Abstract
Methyl-CpG binding proteins (MBDs) mediate histone deacetylase-dependent transcriptional silencing at methylated CpG islands. Using chromatin immunoprecitation (ChIP) we have found that gene-specific profiles of MBDs exist for hypermethylated promoters of breast cancer cells, whilst a common pattern of histone modifications is shared. This unique distribution of MBDs is also characterized in chromosomes by comparative genomic hybridization of immunoprecipitated DNA and immunolocalization. Most importantly, we demonstrate that MBD association to methylated DNA serves to identify novel targets of epigenetic inactivation in human cancer. We combined the ChIP assay of MBDs with a CpG island microarray (ChIP on chip). The scenario revealed shows that, while many genes are regulated by multiple MBDs, others are associated with a single MBD. These target genes displayed methylation- associated transcriptional silencing in breast cancer cells and primary tumours. The candidates include the homeobox gene PAX6, the prolactin hormone receptor, and dipeptidylpeptidase IV among others. Our results support an essential role for MBDs in gene silencing and, when combined with genomic strategies, their potential to ‘catch’ new hypermethylated genes in cancer.
Keywords: ChIP on chip/DNA methylation/epigenetic inactivation/MBD/MeCP2
Introduction
DNA methylation, the major epigenetic modification of mammalian genomes, plays an active role in transcriptional repression (Cedar, 1988). Over the past 10 years, increasing evidence has emerged surrounding the active role of CpG island hypermethylation of tumour suppressor genes in cancer development and progression (Esteller, 2002). One of the necessary steps in the epigenetic pathway to cancer involves methyl-CpG binding proteins (MBDs) that mediate transcriptional silencing of the hypermethylated gene promoters. The mammalian family of MBD proteins is composed of five members, namely MeCP2, MBD1, MBD2, MBD3 and MBD4. With the exception of MBD4, which is involved in DNA repair, all MBD proteins associate with histone deacetylases (HDACs) and couple DNA methylation with transcriptional silencing through the modification of chromatin (Ballestar and Wolffe, 2001; Wade, 2001; Prokhortchouk and Hendrich, 2002). Moreover, recent research has demonstrated the presence of MBD proteins in mediating the transcriptionally silenced state of several promoters of hypermethylated tumour suppressor genes in cancer, and in imprinted and X-chromosome inactivated genes in normal cells (Magdinier and Wolffe, 2001; Nguyen et al., 2001; Bakker et al., 2002; El-Osta et al., 2002; Fournier et al., 2002).
An essential issue concerns the physiological relevance of the existence of four different MBD-containing co-repressor complexes. The most straightforward explanation is that each complex is targeted to a different subset of genes. To understand the specific roles of different MBDs it is of inherent interest to biochemically characterize the MBD-containing complexes. Thus far, MBD3 is the best-characterized member of the MBD family. It has been reported to be an integral component of the Mi-2/NuRD complex (Wade et al., 1999; Zhang et al., 1999) that also contains a nucleosome remodelling ATPase, HDAC1 and HDAC2 and other proteins. Mammalian MBD3, in contrast to its Xenopus homologue, does not selectively bind methylated DNA and in mammals the Mi-2/NuRD complex is targeted to methylated DNA through association with any of the two forms of MBD2: MBD2a or MBD2b. This combination of Mi-2/NuRD and MBD2 may be synonymous of the MeCP1 complex, the first identified to display binding activity towards methylated DNA (Feng and Zhang, 2001). The Mi-2/NuRD complex may also interact with other sequence-specific DNA binding proteins to cause transcriptional repression. In the case of MBD1, although early reports suggested HDAC-dependent repression (Ng et al., 2000), recent data indicate that MBD1 represses transcription in an HDAC-independent manner that involves association with a novel chromatin-associated factor (Fujita et al., 2003).
An alternative approach in understanding the biology of MBD proteins arises from the study of MBD targets. Reports on MBD association to methylated loci have revealed a complex picture. On the one hand, a single association of MBD proteins to methylated loci (Magdinier and Wolffe, 2001; Bakker et al., 2002) has been described. However, multiple recruitment of MBD proteins has been reported in several genes (Fournier et al., 2002; Koizume et al., 2002). Early studies indicated significant differences in the association of MBDs to DNA. For instance, MBD2/MeCP1 is released from nuclei by low salt, suggesting that it is not stably complexed with DNA and seems to require densely methylated DNA. That aside, MBD1 can also affect transcription from unmethylated and hypomethylated promoters (Fujita et al., 2000). The reasons behind the specific role of each MBD-containing complex remain to be explored. The use of chromatin immunoprecipitation (ChIP) with antibodies specific for a particular MBD together with immunocytological analysis potentially provide powerful tools to identify the set of genes controlled by each particular MBD-containing complex and therefore a way to understand the individual roles of each of these complexes.
As a first step towards determining the targeting of MBD proteins in a genome-wide context, we have combined individualized studies for each MBD (i.e. ChIP assays for several promoters of tumour suppressor genes) with global genomic approaches [5-methylcytosine (mC) analysis of the MBD-immunoprecipitated products and comparative genomic hybridization]. Most importantly, we have also combined ChIP analysis with a CpG island microarray to define the global profile of MBD targeting. Our studies show the existence of a unique pattern of MBD-binding sites in transformed cells and unmask a new set of epigenetically silenced genes in cancer.
Results
A gene-specific profile of MBDs exists in hypermethylated CpG island promoters
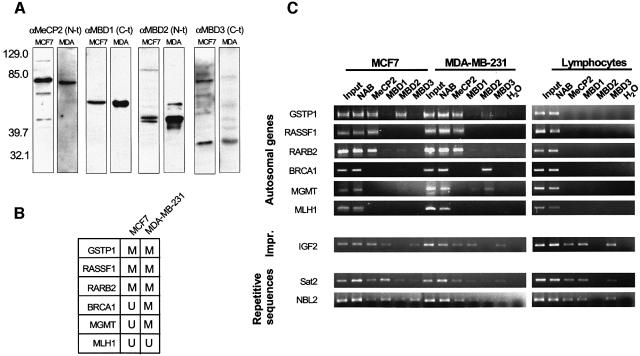
To investigate the involvement of MBDs in epigenetic repression, we initially adopted a candidate gene approach to study genes known to be hypermethylated in a breast tumour model, specifically in the breast cancer cell lines MCF7 and MDA-MB-231. ChIP analyzes were performed using antibodies raised against epitopes unique to each MBD protein (MeCP2, MBD1, MBD2 and MBD3). We have recently validated these antibodies in investigating the recruitment of MBD proteins to imprinted-methylated loci (Fournier et al., 2002). We first performed western blot analysis with these antibodies in all the cell lines studied to ensure that the MBD proteins were indeed expressed (Figure 1A). ChIP assays were then performed in order to identify which particular MBD protein was associated with methylated DNA sequences in MCF7 and MDA-MB-231 cells. Additionally, normal lymphocytes, representing non-tumoural non-cultured cells, and non-transformed lymphoblastoid cell lines were also used as negative controls.
