σE among the YaeL variants tested. Like PDZ3 in PSD90, the YaeL PDZ domain may recognize some protein ligand(s).
We described previously a YaeL derivative (YaeLIns31) with a 31-amino acid insertion at a C-terminal part of PDZ (Kanehara et al., 2001). This mutant form proved to exhibit complementation activity dosage-dependently, while it does not effectively support degradation of intact RseA (K.Kanehara, unpublished data). We now believe that this insertion mutation lowers the overall YaeL activity without specifically compromising the PDZ function. All the other PDZ domain mutants of YaeL except G243Q increased the σE activity up to 2.5-fold over levels observed for unstressed wild-type cells. This activation is still much less pronounced than that observed under physiological stresses. As is the case in DegS (Walsh et al., 2003), the deletion and the sequence alterations of PDZ might structurally destabilize YaeL, leading to partial inactivation. Consistent with this, the mutant proteins underwent significant degradation especially in rich media-grown cells (data not shown).
YaeLΔPDZ(D402N) reproducibly lowered the LacZ activity of the reporter strain even in the presence of chromosomal yaeL+ (Figure 4; circles). The inactive protein may sequester RseA from the wild-type enzyme. Our crosslinking and pull-down experiments indeed showed that YaeLΔPDZ retains the ability to bind to RseA (Figure 3).
Walsh et al. (2003) showed that C-terminal peptides of outer membrane proteins activate DegS through their direct interaction with the DegS PDZ domain. Thus, two PDZ-like domains have been characterized as participating in the regulation of the σE extracytoplasmic stress response. Involvement of multiple PDZ domains, on a single protein or separate proteins, is a feature often seen in biological systems using this regulatory protein fold (Harris and Lim, 2001). Walsh et al. (2003) proposed the following mechanism for the outer membrane peptide activation of DegS. In the resting state, DegS is kept inactive by the PDZ domain that is interacting with the protease domain and occluding both the protease active site and its own peptide binding site. Upon imposition of an extracytoplasmic stress, outer membrane peptide ligands become available for the DegS PDZ domain to bind, leading to its release from the protease active site.
Although the PDZ domains of YaeL and of DegS are similar in that they negatively control protease activities, the mode of YaeL regulation must differ from the regulatory mechanism elucidated for DegS. This is because, in YaeL, PDZ and the protease active site are topologically separated by the membrane (Kanehara et al., 2001). Thus, it is unrealistic to assume that the periplasmic PDZ interacts directly with the protease active sites that are located within or on the cytosolic side of the membrane. While DegS is activated by ligands derived from outer membrane proteins (Walsh et al., 2003), OmpC overproduction in the ΔdegS cells does not elicit stress responses (Ades et al., 1999). Thus, YaeL is not simply activated for the stress response by a ligand commonly recognized by these proteases. The ligands that the PDZ domain of YaeL may interact with remain to be identified. Since human S2P does not contain a PDZ homologous region, PDZ domains might have been recruited to confer the regulatory specificity to the bacterial S2P homologs.
Our results showed that the periplasmic domain of RseA is also required for the negative control of YaeL proteolysis of this substrate. In other words, the periplasmic region of RseA contains (an) element(s) that functions to avoid the enzymatic attack by YaeL. Deletion analyses identified two glutamine-enriched regions, Q1 and Q2, whose simultaneous absence or the total replacement of their glutamines by alanine rendered the protein susceptible to YaeL even in the absence of DegS. Importance of a glutamine-rich region for the regulation was demonstrated clearly by insertion of a Gln8 sequence into RseA190ΔQ1, which lacks both Q1 and Q2. The Gln8 addition restored the YaeL avoidance. Although some outside sequences are also required for certain constructs, the presence of either Q1, Q2 or of artificial Gln8 is essential for RseA to avoid an attack from YaeL. This suggests that the chemical nature of the clustered glutamines, rather than the exact amino acid sequence or a specific three-dimensional conformation of these elements, is important for the YaeL-avoidance function. Glutamine residues outside the Q1 and Q2 segments might also contribute to the regulation. An important factor for the controlled proteolysis may be the existence of a certain total number of closely spaced glutamine residues in the C-terminal region of RseA. Partial and weak phenotypes observed for some of individual small deletions and substitutions seem to be consistent with the above mentioned ‘glutamine content’ hypothesis.
A well-characterized ability of PDZ domains is to recognize C-terminal peptides. YaeL might recognize the C-terminus of RseA for self-inhibition, whereas the site-1 cleavage will generate a new C-terminus of RseAΔP, which is no longer recognized by PDZ for inhibition. While this model can certainly explain the successive actions of DegS and YaeL, our results argue against it; some of the RseA variants that shared the same C-terminal sequences of at least 16 residues showed different YaeL-susceptibility in the absence of DegS.
