Abstract
Sphingosine kinase 1 (SK1) produces sphingosine-1-phosphate (S1P), a potent signaling lipid. The subcellular localization of SK1 can dictate its signaling function. Here, we use artificial targeting of SK1 to either the plasma membrane (PM) or the endoplasmic reticulum (ER) to test the effects of compartmentalization of SK1 on substrate utilization and downstream metabolism of S1P. Expression of untargeted or ER-targeted SK1, but surprisingly not PM-targeted SK1, results in a dramatic increase in the phosphorylation of dihydrosphingosine, a metabolic precursor in de novo ceramide synthesis. Conversely, knockdown of endogenous SK1 diminishes both dihydrosphingosine-1-phosphate and S1P levels. We tested the effects of SK1 localization on degradation of S1P by depletion of the ER-localized S1P phosphatases and lyase. Remarkably, S1P produced at the PM was degraded to the same extent as that produced in the ER. This indicates that there is an efficient mechanism for the transport of S1P from the PM to the ER. In acute labeling experiments, we find that S1P degradation is primarily driven by lyase cleavage of S1P. Counterintuitively, when S1P-specific phosphatases are depleted, acute labeling of S1P is significantly reduced, indicative of a phosphatase-dependent recycling process. We conclude that the localization of SK1 influences the substrate pools that it has access to and that S1P can rapidly translocate from the site where it is synthesized to other intracellular sites.51: 2546–2559.
Keywords: sphingolipid metabolism, lipid kinase, phosphorylated sphingolipid, sphingosine-1-phosphate lyase, sphingosine, ceramide
Sphingosine-1-phosphate (S1P) has emerged as a potent bioactive lipid with established roles in essential cellular processes such as vascular maturity, cell proliferation, trafficking of immune cells, cytoskeletal rearrangement, invasion, and angiogenesis (1–3). Key to such diversity of functions lies in the ability of S1P to act as both an intracellular effector molecule and an extracellular ligand for a family of five G protein-coupled receptors, S1PR1-5 (4–7). S1P made within the cell can be secreted from the cell where it binds S1PRs in an autocrine or paracrine fashion, triggering a wide range of cellular responses, including proliferation, enhanced extracellular matrix assembly, stimulation of adherens junctions, formation of stress fibers, and inhibition of apoptosis induced by either ceramide or growth factor withdrawal (8–11).
Like many signaling molecules, basal levels of S1P are tightly controlled by balancing synthesis with degradation. S1P is generated by the ATP-dependent phosphorylation of sphingosine catalyzed by sphingosine kinase (SK), of which there are two mammalian isoforms: SK1 and SK2. After being produced by SK, there are several possible fates for S1P (1). As stated above, one fate of S1P is secretion from the cell. Additionally, S1P can be dephosphorylated by a number of lipid phosphatases or irreversibly cleaved by an S1P-specific lyase into hexadecenal and phosphoethanolamine, both of which can be incorporated into the cellular lipid pool (2, 12–16). Cleavage of S1P by the lyase is the only way for cellular sphingolipids to exit the metabolic pathway. As sphingosine kinase is responsible for producing S1P, this effectively places SK1 in a unique position to control the flux of sphingolipids in cells. Controlling the flux of sphingolipids in this fashion establishes a possible housekeeping function for this enzyme that is consistent with the high basal activity of SK1 (17).
In addition to a high basal activity, SK1 has been shown to be further activated in response to numerous external stimuli. Agonists that can activate SK1 include platelet-derived growth factor (PDGF), vascular endothelial growth factor (VEGF), tumor necrosis factor-α (TNF-α), phorbol ester (PMA), and cyclic AMP (11, 18–22). When activated by agonists, SK1 translocates from internal sites to the plasma membrane (PM), and an increase in enzyme activity leads to higher intracellular and extracellular levels of S1P (23, 24). Previous work from our laboratory showed that translocation of SK1 to the PM is dependent on phosphorylation of SK1 by ERK1/2 and that this translocation event is essential for mediating prosurvival, proproliferative signaling by SK1 (24, 25). Recently, calcium and integrin binding protein 1 (CIB1), a calmodulin-related protein that responds to changes in intracellular calcium, has been identified as the protein responsible for mediating the agonist-induced translocation of SK1 to the PM following treatment of HeLa cells with either phorbol ester or TNF-α (26). SK1, and its product S1P, have both been linked to calcium mobilization in a number of systems that could provide the molecular trigger needed to activate CIB1 (27, 28).
The current study investigates the role that intracellular localization of SK1 might have in regulating the switch between the housekeeping and signaling tasks of this lipid kinase. Currently, it is not known if the translocation of SK1 to the PM following agonist stimulation serves to place S1P production closer to specific effector molecules or to remove S1P production away from the endoplasmic reticulum (ER)-bound degradative enzymes. Additionally, little is known about the substrate pools available to SK1. Sphingosine kinase can utilize either sphingosine, derived from ceramide degradation, or dihydrosphingosine, a product of de novo sphingolipid synthesis. Intracellular localization of SK1 could potentially alter which product is made, S1P or dihydroS1P, based on substrate availability, and this could profoundly alter signaling events (29, 30).
MATERIALS AND METHODS
Materials
Lipids.
D-erythro-[2-3H]sphingosine was from American Radiolabeled Chemicals (St. Louis, MO). All other lipids were from Avanti Polar Lipids (Alabaster, AL).
Antibodies.
Anti-Na+/K+ ATPase (α subunit) was from Sigma-Aldrich (St. Louis, MO). Anti-calnexin was from BD Transduction Laboratories (San Jose, CA). Anti-KDEL was from Stressgen (Ann Arbor, MI). Anti-human SK1 (SP1621) was from ECM Biosciences (Versailles, KY). Anti-S1P lyase (sc-67368) and anti-LDH-A (sc-27230) were from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor® 488 goat anti-mouse IgM and Alexa Fluor® 594 goat anti-rabbit IgM were from Molecular Probes (Eugene, OR). Polyclonal anti-Tom 20 was prepared in our laboratory.
Antisense oligonucleotides.
Silencer® predesigned siRNAs (#118700; #118701) targeting human S1P lyase (SGPL1, Entrez Gene ID #8879) and negative control #3 siRNA (AM4615) were from Applied Biosystems (Foster City, CA). The following functionally validated siRNA oligos were from Qiagen (Valencia, CA): #S102659300 and #S102659307 targeting human S1P phosphatase 1 (SGPP1, Entrez Gene ID #81537); #S100716975 and #S104320771 targeting human S1P phosphatase 2 (SGPP2, Entrez Gene ID # 130367).
Gene expression assays and PCR supplies.
Taqman® gene expression assays designed for SGPL1 (Hs00900722_m1), SGPP1 (Hs00229266_m1), SGPP2 (Hs00544786_m1), SPHK1 (Hs00184211_m1), and SPHK2 (Hs00219999_m1), along with High Capacity cDNA reverse transcription kit, RNase inhibitor, MicroAmp® optical 96-well reaction plates, eukaryotic 18S rRNA endogenous control (#4319413E, VIC/MGB probe), and Taqman® universal PCR master mix were from Applied Biosystems (Foster City, CA). Phase Lock Gel™ (5 PRIME, 2 ml) tubes were from Fisher Scientific (Pittsburgh, PA). TRIzol® reagent was from Invitrogen (Eugene, OR).
Miscellaneous.
[γ-33P]Adenosine 5′-triphosphate was from Perkin Elmer (Waltham, MA). Digitonin, PMA (phorbol myristate 13-acetate), adenosine 5′-triphosphate, and paraformaldehyde were from Sigma-Aldrich (St. Louis, MO). Lipofectamine™ 2000 and Lipofectamine™ RNAiMAX were from Invitrogen (Eugene, OR). ECL plus (GE Biosciences) Western blotting reagent, Whatman Silica Gel 60A thin layer chromatography plates, Kimble borosilicate glass tubes, and Agilent glass HPLC vials were from VWR International (Arlington Heights, IL). High-glucose Dulbecco's Modification of Eagle's-Medium (DMEM), L-glutamine, G418, and penicillin-streptomycin were from Mediatech (Herndon, VA). All organic solvents were from Fisher Scientific (Pittsburgh, PA). Complete protease inhibitor cocktail (25× tablets) was from Roche Applied Science (Indianapolis, IN). Fluorescence mounting media was from Dako (Hamburg, Germany).
Methods
Plasmids and subcloning.
Wild-type human sphingosine kinase 1 (hSK1wt) was prepared as described previously (31). The phosphorylation-deficient mutant of hSK1, hSK1S225A, and the catalytically inactive mutant, hSK1G82D, were made using site-directed mutagenesis of the hSK1wt template per the manufacturer's protocol (QuikChange Site-Directed Mutagenesis Kit, Stratagene). The membrane-targeted constructs Lck and PL16 were made as described previously (24, 32). A second ER-targeted construct was made by subcloning the 35-amino acid carboxyl terminus of the ER-specific isoform of rat hepatic cytochrome b5 (33) onto the linker sequence at the C terminus of hSK1wt. The resulting fusion protein is referred to as Cb5. Sequences of all plasmid DNA constructs were verified prior to use.
Cell culture and transfection.
HeLa (human cervical carcinoma; ATCC CCL-2.2) and Hek293 (human embryonic kidney; ATCC CRL-1573) cells were maintained at 37°C and 5% CO2 in high-glucose DMEM containing fetal bovine serum (10%), L-glutamine (2 mM), penicillin (50 IU/ml), and streptomycin (50 μg/ml). All experiments were conducted with cells between passages 2 and 30. Transient transfections of all plasmid DNA were carried out using Lipofectamine™ 2000 (Invitrogen) per manufacturer's protocol.
siRNA knockdown of degradative enzymes.