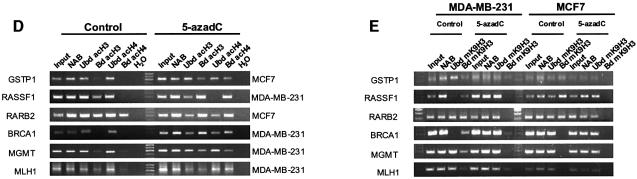
Fig. 1. ChIP analysis of the occupancy by MBD proteins and histone modification status of several hypermethylated promoters. (A) MBD antisera αMeCP2 N-t, αMBD1 C-t, αMBD2 N-t, αMBD3 C-t were tested on MCF7 and MDA-MB-231 nuclear extracts. On the left, molecular size bands of a pre-stained standard (Kaleidoscope, BioRad) are indicated. Comparable results were obtained with MDA-MB-231 nuclear extracts. (B) A summary of the methylation status of the studied promoters in MCF7 and MDA-MB-231 (extracted from Esteller, 2002). (C) MBD occupancy analysed by ChIP assay. Input and ‘unbound’ fraction of the no antibody (NAB) control are shown followed by the four ‘bound’ fractions for each antibody. ChIP assays shown correspond to MCF7 and MDA-MB-231 cells and isolated lymphocytes. Results with lymphoblastoid cell lines are undistinguishable from those obtained with control lymphocytes. Three groups of sequences are shown: CpG islands tumour suppressor genes, an imprinted gene (IGF2) and repetitive sequences (Sat2 and NBL2). (D) Analysis of the acetylation status of each promoter. Commercial anti-acetylH3 and anti-acetylH4 were used (Upstate Biotechnologies). Control cells and 5-azadC treated cells are shown. (E) Analysis of the methylation status of K9 of H3 is studied. An H3 antibody to a branched peptide with four fingers of the K9-dimethylated TARKST sequence (Lachner et al., 2001) was used.
Three types of DNA sequences were analyzed: the CpG islands in the promoter of six tumour suppressor genes, for which DNA methylation and expression status is well characterized (Esteller et al., 2001; Paz et al., 2003a), the CpG island of the imprinted gene IGF2 (insulin-like growth factor 2) and two different repetitive sequences, namely Sat2 (satellite 2) and NBL2 (a non-satellite repeat). The tumour suppressor genes explored were the Ras association domain family 1A gene (RASSF1A), the glutathione S-transferase P1 (GSTP1) gene, the retinoic acid receptor B2 gene (RARB2), the breast cancer 1 gene (BRCA1), the O6-methylguanine-DNA methyltransferase (MGMT) gene and the mutL-homologue 1 (MLH1). A summary of the methylation status of the promoters of these genes is shown in Figure 1B. MBD binding was not observed in any unmethylated promoter. However, a specific profile of MBD occupancy was found for the methylated CpG islands. Whilst MeCP2 and MBD2 were both present in the methylated GSTP1 promoter, for RASSF1A and RARB2 only MeCP2 was bound, and for BRCA1 and MGMT the only MBD present was MBD2 (Figure 1C). In none of the six selected promoters did we find any association with MBD1 or MBD3. In contrast, these six promoters are unmethylated in lymphocytes and lymphoblastoid cell lines and do not exhibit any binding of MBDs in these normal cells (Figure 1C). As a positive control, MBD association to the methylated regions of the IGF2 imprinted gene and the repetitive sequences Sat2 and NBL2 was observed for all the cells analyzed (MDA-MB-231, MCF7, normal lymphocytes and lymphoblastoid cell lines) (Figure 1C).
The presence of DNA methylation and MBD association was accompanied by histone deacetylation and lysine 9 histone H3 methylation at these promoters (Figure 1D and E). The restoration of histone acetylation and demethylation on histone H3 both occurred rapidly with the use of the demethylating agent 5-aza-2-deoxycytidine. Consistent with our results and previous findings (Nguyen et al., 2002), we also found that the promoters that were unmethylated and devoid of MBDs were enriched in acetylated histones and H3 lysine 9 was demethylated (Figure 1D and E).
MBDs immunoprecipitate methylated DNA
The candidate gene approach to identify MBD targets, although necessary, is time consuming. Therefore, to overcome this drawback in a global screen for MBD targets, we employed three independent genomic approaches, using as starting material the immunoprecipitated DNA for each MBD.
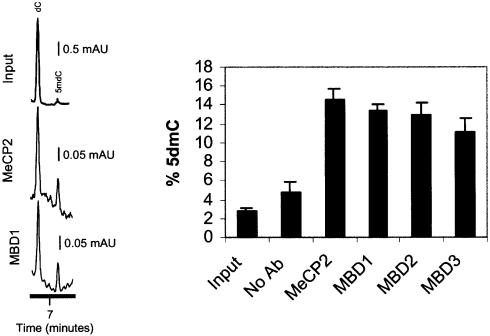
We first analyzed, by high performance capillary electrophoresis (Fraga and Esteller 2002), the total mC DNA content of each MBD-immunoprecipitated DNA in MCF7 cells. An average 3- to 5-fold enrichment in mC DNA in this MBD–ChIP DNA versus the input DNA was observed (Figure 2), strongly supporting the notion that MBD proteins are tightly associated in vivo with methylated DNA sequences. Furthermore, a gradient of mC DNA content versus the overall amount of bound DNA was observed, with MeCP2 being the protein that binds to the highest ratio of methylated cytosines in the human genome, and MBD3 the lowest (Figure 2). These results further support the differential distribution of MBD binding within the genome.
Fig. 2. Analysis of global DNA methylation by HPCE of MBD-immunoprecipitated samples. Two electropherograms, corresponding to the input fraction, the MBD1 and MBD2 immunoprecipitated DNAs from MCF7 cells, are shown. The graph shows the methylcytosine content in the same fractions.