The DegS-dependence of the YaeL proteolysis of RseA can be explained by the removal of the glutamine-rich regions by DegS. As PDZ domains are also known to recognize internal sequences (Hillier et al., 1999; Harris and Lim, 2001), the YaeL PDZ might directly recognize the glutamine-rich regions of RseA, resulting in the inhibition of the protease activity. YaeL and RseA may interact in two different modes, one required for the proteolysis and the other required for the negative regulation. While we detected the former interaction by the experiments shown in Figure 3, the latter mode of interaction is still to be demonstrated experimentally. It is interesting that polyglutamine sequences can form β-sheet structures (Perutz et al., 1994) and that PDZ can recognize an internal sequence with a β-hairpin-like configuration (Hillier et al., 1999; Harris and Lim, 2001). It is also conceivable that the glutamine-rich regions and the PDZ domain on YaeL interact indirectly through a third protein that binds to both of them. RseB could serve as an adaptor protein, as it has an ability to bind to the periplasmic domain of RseA and its deletion accelerates RseA degradation and activates σE, albeit moderately (De Las Peñas et al., 1997; Missiakas et al., 1997; Collinet et al., 2000). Our examinations indicated, however, that RseA in ΔdegS ΔrseB cells was far more stable than the RseA derivative lacking Q1 and Q2 in ΔdegS cells (Y.Akiyama, unpublished data). Thus, the effect of the Q1–Q2 deletion cannot solely be explained by its adverse effect on interaction with RseB.
Whether the interaction between the YaeL PDZ and the RseA glutamine-rich regions is direct or indirect, these enzyme and substrate elements may contribute to a single mechanism of down-regulation of the YaeL activity. We prefer this notion because the periplasmic mutations in RseA alone can give the complete loss of the regulation. Presumably, the concerted actions of the regulatory elements will ensure the substrate-specific regulation of this protease in accordance to the cell’s need to activate the σE-dependent transcription. Biochemical characterization of the molecular details of this system may illuminate a new principle of regulatory interactions between membrane-embedded proteins.
Materials and methods
Bacterial strains and media
Escherichia coli K-12 strains were used in this study. AD16 (Δpro-lac thi/F’ lacIq ZM15 Y+ pro+), AD1811 (AD16, ΔrseA), KK211 (AD16, ΔyaeL ΔrseA), AD1839 (AD16, ΔdegS ΔrseA) and AD1840 (AD16, ΔdegS ΔyaeL ΔrseA) were described previously (Kanehara et al., 2002). KK31 was an AD16 derivative having a chromosomal yaeL-disruption (yaeL::kan) and a complementing plasmid (pKK6) in which yaeL+ had been placed under the control of the arabinose promoter (Kanehara et al., 2002). TR71 (λRS45[rpoHP3-lacZ]) was provided by T.Silhavy.
L medium (Davis, 1980) and M9 medium (Miller, 1972) were used. Ampicillin (50 or 100 µg/ml), chloramphenicol (20 µg/ml), kanamycin (25 µg/ml), spectinomycin (50 µg/ml) and/or tetracycline (12.5 µg/ml) were added for selecting transformants and trasductants as well as for growing plasmid-bearing strains.