HeLa or Hek293 cells were plated in 24-well culture dishes in Penn/Strep-free media overnight. Transient transfections of cells with gene-specific small interfering RNA (siRNA) oligos were carried out using 10 nM siRNA duplexes and Lipofectamine™ RNAiMAX (Invitrogen) per manufacturer's protocol. Either 24 or 48 h posttransfection with siRNA duplexes, cells were cotransfected with 0.4 μg plasmid DNA for each hSK1 construct using Lipofectamine™ 2000 as recommended.
Total RNA isolation and reverse transcription.
Total RNA was isolated using TRIzol® and phase-lock gels (5 PRIME) per manufacturer's protocol. Resulting RNA was quantitated with a NanoDrop 1000 (Thermo Scientific). Prior to performing PCR studies, single strand cDNA was prepared from 1 μg total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions.
Real-time PCR analysis.
Quantitative real-time multiplex PCR was performed using the ABI Prism 7300 system (Applied Biosystems) according to the manufacturer's instructions. TaqMan® Gene Expression Assays were used for all genes to be quantitated and eukaryotic 18S rRNA (ABI #4319413E, VIC/MGB probe) was used as the endogenous control. Essentially, cDNA equivalent to 10 ng of total RNA from experimental samples was used for each PCR reaction performed in duplicate using 96-well optical plates (MicroAMP®, ABI). Standard thermal cycler conditions were used. Analysis of results and fold differences were determined using 7300 SDS v1.3.1 software and the comparative CT method. Fold change was calculated from the ΔΔCT values with the formula (2−ΔΔCT), and data are presented as relative to mRNA expression in scrambled siRNA-transfected cells.
Western blot analyses.
For expression levels of recombinant and endogenous proteins, cells were lysed using multiple passes through a 26-gauge needle in lysis buffer [150 mM NaCl, 10% (v/v) glycerol, 50 mM Tris-HCl (pH 7.4), 0.05% (v/v) Triton X-100, 1 mM DTT, 2 mM Na3VO4, 10 mM NaF, 1 mM EDTA (pH 7.0), and 1× complete protease inhibitor (Roche)]. Total protein concentration was determined using Coomassie Plus Bradford Assay Reagent (Pierce). Proteins were separated by SDS-PAGE electrophoresis, then transferred to PVDF membranes. Membranes were blocked overnight with 5% nonfat dry milk and then incubated with primary antibodies diluted in blocking buffer for 1 h at RT. After washing, membranes were incubated with HRP-conjugated secondary antibodies for 1 h at RT. Blots were visualized using ECL plus reagent (GE Biosciences) and then exposed to film. For Western blot analysis of sucrose gradient fractions, aliquots of each fraction were removed and mixed with equal volumes of 2× Laemmli sample buffer, then separated by SDS-PAGE. Resultant films were scanned and analyzed with ImageQuant software.
Immunofluorescence.
Sterilized glass cover slips (Fisher, 12 mm) were coated in fibronectin and placed in a 24-well culture dish prior to plating cells in complete medium overnight. Cells were transfected with 1 μg plasmid DNA using FuGENE 6® (Roche) per manufacturer's protocol and incubated overnight at 37°C in 5% CO2. Paraformaldehyde fixation, Triton-X100 permeabilization, and subsequent staining was performed as previously described (34). For methanol fixation, media was removed and cells were washed with 1× PBS followed by addition of ice-cold MeOH and incubated at −20°C for 5 min. MeOH was replaced with ice-cold MeOH/acetone (50:50, v/v) and the plate was returned to −20°C for 5 min. MeOH/acetone was removed, and cells were washed three times with cold 1× PBS. All antibodies for methanol-fixed cells were diluted in 3% BSA without detergent. Immunofluorescence was analyzed under oil immersion using the 100× objective of an Olympus BX51WI confocal microscope. Images are presented as representative Z-stacks or individual confocal slices.
Total membrane preparation.
Cells were harvested with trypsin and then pelleted by centrifugation at 1500 rpm for 5 min at 4°C. Total membranes and alkaline extractions were performed exactly as previously described (35).
Sucrose gradient fractionation.
All fractionation experiments started with 10 cm dishes of either untransfected HeLa cells or cells transfected with hSK1 constructs. Cells were harvested with trypsin and broken using nitrogen cavitation to preserve membrane integrity. Total membranes were collected with centrifugation as described above. Total protein concentration was determined using Bradford assay. Total membranes (900 μl) were layered on top of a discontinuous sucrose gradient prepared as follows in a 12 ml tube (Beckman #344059): at bottom 1.0 ml 60%, 1.8 ml 44%, 1.8 ml 42%, 1.8 ml 40%, 1.8 ml 38%, 1.8 ml 34%, 1.0 ml 32%. All sucrose solutions were prepared as w/w with sucrose/resuspension buffer (25 mM Tris, 250 mM Sucrose, 1 mM EDTA) with 1× protease inhibitor cocktail (Roche). Samples were centrifuged using an SW41 rotor spun at 37,000 rpm for, typically, 2.5 h at 4°C. The gradients depicted in supplementary Fig. II were centrifuged for 1 h. Fractions (1 ml) were collected from the top of each gradient, placed in 1.5 ml microfuge tubes, vortexed, and placed on ice. Density of all fractions was determined using a refractometer.
SK1 activity assays.
In vitro and in situ enzyme activity assays for hSK1 were performed as previously described (32). When combining the in situ enzyme assay with siRNA knockdown of gene expression, HeLa cells were plated in 24-well culture dishes in Penn/Strep-free medium overnight. Transfections with gene-specific siRNA oligos were performed as described above 24–48 h prior to cotransfection with hSK1 constructs.
Lipid extraction and measurement of radiolabel incorporated into S1P.
At the end of each in situ enzyme assay, reactions were terminated, and cells were harvested using acidic methanol as described previously (32). Cellular lipids were extracted using a modified version of the method by Bligh and Dyer (36). Samples were dried under nitrogen and then resuspended in chloroform, spotted onto TLC plates (Whatman, Silica gel 60), and resolved using 1-butanol/water/acetic acid (3:1:1, v/v). Resolved lipids were visualized with phosphor screens processed on a Typhoon scanner using ImageQuant software. Radiolabeled S1P was located based on comigration with prepared standards. Known quantities of radiolabeled ATP were spotted onto TLC plates to generate standard curves used to calculate levels of 33P-ATP incorporated into S1P.
In vitro phosphohydrolase activity assay.
Phosphohydrolase activity was determined with a modification of a previously described method (37).
Liquid chromatography tandem mass spectrometry.
To measure steady-state levels of sphingolipids in HeLa cells, liquid chromatography tandem mass spectrometry (LC/MS/MS) was employed exactly as described previously (38). Analysis of sphingoid bases, sphingoid base-1-phosphates, and ceramides was carried out using an API4000 triple quadrupole mass spectrometer with turbo-V ion source (Applied Biosystems, Foster, City, CA) interfaced with an automated Agilent 1100 series liquid chromatograph and autosampler (Agilent Technologies, Wilmington, DE).
Statistical analysis.
Triplicate or quadruplicate samples were measured per independent experiment. Statistical significance of the data was assessed with the two-tailed unpaired Student's t-test; *P ≤ 0.05, ** P≤ 0.01, *** P≤ 0.001.
RESULTS AND DISCUSSION
Constitutive localization of recombinant hSK1
Translocation of SK1 from internal sites to the PM following agonist stimulation has previously been reported (24, 25, 39). We wanted to test the influence that subcellular localization of SK1 has on the immediate metabolism of its product, S1P. To this end, we designed constructs of hSK1 that are constitutively targeted to, and tethered at, specific intracellular membranes. In the absence of agonist activation, SK1 is proposed to be cytosolic and to encounter its substrates in the context of the ER membrane. To mimic the interaction of SK1 with the ER, we designed two targeted constructs, hSK1-PL16 and hSK1-Cb5. Both of these recombinant enzymes contain sequences in the C terminus that act as both an ER-specific targeting signal and a membrane anchoring motif (32, 33). To mimic the agonist-induced translocation of SK1 to the PM, we utilized an SK1 construct that contains the myristoylation/palmitoylation motif of Lck tyrosine kinase (10 amino acids at the N-terminus). This construct has previously been used to show that membrane localization of SK1, in the absence of phosphorylation by ERK1/2, is sufficient to drive oncogenic signaling (24). Characteristics of the SK1 constructs used in this study are summarized in Table 1.
TABLE 1.