MBDs are associated with extensive chromosomal regions
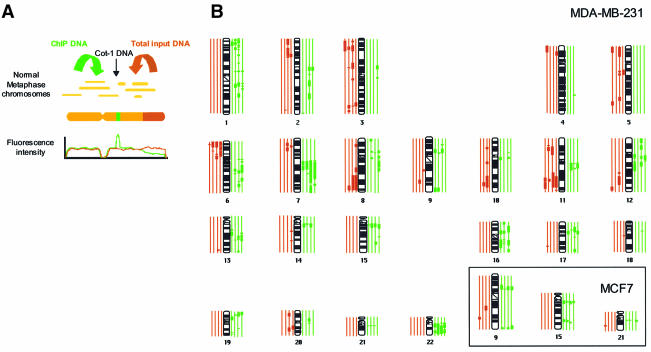
We further explored the distribution of each MBD protein throughout particular chromosomal regions by using a modification of the comparative genomic hybridization (CGH) protocol (Cigudosa et al., 1998). We labelled the DNA from MCF7 and MDA-MB-231 immunoprecipitated with each one of the MBDs antibodies, and then was competitively hybridized against input DNA from these cells (Figure 3A). We found that the MBD-immunoprecipitated sequences, seen as thick green segments in the CGH karyotype, were unevenly distributed along the chromosomes, yielding a characteristic pattern with broad megabase-length bands (Figure 3B). It is noteworthy that this pattern of banding differs from the one obtained when MDA-MB-231 or MCF7 DNA is competitively hybridized against DNA extracted from normal cells (see, for example, Xie et al., 2002), discarding artifacts due to the aneuploidy of these cancer cells. A remarkable finding is that many of the resulting bands are shared between both the different MBDs and also the two cell lines studied (Figure 3B, see inset). For instance, there were common specific regions of chromosomes 1 (at band 1q13), 9 (9p13), 11 (11q13), 13 (13q21), 15 (15q13), 16 (16p11.2), 17 (17q11.2), 19p, 21 (21q11.2), and 22q recurrently enriched in ChIP isolated sequences in both cell lines (Figure 3B, see inset). Quantitative differences in MBD distribution, however, were still very apparent as the number of regions that contained immunoprecipitated sequences was higher for MBD3 and MeCP2 (53 and 48 segments) versus those obtained with MBD1 and MBD2 (34 and 37 segments). Furthermore, a quantitative difference was also observed between both breast cancer cell lines: chromosomes in MDA-MB-231 contained a much higher proportion of MBD–ChIP sequences compared with MCF7, reflecting the higher number of hypermethylated CpG islands present in MDA-MB-231 versus MCF7 (Paz et al., 2003a). Our CGH data suggest that MBDs tend to be clustered at certain chromosomal loci, an observation that is consistent with the appearance of nuclear domains obtained with MBD antisera in immunolocalization experiments (Hendrich and Bird, 1998). We have observed that immunostaining with anti-MBD antisera produces a combined pattern of discrete foci in a context of diffuse nuclear staining (data not shown). The diffuse MBD pattern colocalizes with mC staining (especially in the case of MBD1) and some of the MBD foci are also coincident with mC spots (data not shown). This observation may reflect that some of the observed MBD nuclear distribution is due more to architectural features of the nucleus than to genome sequence patterns.
Fig. 3. Comparative genomic hybridization of MBD-immunoprecipitated DNAs in metaphase chromosomes. (A) Diagram showing the combination of comparative genome hybridization with the ChIP assay. (B) Ideograms showing hybridization of ChIP DNAs on the human karyotype. Thick vertical lines on either side of the chromosome ideogram indicate only recurrent MBD enrichment (green) or exclusion (red) of a chromosome or a chromosomal region. The four green and four red lines correspond, respectively, to MBD1, MBD2, MBD3 and MeCP2 from inside to outside. The analysis shown corresponds to MDA-MB-231 samples. The inset shows three chromosomes from the MCF7 samples.
MBDs identify novel hypermethylated genes in cancer
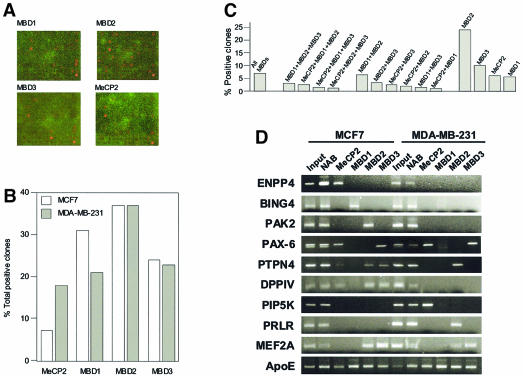
The resolution of CGH and immunolocalization does not allow the identification of specific sequences associated with MBD proteins. To obtain an accurate and comprehensive profile of the genes targeted by MBDs in human breast cancer, we have combined chromatin immunoprecipitation and array technology (ChIP on chip). We have probed the MCF7 and MDA-MB-231 DNA from ChIP assays using the four different MBD antisera described above with a DNA microarray that contains a library of 7777 CpG islands. This CpG island microarray provides a high throughput method for the identification of in vivo nuclear factor targets, as has been demonstrated for E2F (Weinmann et al., 2002). Three independent hybridizations of the CpG island microarray with three independent chromatin-immunoprecipitated samples were performed for each MBD protein in each of the cell lines. From a purely quantitative standpoint, MBD2 produced the highest number of positive clones, supporting the in vitro data that showed that MBD2 has the highest affinity for methylated DNA 8 (Fraga et al., 2003), followed by MBD1, MBD3 and MeCP2 (Figure 4B). The array hybridization results across all spotted CpG islands show that 7.1% of the clones were positive for all four MBDs, while 24.2, 10.3, 9 and 5.8%, were positive singly for MBD2, MBD3, MeCP2 and MBD1, respectively (Figure 4C). Therefore, the data indicate that MBD target sequences tend to be occupied by either a single MBD (preferentially MBD2) or all of them, rather than double or triple occupancy.
Fig. 4. (A) Representative microarray images for different MBD-immunoprecipitated MDA-MB-231 samples. Cy5 labelled chromatin DNAs were hybridized to CpG island microarray as described in the text. The hybridization images were acquired and normalized signal intensities of hybridized spots were compared to those of the control (total input). (B) Graph showing the percentage of total positive clones (relative to the total input control) obtained for each MBD-immunoprecipitated DNA. (C) Graph showing the percentage of positive clones for the different combinations: all MBD proteins, three MBDs, two MBDs or a single MBD. (D) Confirmation of MBD targets by individual ChIP analysis and PCR with primers designed to the individual loci. Input, no antibody (unbound fraction) and MBD immunoprecipitated fractions (bound) are shown.
In order to identify the genes immunoprecipitated by the MBDs, we sequenced 60 CpG island-positive clones that were common to three independent experiments: 10 from the subgroup immunoprecipitated by all MBDs, 10 from each of the four subgroups immunoprecipitated by a unique MBD, and 10 from a subgroup immunoprecipitated by both MBD2 and MBD3. This last subset was chosen upon the basis of reports regarding MBD2 and MBD3 links within the MeCP1 complex (Feng et al., 2001; Hendrich et al., 2001). Table I summarizes the sequencing data and blast search results. The list of novel MBD target genes in MCF7 and MDA-MB-231 breast cancer cells includes excellent candidate genes for the malignant phenotype. Some of these genes have already been associated with cancer, such as the prolactin hormone receptor (PRLR) (Jonathan et al., 2002), the protein- tyrosine phosphatase non-receptor type (PTPN4) (Ogata et al., 1999), the dipeptidyl peptidase IV (DPPIV) (Pethiyagoda et al., 2000) and PIP5K (Chatah and Abrams, 2001). It was essential to confirm that the identified clones were bound by MBDs in vivo and the results from individual ChIPs with MCF7 and MDA-MB-231 cells with the four MBD antibodies confirmed the results from the CpG island microarray (Figure 4D). For sequences immunoprecipitated with MeCP2 antiserum, results were confirmed using the corresponding commercial antibody (data not shown). As a negative control, we performed individual ChIP analysis with isolated lymphocytes and lymphoblastoid cell lines and we found that MBDs were absent in the promoters of these genes.