Plasmids
pKK11 (encoding YaeL+-His6-Myc), pKK28 [YaeL(H22F)-His6-Myc], pKK55 (HA-RseA), pKK58 (HA-RseA140), pSTD630 (YaeL-His6-Myc) and pSTD631 [YaeL(H22F)-His6-Myc] were described previously (Kanehara et al., 2001, 2002). pKK131 (YaeLΔPDZ-His6-Myc) was constructed by deleting from pKK11 a DNA segment corresponding to a region from Glu203 to Gln279 by site-directed mutagenesis and pKK135 [YaeLΔPDZ(D402N)-His6-Myc] was constructed by replacing a 2.3 kb AseI fragment of pKK131 by a corresponding fragment of pKK35 (Kanehara et al., 2001). Plasmids encoding PDZ missense mutant forms of YaeL-His6-Myc was constructed by introducing GGG (Gly214) to CAG (Gln), GCG (Ala234)/GCA (Ala235) to AAG (Lys)/AAG (Lys), GGC (Gly243) to CAG (Gln), GAC (Asp244) to AAG (Lys) and ATC (Ile246) to TAT (Tyr) codon alterations into yaeL-his6-myc on pKK11 by site-directed mutagenesis. pSTD691 (HA-RseA) was constructed as follows. First, a 2 kb HindIII fragment (carrying a spectinomycin-resistance gene) of pHP45Ω (Prentki and Krisch, 1984) was treated with T4 DNA polymerase and ligated with DraI-digested pSTV28 (Takara Bio Inc.), yielding a pSTV28-derived vector with a spectinomycin-resistance gene instead of a chloramphenicol-resistance gene (named pSTD689). Then, a ScaI–HindIII ha-rseA fragment of pKK55 was cloned into pSTD689. pSTD714 (HA-RseA165) was constructed by cloning a PCR-amplified ha-rseA165 fragment into the KpnI–HindIII sites of STD689. Other plasmids encoding RseA derivatives with a C-terminal deletion were constructed by cloning PCR-amplified fragments into the KpnI–HindIII sites of pCH85 (Kanehara et al., 2002). Plasmids encoding RseA derivatives with an internal-deletion, an insertion or an amino substitution were constructed by site-directed mutagenesis using pKK55 as a template. Plasmids encoding RseA190Δ(156–170) derivative carrying a Gln8 or a Ser8 insertion were constructed by inserting a sequence of CAGCAGCAGCAGCAGCAGCAGCAG or TCTTCTTCTTCTTCTTC TTCTTCT, respectively, between the codons for Tyr180 and Glu181. For construction of plasmids encoding RseA variants with Gln to Ala substitutions in Q1 and Q2, all of the 6 Gln codons in these region were changed to GCA. All the site-directed mutagenesis experiments were carried out by the procedures of Sawano and Miyawaki (2000) using appropriately designed primers. Nucleotide sequences of all the mutated genes and PCR-amplified segments were confirmed by DNA sequencing.
Pulse–chase, immunoprecipitation and immunoblotting experiments
[35S]methionine pulse–chase experiments were carried out essentially as described (Taura et al., 1993). Labeled proteins were immunoprecipitated using anti-HA (Y11) (Santa Cruz Biotechnology, Inc), separated by 12.5% SDS–PAGE and visualized by BAS1800 phosphorimage analyzer.
Immunoblotting with monoclonal anti-Myc (AB1) and polyclonal anti-HA(Y11) antibodies (Santa Cruz Biotechnology, Inc.) was carried out as described previously (Shimoike et al., 1995). Antibody-decorated proteins were visualized using ECL detection kit (Amersham Biosciences) and Fuji LAS1000 lumino-image analyzer.
Crosslinking and co-immunoprecipitation of RseA and YaeL
Total membrane vesicles were prepared (Kanehara et al., 2001) from cells of strain AD1840 carrying appropriate combinations of YaeL and RseA plasmids. Plasmids used were pKK28 [YaeL(H22F)-His6-Myc], pKK135[YaeLΔPDZ(D402N)-His6-Myc], pSTD691(HA-RseA), and pSTD714(HA-RseA165). pTWV228 and pSTD689 served as vector controls. Membranes were suspended in buffer A (50 mM HEPES–KOH pH 7.5, 50 mM KCl, 10% glycerol) and treated with 0 or 0.2 mg/ml DSP (in dimethyl sulfoxide) at 4°C for 1 h. As controls, two kinds of the DSP-treated or untreated vesicles with singly expressed YaeL or RseA were mixed. All the samples were then diluted 10-fold with buffer B (50 mM HEPES–KOH pH 7.5, 300 mM KCl, 10% glycerol) and solubilized with 1% N-dodecyl-β-d-maltoside at 0°C for 1 h. Supernatants after ultracentifugation were incubated with agarose-conjugated mouse monoclonal anti-HA (F7) antibodies (Santa Cruz Biotechnology, Inc.) at 4°C for 2 h with rotation. Immunocomplexes were isolated by centrifugation, washed three times with buffer B-0.1% N-dodecyl-β-d-maltoside and dissolved in SDS sample buffer containing 10% mercaptoethanol. Proteins were separated by 12.5% PAGE and analyzed by immunoblotting.
β-galactosidase assay
The σE activity was assayed by monitoring β-galactosidase activity expressed from a chromosomal σE-dependent lacZ reporter gene (rpoHP3-lacZ). The enzymatic activity was measured as described by Miller (1972) and expressed in Miller units.
Acknowledgments
Acknowledgements
We thank T.Silhavy for a bacterial strain, H.Mori and S.Chiba for stimulating discussion, K.Mochizuki, M.Sano and Y.Yoshikaie for technical support. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan and from CREST, Japan Science and Technology Corporation.