Summary of hSK1 constructucts
| Construct | Targeting Sequence | Sequence Placement | Predicted Localization | Correct by Microscopy | Correct by Fractionation |
|---|---|---|---|---|---|
| hSK1wt | None | — | Cytosol | Yes | Yes |
| hSK1S225A | None (phosphorylation deficient mutant) | — | Cytosol | Yes | Yes |
| hSK1G82D | None (catalytically inactive mutant) | — | Cytosol | Yes | Yes |
| Lck-hSK1 | Myristoylation / palmitoylation motif (from Lck tyrosine kinase) | N-terminal | PM | Yes | Yes |
| hSK1-PL16 | Polyleucine (16 leucine residues) | C-terminal | ER | Yes | Yes |
| hSK1-Cb5 | ER-specific Cytochrome b5 carboxyl terminus | C-terminal | ER | Yes | Yes |
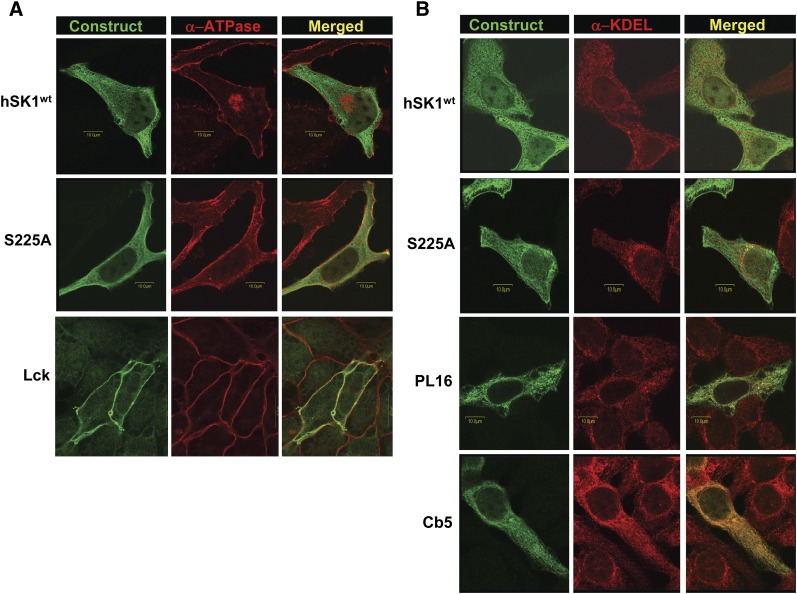
The intracellular localization of these recombinant hSK1 constructs was verified by two separate methods. First, using confocal microscopy, colocalization of each construct with specific intracellular markers was assessed (Fig. 1). The PM-targeted construct Lck colocalized very well with the α subunit of ATPase, a known PM marker (Fig. 1A, bottom panels). Additionally, the ER-targeted constructs PL16 and Cb5 colocalized well with the ER marker, KDEL immunoreactivity (Fig. 1B, bottom panels). In contrast, wild-type hSK1 and the phosphorylation-deficient mutant S225A show a diffuse, cytosolic distribution that did not overlay well with either α-ATPase (Fig. 1A, upper panels) or α-KDEL (Fig. 1B, upper panels). This is in agreement with the current notion, based on the lack of an obvious membrane anchoring sequence, that SK1 is a soluble protein that can readily move about the cytosol (17).
Fig. 1.
Intracellular localization of recombinant hSK1 constructs as assessed by confocal microscopy. HeLa cells were transiently transfected with recombinant hSK1wt; the phosphorylation-deficient mutant hSK1S225A; the plasma membrane-targeted construct Lck; or the endoplasmic reticulum-targeted constructs PL16 and Cb5, as described. Cells were immunostained with α-FLAG antibody (green) for detection of transfected constructs in conjunction with antibodies for specific intracellular markers (red). Immunofluorescence was analyzed by confocal microscopy, and images are presented as representative Z-stacks or individual confocal slices. Merged images (yellow) represent the amount of colocalization of each hSK1 construct with the specified intracellular marker. A: Immunostaining for the plasma membrane marker, the α subunit of ATPase. B: Immunostaining for endoplasmic reticulum as assessed by α-KDEL immunoreactivity. Abbreviations: HeLa, human cervical carcinoma; SK1, sphingosine kinase 1.
To further verify the intracellular localization of our recombinant SK1 constructs, we conducted sucrose density gradient centrifugation studies. Total membranes from HeLa cells that were transiently transfected with each construct were fractionated on discontinuous sucrose gradients as described in “Methods.” Fractions were collected from each gradient, and Western blot analysis for various membrane markers was performed (supplementary Fig. I). Densitometry measurements for each band were recorded and used to generate distribution curves for each intracellular marker. Profiles for each SK1 construct were generated by overlaying the distribution curves (supplementary Fig. IB-F). Similar profiles were seen for the two cytosolic constructs SKwt and S225A (supplementary Fig. IB, C). This distribution is bimodal and complex. These constructs closely overlap with the ER marker in the dense portion of the gradient. Reactivity is also seen in the top-most fraction, consistent with a cytosolic distribution, but also codistributes with the less dense plasma membrane marker. The ER-targeted constructs Cb5 and PL16 had similar profiles to each other and displayed a distribution profile almost identical to the ER marker protein calnexin (supplementary Fig. ID, E). The PM-targeted construct Lck exhibited a distribution profile that mimicked the PM marker protein ATPase (supplementary Fig. IF). To clarify the PM localization of Lck-SK, we modified the gradient conditions to produce a Na+/K+ ATPase distribution with a single peak, well separated from the ER marker (supplementary Fig. IIA). This modification demonstrated unambiguously that Lck-SK is predominantly plasma membrane-targeted while retaining the localization profile of the ER-targeted construct Cb5 (supplementary Fig. IIB).
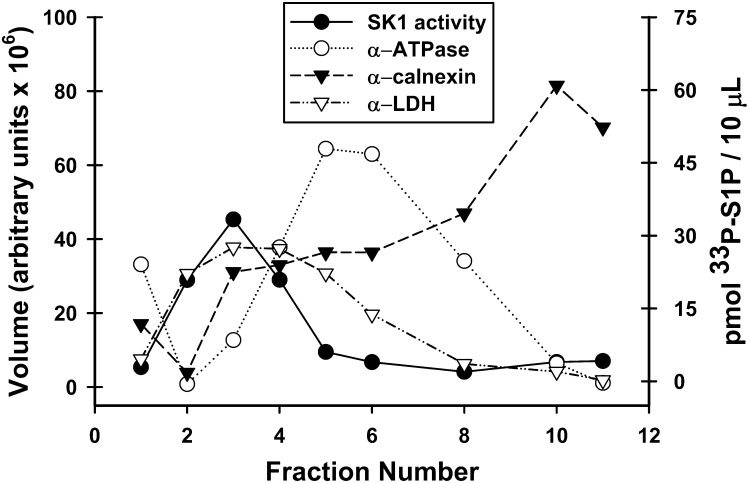
The distribution of endogenous SK1 was determined by conducting in vitro enzyme assays on aliquots of equal volume from each gradient fraction and overlaying these results with the distribution of various intracellular markers (Fig. 2). The enhanced scale of this fractionation (to allow measurement of the low endogenous SK1 activity) slightly altered the distribution of markers relative to that in supplementary Fig. I. This was most notable with the plasma membrane marker Na+/K+ ATPase, which in this fractionation exhibited a single peak. The cytosolic marker lactate dehydrogenase (LDH) was added to this distribution profile to aid in the characterization of endogenous SK1 localization. Conditions that favor SK1 over SK2 were used for the in vitro enzyme assay to ensure that accurate localization of SK1 was being assessed. The endogenous SK1 activity cofractionated with the cytosolic enzyme LDH. This confirms that the majority of endogenous sphingosine kinase is not stably associated with intracellular membranes. Interestingly, when a total membrane fraction is prepared (supplementary Fig. IIIB), a considerable amount of enzyme activity sediments with the total membrane fraction. This finding indicates that although endogenous SK1 has membrane association, it is labile to the conditions employed in sucrose density fractionation. In contrast to the overexpressed wild-type enzyme, there was little overlap of endogenous SK1 activity with either the ER or PM markers. These data suggest that high levels of SK1 expression may force or accentuate a stable association with intracellular membranes, particularly the ER.
Fig. 2.
Localization of endogenous SK1 in HeLa cells as assessed by sucrose density gradient fractionation. Total membranes from nontransfected HeLa cells were fractionated on a discontinuous sucrose density gradient as detailed in “Methods.” For intracellular markers (left axis), equal volumes of each fraction were resolved using SDS-PAGE and immunoblotted with antibodies specific for intracellular markers (α-ATPase for plasma membrane (PM), open circles; α-calnexin for endoplasmic reticulum (ER), filled triangles; α-LDH for cytosol, open triangles). For SK1 localization, in vitro enzyme activity assays (filled circles, right axis) were performed using 10 μl aliquots from each fraction. Abbreviations: HeLa, human cervical carcinoma; LDH, lactate dehydrogenase; SK1, sphingosine kinase 1.
Membrane-targeted hSK1 constructs are intrinsic membrane proteins and are active
Next, we tested whether the targeted hSK1 constructs were anchored to intracellular membranes as intrinsic membrane proteins and whether anchoring altered enzyme activity. HeLa cells were transiently transfected with each of the SK1 constructs, and total membranes were harvested by centrifugation as described. Once harvested, membrane pellets were divided into equal aliquots and subjected to either alkaline treatment (supplementary Fig. IIIA) or in vitro enzyme activity assays (supplementary Fig. IIIB). Alkaline treatment is an established method of distinguishing peripheral membrane proteins from proteins that are inserted into the membrane (40). Following alkaline treatment, equal volumes of each aliquot were separated by SDS-PAGE, and Western blot analysis was performed to detect the presence of SK1, calnexin and Tom20 (integral membrane proteins), and LDH (cytosolic protein). None of the membrane-targeted SK1 constructs had detectable levels of protein in the cytosolic fraction, and the membrane-associated portion of each construct was resistant to alkaline extraction (supplementary Fig. IIIA). In contrast, most of the wild-type SK1 was detected in the cytosolic fraction (80% of total), and the remaining enzyme associated with the membrane fraction was completely susceptible to alkaline treatment. These results, combined with the SK1 activity measurements (supplementary Fig. IIIB), indicate that the membrane-targeted SK1 constructs have been successfully anchored to intracellular membrane locations without any loss of enzyme activity.