Table I. MBD targets identified by ChIP-CpG microarray analysis.
| Gene | Accession No. | Location | MBD | Present in other cell line |
|---|---|---|---|---|
| MCF7 cells | ||||
| clone RP11–403I13 | AL356957 | MeCP2 | Also MDA-MB-231 | |
| clone RP11–99O17 | AC018706 | 7p15.2 | MeCP2 | |
| DKFZp686D16148 | AL701693 | 19q13.42 | MeCP2 | Also MDA-MB-231 |
| enpp4 | AL035701 | 6p21.1 | MeCP2 | |
| FLJ00132 | AK074061 | 11q12.2 | MeCP2 | |
| shoygo (ax09h10) | BG941206 | 4p14 | MBD1 | Also MDA-MB-231 |
| bing4 | NM_005452 | 6p21.32 | MBD1 | |
| clone RP11–576P10 | 7p11.2-p21 | MBD1 | ||
| pak2 | 3p24.3 | MBD2 | ||
| clone RP1-77N19 | Z98886 | 1p36.2–36.3 | MBD2 | |
| tbx19 | AJ010277 | 1q23-q24 | MBD2 | |
| atp5i | BAA78778 | 4p16.3 | MBD2 | Also MDA-MB-231 |
| cox6c | P09669 | 8q22-q23 | MBD3 | Also MDA-MB-231 |
| LOC199704 | XM_113994 | 19q13.13 | MBD3 | |
| leng6 | AF211971 | 19q13.4 | MBD3 | Also MDA-MB-231 |
| clone CTC-454I21 | AC012309 | 19q13.13 | MBD3 | |
| stearoyl-CoA desaturase | BAA93510 | 10q24.33 | MBD3 | |
| Paired box protein pax-6 | CAB05885 | 11p13 | MBD3 | Also MDA-MB-231 |
| mps-1 | A48045 | MBD2 + MBD3 | Also MDA-MB-231 | |
| cox6c | P09669 | 8q22-q23 | MBD2 + MBD3 | |
| ptpn4 | NM 002830 | 2q14.1 | MBD2 + MBD3 | Also MDA-MB-231 |
| efna5 | P52803 | 5q21 | All MBDs | |
| bcat2 | NM_001190 | 19q13 | All MBDs | |
| dppiv | P27487 | 2q24.3 | All MBDs | |
| MDA-MB-231 cells | ||||
| clone: IMAGE:4550691 | BG331429 | 4p16.3 | MeCP2 | |
| pip5k | BC007833 | 1q22-q24 | MeCP2 | |
| FLJ32618 | AK057180 | 16p13.13 | MeCP2 | |
| FLJ32618 | AK057180 | 16p13.13 | MeCP2 | |
| FLJ32760 | AK057322 | 2q37.3 | MeCP2 | |
| CS0DJ015YH07 | AL558734 | 9p13.3 | MeCP2 | |
| oatp-d | AB031050 | 15q26.1 | MBD1 | |
| clone YB67A06 | AF147380 | 1q31.1 | MBD1 | |
| ptprm | NM_002845 | 18p11.23 | MBD1 | |
| clone RP1-207F6 | AL133258 | 6p22.1 | MBD2 | |
| kcnk10 | NM_021161 | 14q31.3 | MBD2 | |
| prlr | NM_000949 | 5p13.3 | MBD2 | |
| clone RP5-947L8 | AL355178 | 1p34.1–36.11 | MBD2 | |
| clone RP13–202B6 | AL591625 | Xp21.1–21.3 | MBD3 | |
| bat5 | NM_021160 | 6p21.33 | MBD3 | |
| DKFZp564E1878 (hspc228) | AL136684 | 12q13.3 | MBD2 + MBD3 | |
| HSPC228 | NM_016485 | 6q24.1 | MBD2 + MBD3 | Also MCF7 |
| HSPC228 | NM_016485 | 6q24.1 | MBD2 + MBD3 | Also MCF7 |
| mef2a | NM_005587 | 15q26.3 | MBD2 + MBD3 | Also MCF7 |
| alox12b | AF038461 | 17p13.1 | All MBDs | |
| Apolipoprotein E precursor (Apo-E) | P02649 | 16p11.2 | All MBDs | Also MCF7 |
| clone: IMAGE:3086912 | BF509821 | 11q12.3 | All MBDs | |
| Beta crystallin B2 (crybb2) | P43320 | 22q11.23 | All MBDs | |
| FLJ14146 | NM_024709 | 1q42.12 | All MBDs | |
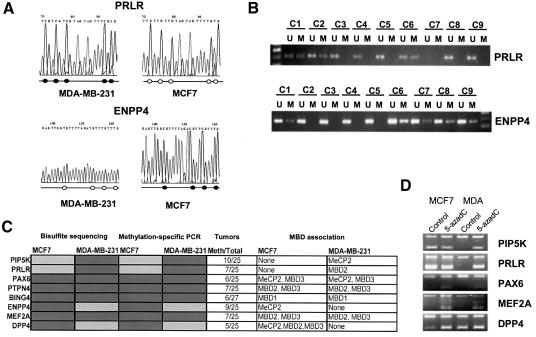
Once their association to MBD proteins was confirmed, this subset of genes was subjected to further characterization for DNA methylation and expression. Bisulfite genomic sequencing spanning their corresponding CpG islands was used to demonstrate hypermethylation in the corresponding breast cancer cell lines where MBD binding had been confirmed by ChIP analysis (MCF7, MDA-MB-231 or both) (Figure 5A and C). Furthermore, these results were confirmed by methylation-specific PCR analysis (Figure 5C). DNA extracted from isolated lymphocytes and lymphoblastoid cell lines was also analyzed for the methylation status of all the above genes by both bisulfite genomic sequencing and methylation-specific PCR. In both cases, all these genes were unmethylated in these normal cells in agreement with the absence of MBD binding previously demonstrated.