References
- Ades S.E., Connolly,L.E., Alba,B.M. and Gross,C.A. (1999) The Escherichia coli σE-dependent extracytoplasmic response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev., 13, 2449–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba B.M., Zhong,H.J., Pelayo,J.C. and Gross,C.A. (2001) degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol., 406, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Alba B.M., Leeds,J.A., Onufryk,C., Lu,C.Z. and Gross,C.A. (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev., 16, 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.S., Ye,J., Rawson,R.B. and Goldstein,J.L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell, 100, 391–398. [DOI] [PubMed] [Google Scholar]
- Campbell E.A., Tupy,J.L., Gruber,T.M., Wang,S., Sharp,M.M., Gross,C.A. and Darst,S.A. (2003) Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell, 11, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Collinet B., Yuzawa,H., Chen,T., Herrera,C. and Missiakas,D. (2000) RseB binding to the periplasmic domain of RseA modulates the RseA:σE interaction in the cytoplasm and the availability of σE·RNA polymerase. J. Biol. Chem., 275, 33898–33904. [DOI] [PubMed] [Google Scholar]
- Davis R.W., Botstein,D. and Roth,J.R. (1980) Advanced Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- De Las Peñas A., Connolly,L. and Gross,C.A. (1997) The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol., 24, 373–385. [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Lee,A., Lewis,J., Kim,E., Sheng,M. and MacKinnon,R. (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell, 85, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Duncan E.A., Dave,U.P., Sakai,J., Goldstein,J.L. and Brown,M.S. (1998) Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem., 273, 17801–17809. [DOI] [PubMed] [Google Scholar]
- Flynn J.M., Neher,S.B., Kim,Y.I., Sauer,R.T. and Baker,T.A. (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell, 11, 671–683. [DOI] [PubMed] [Google Scholar]
- Harris B.Z. and Lim,W.A. (2001) Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci., 114, 3219–3231. [DOI] [PubMed] [Google Scholar]
- Hillier B.J., Christopherson,K.S., Prehoda,K.E., Bredt,D.S. and Lim,W.A. (1999) Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS–syntrophin complex. Science, 284, 812–815. [PubMed] [Google Scholar]
- Kanehara K., Akiyama,Y. and Ito,K. (2001) Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene, 281, 71–79. [DOI] [PubMed] [Google Scholar]
- Kanehara K., Ito,K. and Akiyama,Y. (2002) YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev., 16, 2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Srinivasula,S.M., Chai,J., Li,P., Wu,J.W., Zhang,Z., Alnemri,E.S. and Shi,Y. (2002) Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol., 9, 436–441. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Missiakas D. and Raina,S. (1998) The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol., 28, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Missiakas D., Mayer,M.P., Lemaire,M., Georgopoulos,C. and Raina,S. (1997) Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol., 24, 355–371. [DOI] [PubMed] [Google Scholar]
- Perutz M.F., Johnson,T., Suzuki,M. and Finch,J.T. (1994) Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA, 91, 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P. and Krisch,H.M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene, 29, 303–313. [DOI] [PubMed] [Google Scholar]
- Raivio T.L. and Silhavy,T.J. (1999) The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol., 2, 159–165. [DOI] [PubMed] [Google Scholar]
- Raivio T.L. and Silhavy,T.J. (2001) Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol., 55, 591–624. [DOI] [PubMed] [Google Scholar]
- Sakai J., Rawson,R.B., Espenshade,P.J., Cheng,D., Seegmiller,A.C., Goldstein,J.L. and Brown,M.S. (1998) Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell, 2, 505–514. [DOI] [PubMed] [Google Scholar]
- Sawano A. and Miyawaki,A. (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res., 28, E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike T., Taura,T., Kihara,A., Yoshihisa,T., Akiyama,Y., Cannon,K. and Ito,K. (1995) Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J. Biol. Chem., 270, 5519–5526. [DOI] [PubMed] [Google Scholar]
- Taura T., Baba,T., Akiyama,Y. and Ito,K. (1993) Determinants of the quantity of the stable SecY complex in the Escherichia coli cell. J. Bacteriol., 175, 7771–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller P.R. and Sauer,R.T. (1996) Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol., 178, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh N.P., Alba,B.M., Bose,B., Gross,C.A. and Sauer,R.T. (2003) OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell, 113, 61–71. [DOI] [PubMed] [Google Scholar]
- Weihofen A. and Martoglio,B. (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol., 13, 71–78. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson,R.B., Komuro,R., Chen,X., Dave,U.P., Prywes,R., Brown,M.S. and Goldstein,J.L. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell, 6, 1355–1364. [DOI] [PubMed] [Google Scholar]