Effects of SK1 localization on steady-state levels of sphingolipids
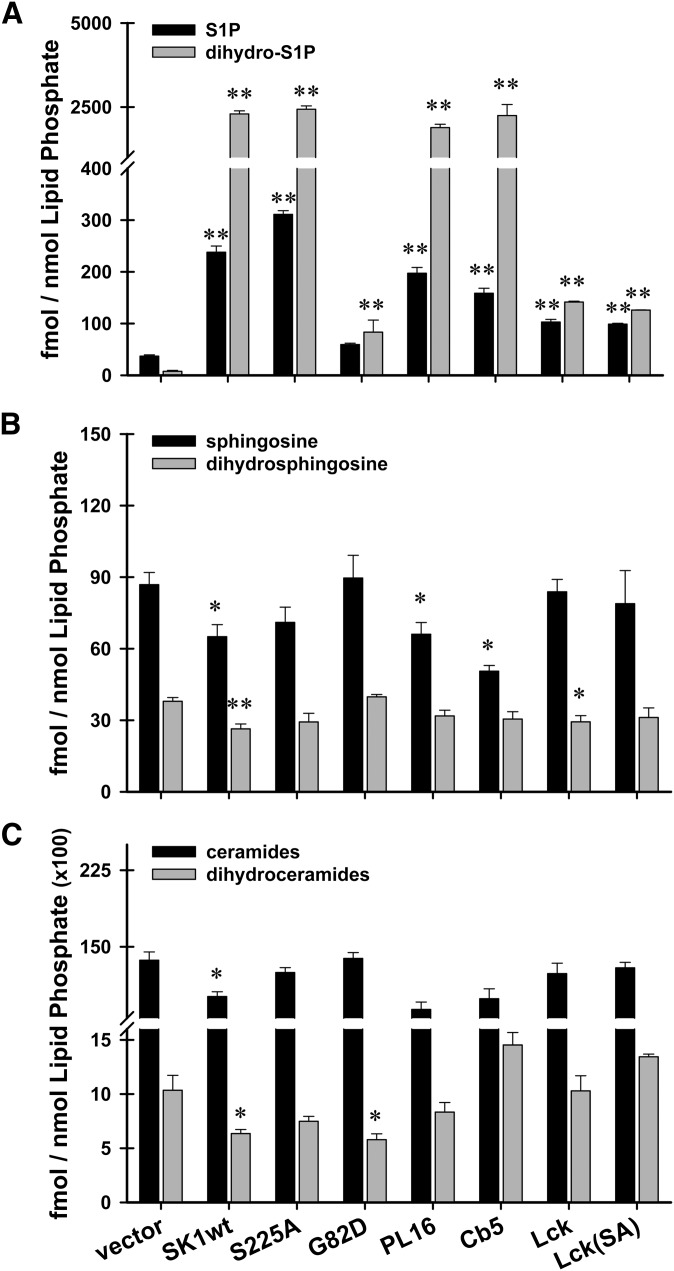
Most of the enzymes mediating the biosynthesis of substrates used by sphingosine kinase, as well as the enzymes responsible for the downstream metabolism of sphingosine-1-phosphate (S1P phosphatases and S1P lyase), are localized to the endoplasmic reticulum (13, 14, 16, 41). To test whether constitutive localization of SK1 to distinct intracellular membranes would affect steady-state levels of sphingolipids, HeLa cells were transiently transfected with our targeted constructs for 24 h prior to harvesting and extraction of total cellular lipids. Samples were then subjected to liquid chromatography tandem mass spectrometry (LC/MS/MS) as previously described (38). Steady-state levels were measured for the sphingoid base phosphates, S1P and dihydroS1P (Fig. 3A); their precursor substrate lipids, sphingosine and dihydrosphingosine (Fig. 3B); and a panel of 12 ceramide species and 7 dihydroceramide species (Fig. 3C). All lipid values were normalized to total phospholipid levels. For each transfectant, SK1 activity levels were verified using in vitro enzyme activity assays (Table 2).
Fig. 3.
Cells overexpressing hSK1 constructs have altered steady-state levels of sphingoid base lipids. HeLa cells were transiently transfected with empty vector, hSK1wt, hSK1S225A, hSK1G82D, or one of the membrane-targeted forms of hSK1. 24 h after transfection, cells were harvested and cellular lipids were extracted. Samples were subjected to LC/MS/MS as described. Results are expressed as fmol sphingolipid/nmol total phospholipid. Data are means ± SEM (n ≥ 3) of independent experiments. Statistical analysis was performed using an unpaired Student's t-test; *P ≤ 0.05, **P ≤ 0.01. A: Steady-state levels of phosphorylated sphingosine (S1P; black bars), and phosphorylated dihydrosphingosine (dihydro-S1P; gray bars). B: Steady-state levels of the precursor lipids sphingosine (black bars) and dihydrosphingosine (gray bars). C: Steady-state levels of total ceramides (black bars) and total dihydroceramides (gray bars). Abbreviations: HeLa, human cervical carcinoma; SK1, sphingosine kinase 1.
TABLE 2.
SK1 activity for LC/MS/MS samples
| Construct | Specific activity (pmol/min/μg protein) | Construct | Specific activity (pmol/min/μg protein) |
|---|---|---|---|
| Vector | 0.09 ± 0.01 | PL16 | 14.90 ± 0.33 |
| SK-wt | 18.38 ± 0.87 | Cb5 | 32.68 ± 0.73 |
| S225A | 21.65 ± 0.25 | Lck | 6.98 ± 0.12 |
| G82D | 0.06 ± 0.01 | Lck(S225A) | 6.82 ± 0.47 |
Surprisingly, overexpression of active SK1 led to greater increases in steady-state levels of dihydrosphingosine-1-phosphate (dihydroS1P) than in levels of S1P (Fig. 3A). These results for wild-type SK1 are in agreement with a recent study in human lung epithelial cells by Berdyshev et al. (42). It is notable that the increase in dihydro-S1P levels found for wild-type sphingosine kinase is mimicked by targeting sphingosine kinase to the endoplasmic reticulum (Fig. 3A, PL16 and Cb5). This suggests that unanchored sphingosine kinase, at least when expressed at high levels, maintains a functional association with the endoplasmic reticulum, the site of dihydrosphingosine synthesis. The utilization of dihydrosphingosine, an obligate precursor of de novo ceramide synthesis, points to a function of sphingosine kinase in controlling ceramide biosynthesis. This is supported by observations, reported here as well as previously, demonstrating that genetic knockdown of sphingosine kinase reduces dihydrosphingosine-1-phosphate production and enhances ceramide de novo biosynthesis (see below) (Fig. 4A–C) (42, 43). Importantly, the significant increases in steady-state levels of dihydroS1P are not accompanied by significant decreases in the precursor lipid dihydrosphingosine (Fig. 3B) or total ceramides (Fig. 3C), as might be expected. This suggests that a compensatory increase in dihydrosphingosine synthesis balances the high level of dihydrosphingosine consumption by SK1.
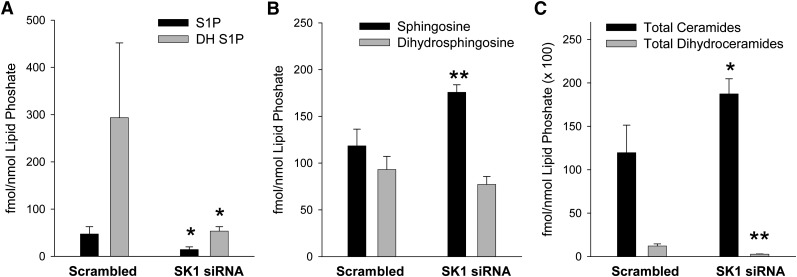
Fig. 4.
Changes in sphingolipid mass levels resulting from knockdown of SK1. HeLa cells were transfected with siRNA against SK1 for 48 h. Cellular lipids were then extracted for sphingolipid analysis using LC/MS/MS as described. Data are means ± SD of four replicates. Statistical analysis was performed using an unpaired Student's t-test comparing the samples treated with scrambled oligonucleotides to the indicated siRNA. *P ≤ 0.05, **P ≤ 0.01. A: Sphingosine-1-phosphate (black bars) and dihydrosphingosine-1-phosphate (gray bars). B: Sphingosine (black bars) and dihydrosphingosine (gray bars). C: Total ceramides (black bars) and total dihydroceramides (gray bars). Statistical analysis was performed using an unpaired Student's t-test comparing the samples treated with scrambled oligonucleotides to the indicated siRNA. *P ≤ 0.05, **P ≤ 0.01. Abbreviations: HeLa, human cervical carcinoma; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
Remarkably, targeting SK1 to the PM (Lck and Lck-SA) drastically reduced the magnitude of increase seen in the steady-state levels of dihydroS1P (Fig. 3A). We confirmed that this was not due to differences in the intrinsic ability of these constructs to utilize sphingosine and dihydrosphingosine as substrates (supplementary Fig. IVA). These results suggest that intracellular pools of dihydrosphingosine are not accessible to SK1 that is localized to the PM, and they could point to an unexplored consequence of the PM translocation of SK1 that has been shown to follow agonist stimulation. Because S1P and dihydroS1P have recently been shown to exert opposite signaling effects in certain cell systems (29, 30), the implications of our results warrant further study.
To complement the observations of the effects of SK overexpression on S1P and dihydroS1P level, we examined the effects of knocking down endogenous SK1 on sphingolipid levels (Fig. 4). SK1 knockdown reduces levels of both S1P and dihydroS1P (Fig. 4A) and increases levels of sphingosine and ceramides (Fig. 4B, C). This finding mirrors the results of a similar knockdown in lung endothelial cells (44), and it confirms that both sphingosine and dihydrosphingosine are substrates for SK1. In this data set, the levels of dihydroS1P in the control cells were elevated relative to levels of S1P, contrary to the data presented in Fig. 3A and levels commonly found by other investigators in various cell types. This presumably is a result of relatively high levels of dihydrosphingosine found in these cultures (compare Fig. 3B, vector with Fig. 4B, scramble). The underlying reason for this difference is unclear. The data presented here and by other groups (42, 43) demonstrating that depletion of SK1 enhances ceramide levels suggests that SK1 may control de novo synthesis of ceramide by controlling the flux of the precursor dihydrosphingosine into the biosynthetic pathway. As noted previously (42), the increase in dihydroS1P is far greater than can be accounted for by the steady-state decrease in dihydrosphingosine. Clearly there must be an accompanying increase in dihydrosphingosine de novo synthesis. Weissman et al. (45) have recently identified the Orm proteins as regulators of serine-palmitoyl transferase (SPT), the initiating step in the de novo sphingolipid pathway. It will be interesting to determine if Orm protein regulation of SPT is responding to the depletion of dihydrosphingosine by sphingosine kinase action.