Fig. 5. DNA methylation and expression analysis of the particular genes found using the ChIP on chip approach. (A) Bisulfite genomic sequencing of the PRLR CpG island demonstrating hypermethylation in MDA-MB-231 cells and hypomethylation in MCF7 cells. On the contrary, the ENPP4 CpG island is hypermethylated in MCF7 cells but not in MDA-MB-231 cells. A fragment of the sequence is shown. Unmethylated Cs become Ts upon bisulfite modification. Below, a schematic representation of some of the CpG sites included in the PCR fragment is shown. CpG sites are represented as circles that are black when methylated. (B) Methylation-specific PCR confirms the presence of hypermethylation in ENPP4 and PRLR primary breast tumours. A number of cases are shown, indicated as c1–c9. (C) Summary table with the bisulfite sequence, methylation-specific PCR results for MCF7, MDA-MB-231 and primary breast tumours. Dark and light shading indicates methylation or no methylation, respectively. (D) Expression analysis monitored by RT–PCR after co-amplification with GAPDH. In all panels, the top band (400 bp) is GAPDH and the bottom band (of ∼200 bp) is each of the specific genes.
To address the functional consequences of the CpG island promoter hypermethylation in association with the binding of MBDs, we performed expression analysis of the described genes. In all analyzed cases, we found that genes with unmethylated sequences and no MBD binding were expressed, while hypermethylated MBD-associated genes were transcriptionally silenced. In these latter cases, the use of 5-aza-2′-deoxycytidine was able to restore gene expression (Figure 5D). Furthermore, we demonstrated that this CpG island hypermethylation was not merely a feature of these particular breast cancer cell lines, but was present in a significant proportion of human primary breast carcinomas (Figure 5B and C).
Two MBD-associated genes, PAX6 and PRLR, inhibit growth in MDA-MB-231 cells
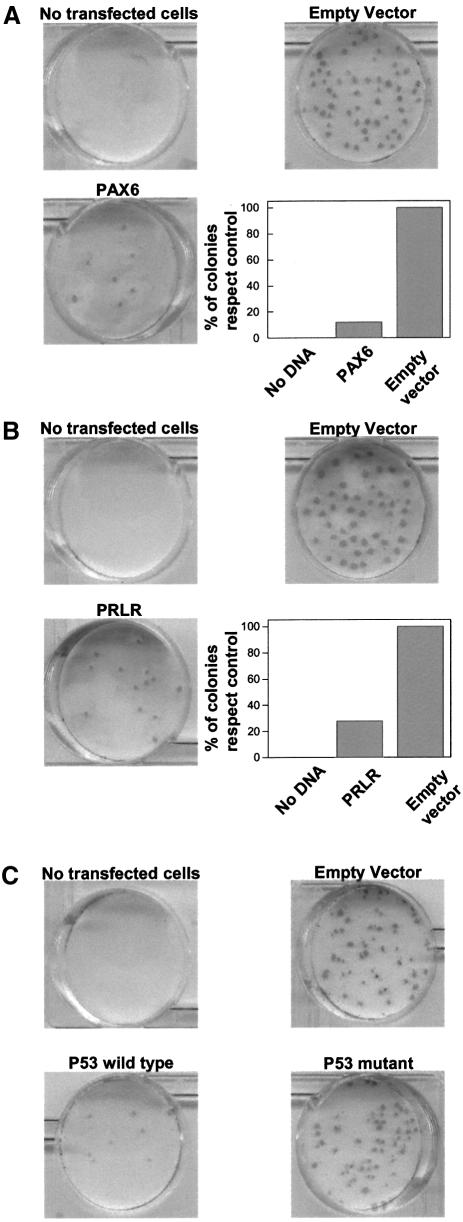
To gain further insight into the potential role of some hypermethylated genes identified by our ChIP on chip strategy of genomic screening, we decided to reintroduce some of these genes into the MDA-MB-231 cell line, as carried out for other hypermethylated tumour suppressor genes (Tomizawa et al., 2001; Yoshikawa et al., 2001). Expression vectors containing PAX6 and PRLR were independently used to transfect MDA-MB-231 cells, in which both genes were silenced in association with methylation. As positive control for growth inhibition, we used wild-type p53. A mutant form of p53 (N175H) was used as an additional negative control, besides empty vector. Forty-eight hours after transfection, cells were selected with neomycin, and resistant colonies developing after 16 days were stained. As expected, the wild-type p53 control dramatically suppressed colony formation while mutant p53 produced a similar number of colonies to that of the empty vector (Figure 6C). Interestingly, we found that the number of neomycin-resistant colonies transfected with PAX6 and PRLR was reduced by 84 and 70% when compared with the control vector in three independent experiments (Figure 6A and B). Thus, these data support the hypothesis that the loss of the expression of these genes in breast cancer cells is related to cell growth.
Fig. 6. The effect of exogenous expression of PAX6 and PRLR on the colony formation of MDA-MB-231 cells. PAX6 and PRLR methylation-silenced cells (i.e. MDA-MB-231) were transfected with a PAX6 (A) and PRLR (B) expression vector or the empty vector as indicated in Materials and methods. Relative number of resistant colonies after transfection, calculated as the percentage of colonies with respect to the untransfected cells, are shown in the inset. (C) Effect of the exogenous expression of wild-type and mutant p53.
Discussion
It has been estimated that 70% of all CpG dinucleotides in the mammalian genome are methylated. The majority of the remaining unmethylated CpG sites are located within the CpG islands found frequently near the promoter and first exons regions of protein-coding genes. In cancer, inhibition of transcription by hypermethylation of the CpG islands contributes to the inactivation of regulatory or DNA repair genes (Jones and Baylin, 2002). It has been proposed that MBD proteins serve as the bridge between histone modification enzymes and hypermethylated DNA associated with gene inactivation (Bird and Wolffe, 1999).
Our results support the notion that the presence of MBD proteins is an essential and general feature in the methylation-mediated silencing of tumour suppressor and DNA repair genes. First, we have identified MBD proteins in all selected promoters previously characterized for their hypermethylated status. Secondly, in a general screening of MBD targets by using chromatin immunoprecipitated as a probe in a CpG island microarray we have found that all the positive clones studied were methylated.
Interestingly, our candidate gene and genome-wide analyzes of the distribution of MBDs throughout the genome reveal a complex picture. For instance, the MBD pattern of association appears to be gene specific. Whilst some sequences associate with only one specific MBD, others associate several or all of them (see Figure 1C; Table I). For example, MeCP2 is the only MBD that we have identified in association with the RASSF1A, RARB2, ENPP4 and PIP5K promoters, among others. For other sequences, such as that for the ApoE promoter or the NBL2 non-satellite repeat, all MBDs were found to be associated (Figures 1C and 4D). The data from the CpG microarray hybridization suggest that there is a slight tendency for single association or association of all MBDs rather than intermediate states of double or triple occupancy.
Comparison of the association of MBD2 and MBD3 with DNA constitutes a particularly interesting case. Although mammalian MBD3 does not directly bind methylated DNA (Fraga et al., 2003), it has been proposed that the MBD3-containing Mi-2/NuRD complex is recruited to methylated sequences through the recruitment of MBD2 (Feng and Zhang, 2001). For this reason, we investigated whether the pattern of distribution in chromosomes and throughout the CpG microarrays clones was similar. However, the antisera to MBD2 only recognizes the MBD2a subfraction, as the antibody is developed the N-terminus of MBD2a, which is absent in the MBD2b form. Therefore, the partial coincidence in the distribution of MBD2 and MBD3 could be due to the fact that our antibodies do not immunoprecipitate MBD2b-interacting sequences.