Inhibition of enzymes that degrade S1P
S1P can be degraded through either a dephosphorylation event or an irreversible cleavage event, or it can be secreted from the cell where it acts as a ligand for a family of G protein-coupled receptors (1). There are three known families of enzymes that utilize S1P and dihydroS1P as substrates. The first is an S1P-specific lyase (SPL), the second is an S1P-specific group of phosphatases (SPP1 and SPP2), and the third is the broad-specificity family of lipid phosphate phosphohydrolases (LPP) (2, 12, 13, 15, 41, 46). S1P lyase and S1P-specific phosphatases 1 and 2 are known to be integral membrane proteins that reside at the ER. We therefore wished to test whether localization of sphingosine kinase specifically to the site of S1P degradation (ER) would accelerate degradation of newly made S1P and whether localization of SK1 away from the degradative enzymes (to the PM) would render S1P generated at the PM refractory to degradation.
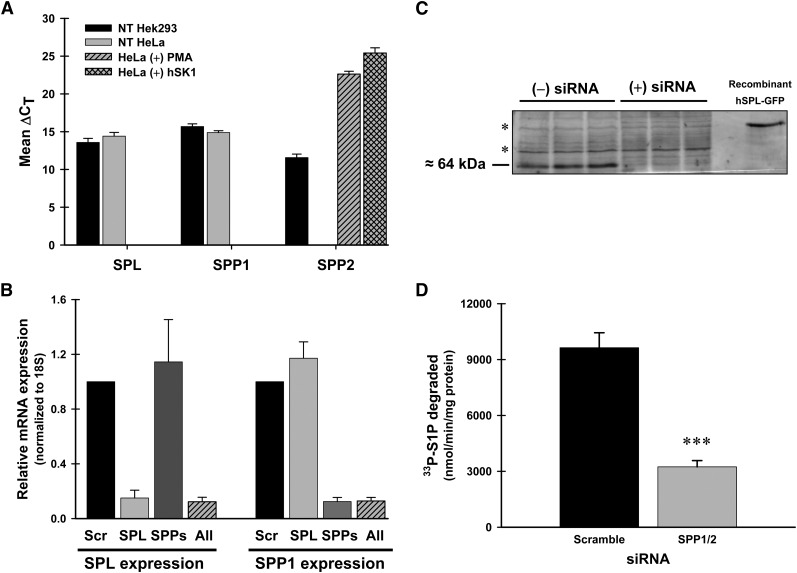
To make this measurement, we combined siRNA-mediated depletion of the degradative enzymes with the targeted localization of S1P production. Initially, we used quantitative PCR to measure endogenous levels of mRNA for the lyase (SPL) and the S1P-specific phosphatases, SPP1 and SPP2, in Hek293 and HeLa cells (Fig. 5A). Endogenous levels of mRNA for SPL and SPP1 were readily detectable in unstimulated Hek293 and HeLa cells. SPP2 mRNA levels could be detected in Hek293 cells but were not detectable in HeLa cells unless the cells were either stimulated with PMA or transfected with recombinant SK1 prior to RNA isolation (Fig. 5A, hashed bars). Given the fact that most of our experiments would involve the use of recombinant SK1, we targeted both SPP1 and SPP2 for our inhibition studies.
Fig. 5.
Endogenous mRNA levels of SPL and SPP1, but not SPP2, are readily detected in unstimulated HeLa and Hek293 cells. Efficient knockdown of SPL and SPP1 at the mRNA and protein levels. A: Endogenous mRNA levels. Hek293 (black) and HeLa (gray) cells were grown to confluency, and then total RNA was isolated. Additionally, subconfluent HeLa cells were either treated with PMA (300 nM, gray hashed) for 4 h or transiently transfected with hSK1wt for 24 h (gray lattice) prior to RNA isolation. cDNA generated from 1 μg of total RNA was used to perform real-time RT-PCR. Data are means of ΔCT (CT gene − CT 18S) values ± SD for six independent RNA isolations. B: siRNA knockdown of SPL and SPP1/2 mRNA. HeLa cells were transfected with scrambled siRNA oligos (Scr), gene-specific oligos for S1P lyase (SPL), S1P-specific phosphatases (SPP), or a combination of oligos for both SPL and SPPs (All) for 48 h. Total RNA was isolated and real-time RT-PCR was performed. Data are relative expression levels of each gene compared with cells transfected with scrambled siRNAs determined by the ΔΔCT method. C: Western blot analysis of total membranes (50 μg/lane) from HeLa cells transfected with scrambled oligos [(−) siRNA] or SPL-specific oligos [(+) siRNA]. Endogenous hSPL is approximately 64 kDa as indicated. The asterisk (*) indicates nonspecific protein bands. D: Phosphohydrolase activity assay for total membranes (50 μg) from HeLa cells transfected with gene-specific siRNAs for SPP1/2. 33P-S1P was used as the substrate as described. Data are means ± SD for n = 4 samples and are representative of two independent experiments. Statistical significance compared with NT HeLa cells was determined by a two-tailed, unpaired Student's t-test. ***P ≤ 0.001. Abbreviations: Hek293, human embryonic kidney; HeLa, human cervical carcinoma; PMA, phorbol ester; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
We utilized commercially available siRNA oligonucleotides as described in “Materials.” Specific knockdown of each gene was verified by quantitative PCR and by protein expression or enzyme activity. As seen in Fig. 5B, when SPP1/2 were inhibited (SPPs), the lyase expression level (SPL) was not affected, and when the lyase was inhibited, no effect was seen on SPP1 expression. SPP2 expression was undetectable under these conditions (data not shown). For S1P lyase, Western blot analysis shows a significant reduction in detectable protein levels after 48 h of siRNA treatment (Fig. 5C). For SPP1/2 inhibition, phosphohydrolase activity was reduced by almost 70% following 48 h of siRNA treatment (Fig. 5D). Collectively, these results show an effective inhibition of both the lyase- and SPP-mediated degradative pathways at the mRNA and protein levels in HeLa cells.
Effects of sphingosine-1-phosphate lyase and phosphatase knockdowns on acute S1P production
To assess the relative contributions made by each of the degradative pathways on the metabolism of localized pools of S1P, we combined selective siRNA-mediated inhibition of degradation with a robust and sensitive in situ enzyme assay developed in our laboratory (32). This assay allowed us to preserve the intracellular localization of S1P production and degradation through the use of intact monolayers of cells.
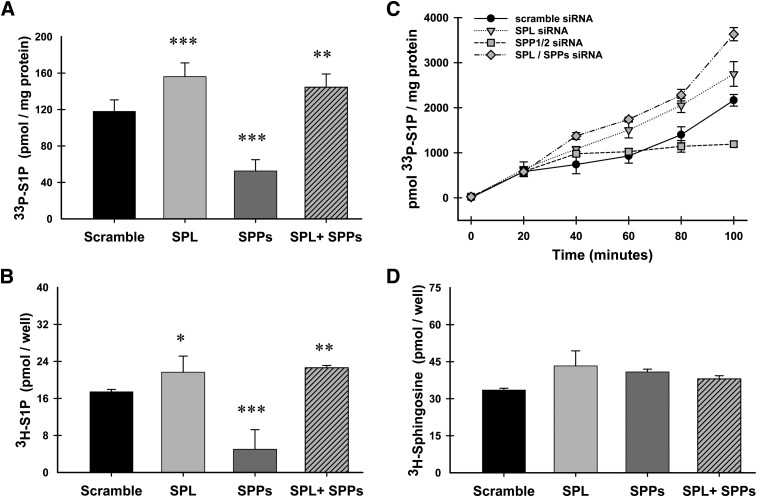
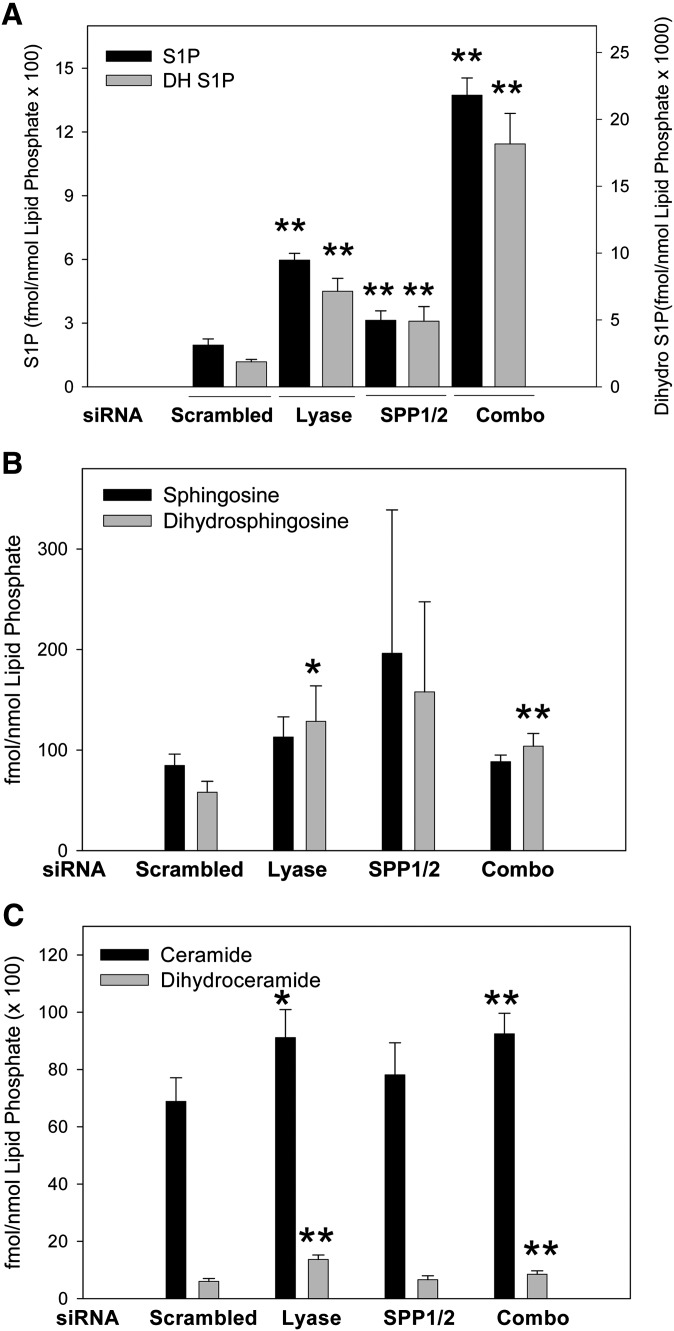
We chose to initially characterize this system with the plasma membrane-targeted form of SK1, Lck-SK (Fig. 6). Newly produced S1P was labeled by using either 33P-ATP (Fig. 6A, C) or 3H-sphingosine (Fig. 6B, D) as substrates, and all reactions were terminated at 100 min except for the time course study depicted in Fig. 6C. As expected, when lyase expression was inhibited (SPL), levels of newly produced S1P were significantly increased over the levels seen when scrambled siRNAs were used ( ). This result was identical when using either 33P-ATP or 3H-sphingosine as a substrate. For subsequent experiments, we utilized 33P-ATP labeling because of its ease of use and sensitivity. The time-course of this reaction (Fig. 6C) illustrates that the enhanced accumulation of sphingosine-1-phosphate observed with lyase knockdown becomes apparent as early as 40 min after starting the assay. Surprisingly, knockdown of the S1P- specific phosphatases (SPPs) profoundly inhibits the accumulation of newly produced S1P rather than enhancing its accumulation (Figs. 6, 7, 8). The marked reduction in S1P production induced by knocking down SPP1/2 is dependent on degradation of S1P by the S1P lyase. S1P lyase knockdown completely reverses the effect of SPP1/2 knockdown, and yields levels of S1P similar to that of S1P lyase knockdown alone. These results are reflected in both the time-course experiment depicted in Fig. 6C as well as the single, 100-min time point studies depicted in Figs. 6–8.
Fig. 6.
Effects of siRNA-mediated inhibition of degradative enzymes on acute production of S1P. HeLa cells were transfected with gene-specific siRNAs for either lyase (SPL), the S1P-specific phosphatases (SPP), or both in combination (SPL+SPP). Following siRNA treatment, cells were transiently transfected with the PM-targeted construct Lck. 24 h after transfection, in situ enzyme assays were performed using either 33P-ATP (A and C) or 3H-sphingosine (B and D) to label newly produced S1P. A-B, D: Reactions were quenched at 100 min, lipids were extracted and resolved using TLC. C: A time course was performed, and reactions were quenched at indicated times. Data are means ± SD for triplicate measurements and are representative of at least two independent experiments. Statistical significance compared with scrambled siRNA treatment was determined by the use of two-tailed, unpaired Student's t-test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Abbreviations: HeLa, human cervical carcinoma; PM, plasma membrane; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
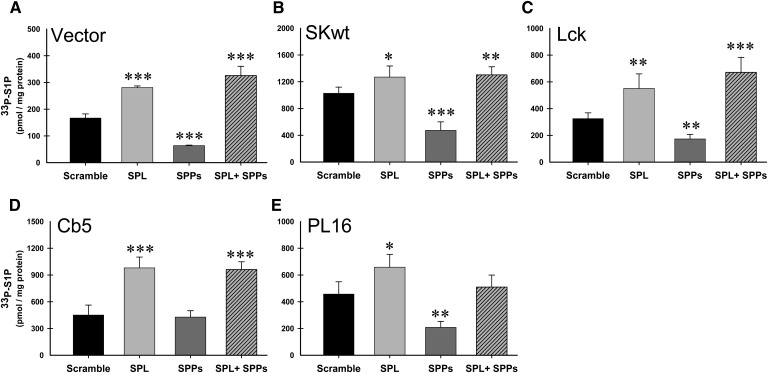
Fig. 7.
Intracellular localization of hSK1 does not markedly affect degradation of S1P. HeLa cells were transfected with gene-specific siRNAs for the lyase (SPL), the S1P-specific phosphatases (SPP), or both in combination (SPL+SPP). Following siRNA treatment, cells were transiently transfected with either empty vector (A) or the recombinant hSK1 constructs as indicated (B-E). 24 h after transfection with constructs, in situ enzyme assays were carried out for 100 min as described. Lipid values were normalized to total protein and data are means ± SD for n = 4 samples and are representative of at least three independent experiments. Statistical significance compared with scrambled siRNA treatment was determined by the use of a two-tailed, unpaired Student's t-test; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Abbreviations: HeLa, human cervical carcinoma; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
Fig. 8.
siRNA-mediated knockdown of SPPs does not inhibit SK1 activity. Two 24-well plates of HeLa cells were transfected with gene-specific siRNAs for the lyase (SPL), the S1P-specific phosphatases (SPP), or both in combination (SPL+SPP). Following siRNA treatment, cells were transiently transfected with hSK1-Cb5. A: 24 h after SK1 transfection, one 24-well plate was used for an in situ enzyme assay as previously described. Data are means ± SD for n = 4 samples. Statistical significance compared with scrambled siRNA treatment was determined by the use of a two-tailed, unpaired Student's t-test; ***P ≤ 0.001. B: The second 24-well plate was used to conduct an in vitro kinase assay using TX-100 in the assay buffer as described. Data are means ± SD for n = 6 samples and are representative of at least two independent experiments. Abbreviations: HeLa, human cervical carcinoma; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
Localization of SK1 does not markedly affect degradation of S1P
To further explore the specific effects that intracellular localization of SK1 has on the degradative fate of acutely produced S1P, we used multiple sets of HeLa cells that each received identical siRNA treatments. Following siRNA treatment, cells were transfected with either empty vector (Fig. 7A) or recombinant hSK1 constructs (Fig. 7B–E) for 24 h prior to conducting in situ enzyme assays as described. As seen in Fig. 7, the degradative fate of newly made S1P in this system is not significantly altered by the intracellular localization of its production. When lyase expression is inhibited (SPL), accumulation of newly produced S1P is significantly increased compared with scrambled siRNAs, with only small variations in the extent of this increase for each construct. In general, for each independent experiment we conducted, S1P levels were further increased when both the lyase and phosphatase pathways were knocked down (SPL+SPPs). This is well exemplified by vector and Lck transfected cells (Fig. 7A, C, respectively). In stark contrast to these results, when SPP1 and SPP2 expression levels are knocked down, levels of newly produced S1P are significantly decreased as compared with scrambled controls. As with the lyase inhibition, this effect does not seem to be dependent on the intracellular localization of SK1 and is seen consistently for all experiments (Fig. 7, SPPs).
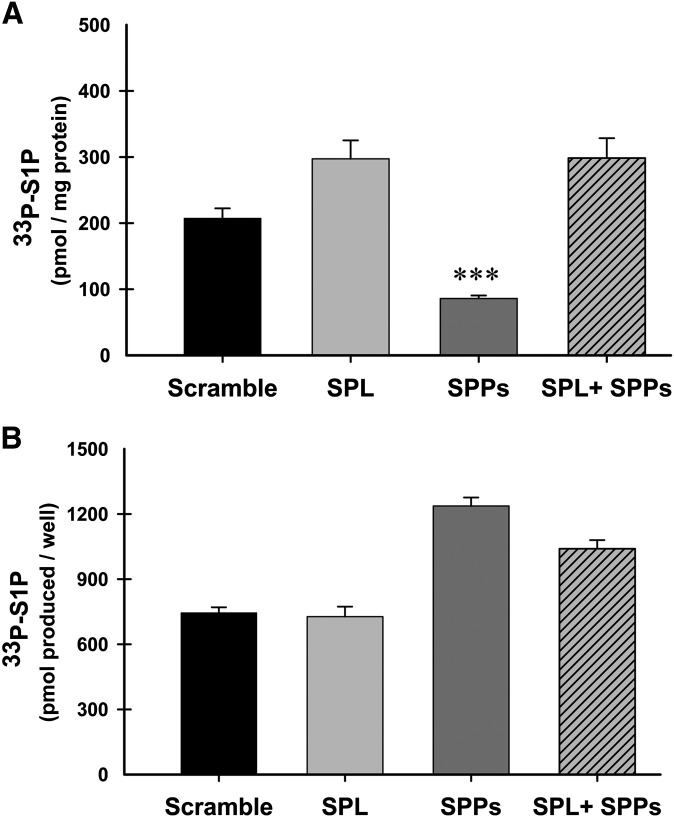
SPP1 and SPP2 knockdown does not inhibit SK1 activity
Because the levels of newly made S1P were reduced when SPP1 and SPP2 were both inhibited, it was important to confirm that siRNA-mediated inhibition of these enzymes was not affecting SK1 activity. To address this issue, we set up separate 24-well plates of HeLa cells that were transfected exactly the same for siRNAs to inhibit the degradative enzymes, followed by transfection with the ER-targeted SK1 construct Cb5. Twenty-four h after transfection with Cb5, one plate was used for a standard in situ assay as described above (Fig. 8A), and the other plate was used for an in vitro kinase assay (Fig. 8B). Even though the in situ assay still results in a significant decrease in S1P levels after SPP1 and SPP2 are knocked down (Fig. 8A, SPPs), the in vitro assay results clearly show that SK1 activity has not been impaired in this system (Fig. 8B, SPPs).