Our global methylcytosine quantitation of chromatin immunoprecipitated with MBD antibodies and immunolocalization studies strongly support the notion that these proteins are truly associated in vivo with methylated sequences. As mentioned above, even MBD3 pulls down methylated DNA, probably due to the interaction of MBD2 (which possesses high specificity for methylated DNA) with the MBD3-containing complex. The ability of these proteins to associate with methylated DNA is corroborated by the methylation analysis of MBD targets identified by ChIP on chip. Interestingly, the finding that MBD proteins only partially colocalize with methylcytosine in vivo suggests that other factors may also participate in the nuclear localization of MBD proteins. This is also supported by the distribution of MBDs over metaphasic chromosomes. When combining the banded pattern obtained by CGH with the sequences obtained by ChIP on chip (see Table I) or known methylated genes, for which MBD association has been demonstrated, several MBD-targeted loci are confirmed. However the degree of overlapping is not complete. For instance, genes like RARB2 or BRCA1 do not coincide with any of the MBD-associated bands identified by CGH. On the other hand some of the conserved chromosomal bands, like 15q11 in which all MBD proteins appear to be associated, coincide with the Prader–Willi imprinting centre, which has a well-known methylation pattern (Perk et al., 2002). Thus, the banded pattern on chromosomes obtained by CGH may reflect that this distribution is in part due to architectural features of nuclear organization.
We have also shown that chromatin immunoprecipitated with antibodies to different MBDs and used as a hybridization probe for a CpG island microarrays is an ideal material to identify novel genes that become methylated in cancer. The ChIP on chip approach provides a high-throughput method of identifying MBD-associated CpG islands. It is also worth noting that all the genes we have selected for further characterization of their methylation and expression status were actually epigenetically silenced. The success of the CpG island microarray leads us to believe that this approach can be utilized to systematically identify targets of hypermethylation in cancer (see also Paz et al., 2003b). The discovery of novel hypermethylated genes can be used to identify potential genes involved in cancer. For instance, HIC1, the first hypermethylated gene defined as a candidate tumour suppressor in the absence of mutations, has been shown recently to possess such function through a mouse knockout model (Chen et al., 2003).
In a cancer cell system, such as the one we utilized here, a high number of sequences become methylated. Not every gene is methylated in every tumour type (Esteller et al., 2001). Instead, a clear specificity in the pattern of methylated genes is observed depending on the tumour type. It has been hypothesized, as with genetic mutations, that preferential methylation of certain genes confers a Darwinian evolutionary advantage to different cancer cells (Esteller, 2002). Supporting this idea, we have shown that the reintroduction of some of these identified genes inhibited colony formation. It is, therefore, plausible that many of the novel methylated genes that can be identified by our approaches are good candidates to provide an advantage to the cancer cells when expression is lost. This is the case for some of the genes we have been able to identify by our approach. For instance, the prolactin receptor, identified here for its association to MBD2 in MDA-MB-231 cells, has been previously reported to be associated to mammary gland differentiation (Kelly et al., 2002).
In summary, our integrated approach reveals the existence of a specific profile of MBD differential occupancy for hypermethylated CpG islands—promoters of tumour suppressor genes in transformed cells. This singular pattern of MBD binding, determined first at single loci and later through whole genomic approaches, provides the first evidence of the ubiquitous presence of MBDs in the methylated CpG island promoters and demonstrates their usefulness in identifying newly epigenetically inactivated genes in cancer cells.
Materials and methods
Antibodies and cell lines
Polyclonal rabbit antisera were made against linear peptides corresponding to amino acids 6–25 of human MeCP2 (antiserum αMeCP2 N-t), 2–20 of human MBD2 (αMBD2 N-t), 254–270 of human MBD3 (αMBD3 C-t) and the KQEPPDPEEDKEENKDDSAS peptide common to all the MBD1 isoforms (αMBD1 C-t). These antibodies were tested for their specific immunoprecipitating ability (Lefkovits and Kohler, 1997). A comparison of targets immunoprecipitated with these antibodies and commercial αMeCP2 and αMBD2 was also performed (Fournier et al., 2002). Commercial antibodies against MeCP2 (Upstate Biotechnologies) were also used for ChIP assays.
MCF7 and MDA-MB-231 cells were cultured in Dulbecco’s modified Eagle’s medium with 4.5 g/l glucose and l-glutamine supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. B-lymphoblastoid cell lines were established by isolating lymphocytes from whole blood by gradient centrifugation with Ficoll/Hypaque (Amershan). Two million cells were resuspended in 2 ml of the supernatant of B95.8 cell line (that produces Epstein–Barr virus), 1 ml of RPMI medium containing 20% fetal bovine serum and phytohemagglutinin PHA.P. After 7 days of primary culture, the medium was changed every 3 days.
Chromatin immunoprecipitation analysis
The ChIP assay was performed as previously described (Fournier et al., 2002). Preliminary time-course experiments between 5 min and 1 h of formaldehyde fixation were performed to yield the best combination of in vivo fixed chromatin, high DNA recovery and small average size of chromatin fragments. In our analyses, chromatin was sheared to an average length of 0.25–1 kb. PCR amplification was performed in 25 µl with specific primers for each of the analysed promoters. For each promoter, the sensitivity of PCR amplification was evaluated on serial dilutions of total DNA collected after sonication (input fraction). Three independent ChIP experiments were performed for each cell line. In PCR amplifications, the input DNA, ‘unbound’ (wash) fraction of the no antibody control and, finally, ‘unbound’ and ‘bound’ fractions for each antibody were run in 2% agarose gels. Primers and conditions for each promoter are available upon request.
Global 5-methylcytosine quantification
Quantification of the mC content was carried out by high performance capillary electrophoresis as previously described (Fraga and Esteller, 2002). In brief, immunoprecipitated DNA samples were speed-back pre-concentrated to 0.1 µg/µl and enzymatically hydrolyzed in a final volume of 5 µl. Samples were then directly injected in a Beckman MDQ high performance capillary electrophoresis apparatus and mC content was determined as the percentage of mC of total cytosines: mC peak area × 100/(C peak area + mC peak area).