Previous studies demonstrated that SPP knockdown resulted in a modest increase in S1P accumulation in lung endothelial cells (43). This conflicts with our observation that in acute labeling studies, S1P production is diminished. We therefore also measured the effects of SPL and SPP knockdown on mass levels of sphingolipids under the same conditions used for the acute labeling experiments in cells ectopically expressing wild-type SK (Fig. 9) to compare with the acute labeling data using overexpressed SK constructs. Consistent with previously reported results (43), we found that knockdown of SPP1/2 slightly increased S1P levels (Fig. 9A). SPL knockdown also increased S1P and dihydroS1P levels and synergized with SPP1/2 knockdown (Fig. 9A). Sphingosine and dihydrosphingosine levels were only slightly affected by these knockdowns (Fig. 9B). Total ceramide levels were also slightly increased by SPL knockdown (Fig. 9C). Similar results were seen with untransfected cells (supplementary Fig. V). The comparison of the acute labeling studies and the steady-state accumulation measurements may be reconciled by considering that steady-state labeling can be strongly influenced by the turnover of labeled pools. We speculate that there is a rapidly turned over, and thus highly labeled, recycled pool of S1P that depends on SPP1/2 for turnover.
Fig. 9.
Changes in sphingolipid mass levels resulting from knockdown of sphingosine-1-phosphate lyase and sphingosine-1-phosphatases 1 and 2 in cells overexpressing recombinant SK1wt. HeLa cells were transfected with siRNA oligos against the indicated genes for 24 h prior to transfection with wild-type SK1. Cellular lipids were extracted for sphingolipid analysis using LC/MS/MS as described. Data are means ± SD of four replicates. Statistical analysis was performed using an unpaired Student's t-test compared with samples treated with scrambled siRNAs. *P ≤ 0.05, **P ≤ 0.01. A: Sphingosine-1-phosphate (black bars) and dihydrosphingosine-1-phosphate (gray bars). B: Sphingosine (black bars) and dihydrosphingosine (gray bars). C: Total ceramides (black bars) and total dihydroceramides (gray bars). Abbreviations: HeLa, human cervical carcinoma; siRNA, small interfering RNA; SK1, sphingosine kinase 1.
This set of experiments has yielded two surprising results. First, we fully expected localization of sphingosine kinase to impact on the downstream metabolism of sphingosine-1-phosphate given the distinct localization of the lyase and phosphatases to the endoplasmic reticulum. However, we can detect no difference in the degradative fate of sphingosine-1-phosphate generated by a plasma membrane localized SK1 construct (Lck) with the fate of sphingosine-1-phosphate made by an SK1 construct localized to the endoplasmic reticulum (Cb5 and PL16). This result implies that sphingosine-1-phosphate can rapidly move between membrane compartments. Intracellular transport of sphingosine-1-phosphate is a completely unexplored area. Second, we find that depletion of the sphingosine-1-phosphate phosphatases (SPPs) diminishes accumulation of S1P in a sphingosine-1-phosphate lyase-dependent fashion. Although this appears counterintuitive, there is considerable evidence that sphingosine, the product of phosphatase activity on sphingosine-1-phosphate, is compartmentalized. For example, the utilization of exogenous sphingosine in the salvage pathway of ceramide production requires the combined activities of sphingosine kinase and sphingosine-1-phosphate phosphatases in both yeast and mammalian cells (27, 47). Our results suggest a similar recycling pathway exists for newly synthesized sphingosine-1-phosphate. In such a scenario, rapid dephosphorylation of S1P by SPP1 and/or SPP2 keeps the resultant sphingosine in close proximity to SK1 for rephosphorylation. In the absence of SPP1 and SPP2, the S1P is able to escape the protected pool and is quickly degraded by the S1P-specific lyase. Further experiments will be needed to validate this theory and to elucidate possible mechanisms.
CONCLUSIONS
Data presented here establish several new points regarding sphingosine kinase 1 metabolism. First, we show that cytosolic and ER-targeted SK1 are able to draw dihydrosphingosine from the sphingolipid biosynthetic pathway to produce dihydrosphingosine-1-phosphate, confirming earlier results for the cytosolic enzyme (42). This result fits well with a proposed housekeeping role for SK1, whereby basal SK1 activity acts to provide a means for sphingoid bases to be removed from the sphingolipid metabolic network. Additionally, as dihydrosphingosine is the direct precursor for ceramide biosynthesis, this ability of cytosolic and ER-targeted SK1 to preferentially utilize dihydrosphingosine as a substrate could have major implications for maintaining cellular levels of ceramide, a known proapoptotic lipid.
Furthermore, we show that localization of SK1 to the plasma membrane blocks the ability of SK1 to utilize dihydrosphingosine, suggesting that intracellular pools of dihydrosphingosine are not accessible to PM-targeted SK1. As the plasma membrane is the site of translocation for SK1 following stimulation of cells with multiple agonists, the inability of SK1 to access dihydrosphingosine while at the PM could point to an unexplored consequence of translocating SK1 to the PM. As S1P and dihydroS1P have been shown to exert opposite signaling effects in certain cell systems (29, 30), having distinct intracellular pools of the substrate lipids sphingosine and dihydrosphingosine would create yet another level of complexity regarding regulation of SK1. Substrate compartmentalization as a means of SK1 regulation has not been studied.
In contrast to the biosynthetic differences discussed above, our results show no significant variations in the degradative fate of newly made S1P when SK1 was localized to distinct intracellular membranes. We found that when S1P lyase was inhibited using siRNAs, the S1P made at the plasma membrane accumulated to the same extent as the S1P that was made at the endoplasmic reticulum compared with the scrambled siRNA control cells. This was a surprise to us as we expected any newly made S1P that was produced at the plasma membrane to be completely protected from degradation by the lyase pathway. This result implies that S1P can move rapidly (our assays were all complete by 100 min) between intracellular membrane sites, either on its own or with the help of a transport protein. We further show that this degradation is mostly driven by lyase cleavage of S1P. When S1P-specific phosphatases are inhibited by siRNAs, the degradation is significantly enhanced. This result suggests that the concerted actions of SK1 and S1P-specific phosphatases work to create intracellular pools of S1P that are protected from access by the lyase. In the absence of phosphatase activity, these pools of S1P are completely accessible to degradation. This model is driven by strong evidence that in both yeast and mammalian cells, SK and SPP are known to act in concert to deliver sphingosine into a metabolically distinct recycling pool (47, 48).
The concept of compartmentalization of enzymes and lipid pools is yet another layer of complexity in sphingolipid metabolism that is poorly understood. Collectively, our results offer new insights into the role that intracellular localization of sphingosine kinase 1 plays in the regulation of this lipid enzyme, and how differential localization of SK1 might be a way for the enzyme to switch from a housekeeping enzyme to a kinase with signaling roles.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr. Amy Massey and Dr. Christine Simmons for their cloning expertise, which was instrumental in the preparation of our targeted SK1 constructs. We would also like to acknowledge the support of Dr. Viswanathan Natarajan for the LC/MS/MS work done at his facility.
Footnotes
Abbreviations:
- CIB1
- calcium and integrin binding protein 1
- ER
- endoplasmic reticulum
- Hek293
- human embryonic kidney cell
- HeLa
- human cervical carcinoma cell
- LDH
- lactate dehydrogenase
- PM
- plasma membrane
- PMA
- phorbol ester
- siRNA
- small interfering RNA
- SK
- sphingosine kinase
- S1P
- sphingosine-1-phosphate
This work was supported by a predoctoral fellowship from the American Heart Association (D.L.S.); a senior research fellowship from the National Health and Medical Research Council of Australia (S.M.P.); and National Institutes of Health Grant R01 CA-111987 (B.W.W.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of fi ve fi gures.