Competitive hybridization of ChIP products to metaphase chromosomes
To study the distribution along the chromosomes of the immunoprecipitated DNA for each one of the proteins, we designed a modified version of the CGH strategy (Cigudosa et al., 1998). Four hybridizations were performed with each MCF7 and MDA-MB-231 cell line. In each hybridization, we compared the input DNA fraction from each ChIP experiment versus the immunoprecipitated DNA with the specific protein (MBD1, MBD2, MBD3 and MeCP2). Input DNA was labelled with Spectrum Red dUTP by CGH nick-translation kit (Vysis, Inc., IL, USA) and the immunoprecipitated fractions were labelled with Spectrum Green. Control experiments (conventional CGH assays) were carried out with input DNAs from the cell lines. The metaphases were captured through a fluorescent microscope (Olympus BX60) and a CCD camera (Photometrics Sensys camera), then analysed using the chromofluor image analysis system (Cytovision; Applied Imaging Ltd, Newcastle, UK). For each hybridization a high number of chromosomes (13–25) were analysed.
Immunostaining and confocal fluorescence microscopy
Isolated nuclei of MCF7 were resuspended in 3 volumes of methanol and fixed for 5 min at room temperature. Drops of fixed nuclei were spotted on glass coverslips and air dried for 10 min at 37°C. For chromatin decondensation, coverslips were incubated in 1 N HCl for 30 min at 37°C, briefly washed in water and incubated for 5 min in standard Tris–borate buffer. Nuclei were rinsed in PBS, blocked with 2% BSA in PBS, and double-stained with mouse monoclonal antibodies against mC in conjunction with rabbit polyclonal antibodies against the indicated MBDs (see above). Confocal optical sections were obtained using a Leica TCS SP confocal microscope (Leica Microsystems, Heidelberg GMbH, Germany) equipped with krypton and argon lasers, and images were processed using Adobe Photoshop 5.0 (Adobe Systems Inc., Mountain View, CA).
CpG island microarray analysis
The immunoprecipitated fractions from ChIP experiments were labelled and used to probe a CpG microarray as previously described (Jonathan et al., 2002). Microarray slides containing the 7776 CpG island loci and the DNA samples were processed essentially as described previously (Yan et al., 2002). Each test sample, including the no antibody controls, was labelled with Cy5 (red) and was hybridized onto the slides, which were processed according to the method of De Risi et al. (http://www.microarrays.org). We then scanned the slides using GenePix 4000A (Axon) and analysed the images with the GenePix Pro4.0 program. Three hybridizations from independent ChIP experiments were performed for each MBD protein and cell line. Normalization was applied using the global average intensity of hybridized loci for each slide. Selection of positive clones was done from the common positive clones among the three independent hybridization experiments.
DNA methylation analysis of particular genes
We carried out bisulfite modification of genomic DNA as described previously (Herman et al., 1996). We established methylation status by PCR analysis of bisulfite-modified genomic DNA using two procedures. First, all genes studied were analysed by bisulfite genomic sequencing of their corresponding CpG islands, as described elsewhere (Clark and Warnecke, 2002). Both strands were sequenced. The second analysis used methylation-specific PCR for all genes analysed in MCF7, MDA-MB-231 and tissue samples, as previously described (Herman et al., 1996). Placental DNA treated in vitro with SssI methyltransferase was used as positive control for all methylated genes. We designed all of the bisulfite genomic sequencing and methylation-specific PCR primers according to genomic sequences around presumed transcription start sites of investigated genes. Primer sequences and PCR conditions for methylation analysis are available upon request.
Semiquantitative RT–PCR expression analysis
We reverse-transcribed total RNA (2 µg) treated with DNase I (Ambion) using oligo-dT primer with Superscript II reverse transcriptase (Gibco BRL). We carried out PCRs in a 25-µl volume containing 1× PCR buffer (Gibco BRL), 1.5 mM of MgCl2, 0.3 mM of dNTP, 0.25 µM of each primer and 2 U of Taq polymerase (Gibco BRL). We used 100 ng of cDNA for PCR amplification, and we amplified all of the genes with multiple cycle numbers (20–35 cycles) to determine the appropriate conditions for obtaining semi-quantitative differences in their expression levels. RT–PCR primers were designed between different exons to avoid any amplification of DNA. PCRs were performed simultaneously with two sets of primers, with GAPDH as internal control to ensure cDNA quality and loading accuracy. Primer sequences are available upon request.
Cell culture and transfection
MDA-MB-231 was grown in DMEM (4.5 g/l glucose) supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin and 2.5 mg/ml amphotericin in a humidified 37°C, 5% CO2 incubator, and maintained in log phase growth.
The transfection of the pSV-β-galactosidase control vector, pcDNA-3-PRLR and pcDNA-PAX6, also containing neomycin resistance gene into the MDA-MB-231 cells was performed by the lipofection method. Briefly, cells were seeded in a six-well plate a day before transfection at a density of 2 × 105 cells/well. Purified plasmid DNA (2 µg) was transfected with LipofectAmine Plus reagent, according to the manufacturer’s recommendations. The experiment was repeated three times. Twenty-four hours after transfection, βgal activity was measured using a βgal activity kit, in the cells carrying the control vector. Clones expressing the proteins transfected were selected in complete medium supplemented with 1 mg/ml geneticin (G418) 48 h post-transfection. Stable clones were maintained in complete medium with G418 (800 µg/ml). Total RNA from individual clones was extracted and RT–PCR was performed to confirm that the clones were expressing the transfected genes. MDA-MB-231 cells were also transfected with the vector containing no inserts and stable clones isolated. After ∼16 days of selection, stable G418-resistant colonies were fixed and stained with 2% methylene blue in 60% methanol. Wild-type p53 (pWZL-hp53 wt; Hygro) and mutant p53 (pWZL-hp53-N175H; Hygro) were used as additional positive and negative controls.
Acknowledgments
Acknowledgements
We are grateful to Thomas Jenuwein for antiserum to H3-K9 methylation and for providing details about its specificity. We also thank Alain Niveleau for antiserum to mC. We would like to thank Manuel Serrano, Charles Clevenger and Ales Cvekl for providing us with expression vectors for transfection experiments. This work was supported by I+D+I project SAF 2001-0059 and the International Rett Syndrome Association. E.B. is funded by the Ramón y Cajal Program.
References
- Bakker J., Lin,X. and Nelson,W.G. (2002) Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J. Biol. Chem., 277, 22573–22580. [DOI] [PubMed] [Google Scholar]
- Ballestar E. and Wolffe,A.P. (2001) Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem., 268, 1–6. [DOI] [PubMed] [Google Scholar]
- Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression: belts, braces and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Cedar H. (1988) DNA methylation and gene activity. Cell, 53, 3–4. [DOI] [PubMed] [Google Scholar]
- Chatah N.E. and Abrams,C.S. (2001) G-protein-coupled receptor activation induces the membrane translocation and activation of phosphatidylinositol-4-phosphate 5-kinase I α by a Rac- and Rho-dependent pathway. J. Biol. Chem., 276, 34059–34065. [DOI] [PubMed] [Google Scholar]
- Chen W.Y., Zeng,X., Carter,M.G., Morrell,C.N., Chiu Yen,R.W., Esteller,M., Watkins,D.N., Herman,J.G., Mankowski,J.L. and Baylin,S.B. (2003) Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat. Genet., 33, 197–202. [DOI] [PubMed] [Google Scholar]
- Cigudosa J.C., Rao,P.H., Calasanz,M.J., Odero,M.D., Michaeli,J., Jhanwar,S.C. and Chaganti,R.S. (1998) Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood, 91, 3007–3010. [PubMed] [Google Scholar]
- Clark S.J. and Warnecke,P.M. (2002) DNA methylation analysis in mammalian cells. Methods, 27, 99–100. [DOI] [PubMed] [Google Scholar]
- El-Osta A., Kantharidis,P., Zalcberg,J.R. and Wolffe,A.P. (2002) Precipitous release of methyl-CpG binding protein 2 and histone deacetylase 1 from the methylated human multidrug resistance gene (MDR1) on activation. Mol. Cell. Biol., 22, 1844–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2002) CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene, 21, 5427–5440. [DOI] [PubMed] [Google Scholar]
- Esteller M., Corn,P.G., Baylin,S.B. and Herman,J.G. (2001) A gene hypermethylation profile of human cancer. Cancer Res., 61, 3225–3229. [PubMed] [Google Scholar]
- Feng Q. and Zhang,Y. (2001) The MeCP1 complex represses transcription through preferential binding, remodeling and deacetylating methylated nucleosomes. Genes Dev., 15, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C., Goto,Y., Ballestar,E., Delaval,K., Hever,A., Esteller,M. and Feil,R. (2002) Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J., 21, 6560–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M.F. and Esteller,M. (2002) DNA methylation: a profile of methods and applications. Biotechniques, 33, 632–649. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar,E., Montoya,G., Taysavang,P., Wade,P.A. and Esteller,M. (2003) The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res., 31, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Shimotake,N., Ohki,I., Chiba,T., Saya,H., Shirakawa,M. and Nakao,M. (2000) Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol., 20, 5107–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Watanabe,S., Ichimura,T., Ohkuma,Y., Chiba,T., Saya,H. and Nakao,M. (2003) MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol., 23, 2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Guy,J., Ramsahoye,B., Wilson,V.A. and Bird,A. (2001) Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev., 15, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J.G., Graff,J.R., Myohanen,S., Nelkin,B.D. and Baylin,S.B. (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA, 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonathan N., Liby,K., McFarland,M. and Zinger,M. (2002) Prolactin as an autocrine/paracrine growth factor in human cancer. Trends Endocrinol. Metab., 13, 245–250. [DOI] [PubMed] [Google Scholar]
- Jones P.A. and Baylin,S.B. (2002) The fundamental role of epigenetic events in cancer. Nat. Rev. Genet., 3, 415–428. [DOI] [PubMed] [Google Scholar]
- Kelly P.A., Bachelot,A., Kedzia,C., Hennighausen,L., Ormandy,C.J., Kopchick,J.J. and Binart,N. (2002) The role of prolactin and growth hormone in mammary gland development. Mol. Cell. Endocrinol., 197, 127–131. [DOI] [PubMed] [Google Scholar]
- Koizume S., Tachibana,K., Sekiya,T., Hirohashi,S. and Shiraishi,M. (2002) Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic Acids Res., 30, 4770–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine-9 creates a binding site for Hp1 protein. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Lefkovits I. and Kohler,G. (1997) Immunology Methods Manual: The Comprehensive Sourcebook of Techniques. Academic Press, Orlando, FL, USA. [Google Scholar]
- Magdinier F. and Wolffe,A.P. (2001) Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl Acad. Sci. USA, 98, 4990–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Jeppesen,P. and Bird,A. (2000) Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol., 20, 1394–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.T., Gonzales,F.A. and Jones,P.A. (2001) Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res., 29, 4598–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C.T., Weisenberger,D.J., Velicescu,M., Gonzales,F.A., Lin,J.C., Liang,G. and Jones,P.A. (2002) Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res., 62, 6456–6461. [PubMed] [Google Scholar]
- Ogata M., Takada,T., Mori,Y., Oh-hora,M., Uchida,Y., Kosugi,A., Miyake,K. and Hamaoka,T. (1999) Effects of overexpression of PTP36, a putative protein tyrosine phosphatase, on cell adhesion, cell growth and cytoskeletons in HeLa cells. J. Biol. Chem., 274, 12905–12909. [DOI] [PubMed] [Google Scholar]
- Paz M.F., Fraga,M.F., Avila,S., Mingzhou,G., Herman,J.G. and Esteller,M. (2003a) A systematic profile of DNA methylation in human cancer cell lines. Cancer Res., 63, 1114–1121. [PubMed] [Google Scholar]
- Paz M.F., Wei,S., Cigudosa,J.C., Rodriguez-Perales,S., Peinado,M.A., Huang,T.H. and Esteller,M. (2003b) Genetic unmasking of epigenetically silenced tumor suppressor genes in colon cancer cells deficient in DNA methyltransferases. Hum. Mol. Genet., 12, 2209–2219. [DOI] [PubMed] [Google Scholar]
- Perk J., Makedonski,K., Lande,L., Cedar,H., Razin,A. and Shemer,R. (2002) The imprinting mechanism of the Prader–Willi/Angelman regional control center. EMBO J., 21, 5807–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethiyagoda C.L., Welch,D.R. and Fleming,T.P. (2000) Dipeptidyl peptidase IV (DPPIV) inhibits cellular invasion of melanoma cells. Clin. Exp. Metastasis, 18, 391–400. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk E. and Hendrich,B. (2002) Methyl-CpG binding proteins and cancer: are MeCpGs more important than MBDs? Oncogene, 21, 5394–5349. [DOI] [PubMed] [Google Scholar]
- Tomizawa Y. et al. (2001) Inhibition of lung cancer cell growth and induction of apoptosis after reexpression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proc. Natl Acad. Sci. USA, 98, 13954–13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P.A. (2001) Methyl CpG binding proteins and transcriptional repression. BioEssays, 23, 1131–1137. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Yan,P.S., Oberley,M.J., Huang,T.H. and Farnham,P.J. (2002) Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev., 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D., Jauch,A., Miller,C.W., Bartram,C.R. and Koeffler,H.P. (2002) Discovery of over-expressed genes and genetic alterations in breast cancer cells using a combination of suppression subtractive hybridiz ation, multiplex FISH and comparative genomic hybridization. Int. J. Oncol., 21, 499–507. [PubMed] [Google Scholar]
- Yan P.S., Efferth,T., Chen,H.L., Lin,J., Rodel,F., Fuzesi,L. and Huang,T.H. (2002) Use of CpG island microarrays to identify colorectal tumors with a high degree of concurrent methylation. Methods, 27, 162–169. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Matsubara,K., Qian,G.S., Jackson,P., Groopman,J.D., Manning,J.E., Harris,C.C. and Herman,J.G. (2001) SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet., 28, 29–35. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation.Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]