REFERENCES
- 1.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 2.Le Stunff H., Mikami A., Giussani P., Hobson J. P., Jolly P. S., Milstien S., Spiegel S. 2004. Role of sphingosine-1-phosphate phosphatase 1 in epidermal growth factor-induced chemotaxis. J. Biol. Chem. 279: 34290–34297. [DOI] [PubMed] [Google Scholar]
- 3.Pyne S., Lee S. C., Long J., Pyne N. J. 2009. Role of sphingosine kinases and lipid phosphate phosphatases in regulating spatial sphingosine 1-phosphate signalling in health and disease. Cell. Signal. 21: 14–21. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez S. E., Milstien S., Spiegel S. 2007. Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol. Metab. 18: 300–307. [DOI] [PubMed] [Google Scholar]
- 5.Rosen H., Goetzl E. J. 2005. Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat. Rev. Immunol. 5: 560–570. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel S., Milstien S. 2000. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 476: 55–57. [DOI] [PubMed] [Google Scholar]
- 7.Takabe K., Paugh S. W., Milstien S., Spiegel S. 2008. “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol. Rev. 60: 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kluk M. J., Hla T. 2002. Signaling of sphingosine-1-phosphate via the S1P/EDG-family of G-protein-coupled receptors. Biochim. Biophys. Acta. 1582: 72–80. [DOI] [PubMed] [Google Scholar]
- 9.Kupperman E., An S., Osborne N., Waldron S., Stainier D. Y. 2000. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 406: 192–195. [DOI] [PubMed] [Google Scholar]
- 10.Lee M. J., Thangada S., Claffey K. P., Ancellin N., Liu C. H., Kluk M., Volpi M., Sha'afi R. I., Hla T. 1999. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell. 99: 301–312. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeldt H. M., Hobson J. P., Maceyka M., Olivera A., Nava V. E., Milstien S., Spiegel S. 2001. EDG-1 links the PDGF receptor to Src and focal adhesion kinase activation leading to lamellipodia formation and cell migration. FASEB J. 15: 2649–2659. [DOI] [PubMed] [Google Scholar]
- 12.Fyrst H., Saba J. D. 2008. Sphingosine-1-phosphate lyase in development and disease: sphingolipid metabolism takes flight. Biochim. Biophys. Acta. 1781: 448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda M., Kihara A., Igarashi Y. 2004. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5′-phosphate binding domain exposed to the cytosol. Biochem. Biophys. Res. Commun. 325: 338–343. [DOI] [PubMed] [Google Scholar]
- 14.Le Stunff H., Galve-Roperh I., Peterson C., Milstien S., Spiegel S. 2002. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J. Cell Biol. 158: 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long J., Darroch P., Wan K. F., Kong K. C., Ktistakis N., Pyne N. J., Pyne S. 2005. Regulation of cell survival by lipid phosphate phosphatases involves the modulation of intracellular phosphatidic acid and sphingosine 1-phosphate pools. Biochem. J. 391: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiss U., Oskouian B., Zhou J., Gupta V., Sooriyakumaran P., Kelly S., Wang E., Alfred H., Saba J. D. 2004. Sphingosine phosphate lyase enhances stress-induced ceramide generation and apoptosis. J. Biol. Chem. 279: 1281–1290. [DOI] [PubMed] [Google Scholar]
- 17.Wattenberg B. W., Pitson S. M., Raben D. M. 2006. The sphingosine and diacylglycerol kinase superfamily of signaling kinases: localization as a key to signaling function. J. Lipid Res. 47: 1128–1139. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi J., Erwin P. A., Dantas A. P., Chen H., Michel T. 2003. VEGF induces S1P1 receptors in endothelial cells: implications for cross-talk between sphingolipid and growth factor receptors. Proc. Natl. Acad. Sci. USA. 100: 10664–10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osawa Y., Banno Y., Nagaki M., Brenner D. A., Naiki T., Nozawa Y., Nakashima S., Moriwaki H. 2001. TNF-alpha-induced sphingosine 1-phosphate inhibits apoptosis through a phosphatidylinositol 3-kinase/Akt pathway in human hepatocytes. J. Immunol. 167: 173–180. [DOI] [PubMed] [Google Scholar]
- 20.Waters C., Sambi B., Kong K. C., Thompson D., Pitson S. M., Pyne S., Pyne N. J. 2003. Sphingosine 1-phosphate and platelet-derived growth factor (PDGF) act via PDGFbeta receptor-sphingosine 1-phosphate receptor complexes in airway smooth muscle cells. J. Biol. Chem. 278: 6282–6290. [DOI] [PubMed] [Google Scholar]
- 21.Xia P., Gamble J. R., Rye K. A., Wang L., Hii C. S., Cockerill P., Khew-Goodall Y., Bert A. G., Barter P. J., Vadas M. A. 1998. Tumor necrosis factor-alpha induces adhesion molecule expression through the sphingosine kinase pathway. Proc. Natl. Acad. Sci. USA. 95: 14196–14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia P., Wang L., Gamble J. R., Vadas M. A. 1999. Activation of sphingosine kinase by tumor necrosis factor-alpha inhibits apoptosis in human endothelial cells. J. Biol. Chem. 274: 34499–34505. [DOI] [PubMed] [Google Scholar]
- 23.Pitson S. M., Moretti P. A., Zebol J. R., Xia P., Gamble J. R., Vadas M. A., D'Andrea R. J., Wattenberg B. W. 2000. Expression of a catalytically inactive sphingosine kinase mutant blocks agonist-induced sphingosine kinase activation. A dominant-negative sphingosine kinase. J. Biol. Chem. 275: 33945–33950. [DOI] [PubMed] [Google Scholar]
- 24.Pitson S. M., Xia P., Leclercq T. M., Moretti P. A. B., Zebol J. R., Lynn H. E., Wattenberg B. W., Vadas M. A. 2005. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. J. Exp. Med. 201: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitson S. M., Moretti P. A. B., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., Wattenberg B. W. 2003. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 22: 5491–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarman K. E., Moretti P. A., Zebol J. R., Pitson S. M. 2010. Translocation of sphingosine kinase 1 to the plasma membrane is mediated by calcium and integrin binding protein 1. J. Biol. Chem. 285: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birchwood C. J., Saba J. D., Dickson R. C., Cunningham K. W. 2001. Calcium influx and signaling in yeast stimulated by intracellular sphingosine 1-phosphate accumulation. J. Biol. Chem. 276: 11712–11718. [DOI] [PubMed] [Google Scholar]
- 28.Gratschev D., Lof C., Heikkila J., Bjorkbom A., Sukumaran P., Hinkkanen A., Slotte J. P., Tornquist K. 2009. Sphingosine kinase as a regulator of calcium entry through autocrine sphingosine 1-phosphate signaling in thyroid FRTL-5 cells. Endocrinology. 150: 5125–5134. [DOI] [PubMed] [Google Scholar]
- 29.Bu S., Yamanaka M., Pei H., Bielawska A., Bielawski J., Hannun Y. A., Obeid L., Trojanowska M. 2005. Dihydrosphingosine 1-phosphate stimulates MMP1 gene expression via activation of ERK1/ 2-Ets1 pathway in human fibroblasts. FASEB J. 20: 184–186. [DOI] [PubMed] [Google Scholar]
- 30.Bu S., Kapanadze B., Hsu T., Trojanowska M. 2008. Opposite effects of dihydrosphingosine 1-phosphate and sphingosine 1-phosphate on transforming growth factor-beta/Smad signaling are mediated through the PTEN/PPM1A-dependent pathway. J. Biol. Chem. 283: 19593–19602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitson S. M., D'Andrea R. J., Vandeleur L., Moretti P. A., Xia P., Gamble J. R., Vadas M. A., Wattenberg B. W. 2000. Human sphingosine kinase: purification, molecular cloning and characterization of the native and recombinant enzymes. Biochem. J. 350 (Pt. 2): 429–441. [PMC free article] [PubMed] [Google Scholar]
- 32.Siow D. L., Wattenberg B. W. 2007. An assay system for measuring the acute production of sphingosine 1-phosphate in intact monolayers. Anal. Biochem. 371: 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu W., Cowie A., Wasfy G. W., Penn L. Z., Leber B., Andrews D. W. 1996. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 15: 4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 34.Wattenberg B. W., Clark D., Brock S. 2007. An artificial mitochondrial tail signal/anchor sequence confirms a requirement for moderate hydrophobicity for targeting. Biosci. Rep. 27: 385–401. [DOI] [PubMed] [Google Scholar]
- 35.Brock S. E., Li C., Wattenberg B. W. 2010. The Bax carboxy-terminal hydrophobic helix does not determine organelle-specific targeting but is essential for maintaining Bax in an inactive state and for stable mitochondrial membrane insertion. Apoptosis. 15: 14–27. [DOI] [PubMed] [Google Scholar]
- 36.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 37.Maceyka M., Milstien S., Spiegel S. 2007. Measurement of mammalian sphingosine-1-phosphate phosphohydrolase activity in vitro and in vivo. Lipidomics and Bioactive Lipids: Lipids and Cell Signaling. Methods in Enzymology (series). Brown H. A., editor Academic Press, San Diego, CA: 243–256. [DOI] [PubMed] [Google Scholar]
- 38.Berdyshev E. V., Gorshkova I. A., Garcia N., Natarajan V., Hubbard W. C. 2005. Quantitative analysis of sphingoid base-1-phosphates as bisacetylated derivatives by liquid chromatography-tandem mass spectrometry. Anal. Biochem. 339: 129–136. [DOI] [PubMed] [Google Scholar]
- 39.Johnson K. R., Becker K. P., Facchinetti M. M., Hannun Y. A., Obeid L. M. 2002. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate (PMA). J. Biol. Chem. 277: 35257–35262. [DOI] [PubMed] [Google Scholar]
- 40.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson K. R., Johnson K. Y., Becker K. P., Bielawski J., Mao C., Obeid L. M. 2003. Role of human sphingosine-1-phosphate phosphatase 1 in the regulation of intra- and extracellular sphingosine-1-phosphate levels and cell viability. J. Biol. Chem. 278: 34541–34547. [DOI] [PubMed] [Google Scholar]
- 42.Berdyshev E. V., Gorshkova I. A., Usatyuk P., Zhao Y., Saatian B., Hubbard W., Natarajan V. 2006. De novo biosynthesis of dihydrosphingosine-1-phosphate by sphingosine kinase 1 in mammalian cells. Cell. Signal. 18: 1779–1792. [DOI] [PubMed] [Google Scholar]
- 43.Taha T. A., Kitatani K., El-Alwani M., Bielawski J., Hannun Y. A., Obeid L. M. 2005. Loss of sphingosine kinase-1 activates the intrinsic pathway of programmed cell death: modulation of sphingolipid levels and the induction of apoptosis. FASEB J. 20: 482–484. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y., Kalari S. K., Usatyuk P. V., Gorshkova I., He D., Watkins T., Brindley D. N., Sun C., Bittman R., Garcia J. G. N., et al. 2007. Intracellular generation of sphingosine 1-phosphate in human lung endothelial cells. J. Biol. Chem. 282: 14165–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigal Y. J., McDermott M. I., Morris A. J. 2005. Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem. J. 387: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Stunff H., Giussani P., Maceyka M., Lépine S., Milstien S., Spiegel S. 2007. Recycling of sphingosine is regulated by the concerted actions of sphingosine-1-phosphate phosphohydrolase 1 and sphingosine kinase 2. J. Biol. Chem. 282: 34372–34380. [DOI] [PubMed] [Google Scholar]
- 48.Futerman A. H., Riezman H. 2005. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15: 312–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.