Abstract
Conjugated linoleic acids (CLA) are dietary fatty acids. Whereas cis-9,trans-11-(c9,t11)-CLA can be found in meat and dairy products, trans-9,trans-11-(t9,t11)-CLA is a constituent of vegetable oils. Previous studies showed that these two isomers activate different nuclear receptors and, thus, expression of genes related to lipid metabolism. Here we show that these CLA isomers are differentially elongated and desaturated in primary monocyte-derived macrophages isolated from healthy volunteers by using gas chromatography-mass spectrometry (GC-MS). We further demonstrate that c9,t11-CLA incorporates in phosphatidylcholine (PC) and phosphatidylethanolamine (PE) species and activates de novo glycerophospholipid synthesis by quantitative electrospray ionization-tandem mass spectrometry (ESI-MS/MS). c9,t11-CLA leads to strong shifts of the species profiles to PC 18:2/18:2 and PE 18:2/18:2, which are due to de novo synthesis and fatty acid remodeling. In contrast, t9,t11-CLA is preferentially bound to neutral lipids, including triglycerides and cholesterol esters. Taken together our results show that c9,t11-CLA and t9,t11-CLA have differential effects on PC and PE metabolism. Moreover, these data demonstrate that the structure of fatty acids not only determines their incorporation into lipid classes but also modulates the kinetics of lipid metabolism, particularly PC synthesis.
Keywords: fatty acid, mass spectrometry, phosphatidylcholine, phosphatidylethanolamine, phospholipid metabolism
Dietary fatty acids (FA) are reported to have numerous pleiotropic effects on cellular metabolism and function, including modulation of gene expression through activation of nuclear receptors and alteration of membrane phospholipid composition (1).
Conjugated linoleic acid (CLA) refers to a group of positional and geometrical (cis/trans) isomers of linoleic acid (c9,c12-FA 18:2). Many CLA isomers have been described; however, the major isomer occurring in meat and dairy products is c9,t11-CLA (2–4). It is produced in the rumen through microbial biohydrogenation of linoleic acid and in tissues by delta-9 desaturation of rumen-derived trans-vaccenic acid (t11-FA 18:1) (5). In contrast, t9,t11-CLA is the predominant isomer found in dietary oils, as it is generated during partial hydrogenation of vegetable oils and oil refining (6, 7). Several data from in vitro and animal studies show that dietary CLAs are beneficial and influence the progression of several diseases, including cardiovascular and inflammatory diseases and cancer (8–11).
We have previously reported that two isomers in particular, c9,t11- and t9,t11-CLA, have specific and even contrasting effects on gene expression associated with lipid metabolism of human macrophages (12, 13). Concerning CLA metabolism, it has been shown that c9,t11-CLA incorporates into phospholipids of leukemia cells and can be found in plasma and cellular lipids of healthy men supplemented with a diet enriched with this CLA isomer (14, 15). However, so far the effects of c9,t11-CLA on individual phospholipid classes, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and its cellular distribution are not precisely described, and data on the metabolism of the t9,t11-CLA isomer are completely unavailable.
Therefore we investigated c9,t11-CLA and t9,t11-CLA metabolism by quantitative lipid mass spectrometry. As a cellular model, human primary monocyte-derived macrophages were chosen because these cells are highly active with respect to fatty acid and phospholipid metabolism (16). Moreover macrophages, which are multifunctional cells present in all tissues of the human body, play important roles in several metabolic diseases, including atherosclerosis and obesity (17). We found that these two CLA isomers differentially affect macrophage phospholipid metabolism. c9,t11-CLA in contrast to t9,t11-CLA activates PC and PE synthesis and incorporates into these membrane lipids.
EXPERIMENTAL PROCEDURES
Reagents and materials
t9,t11-CLA and c9,t11-CLA were purchased from Cayman Chemicals. 13C3-serine, D4-ethanolamine, and D9-choline were obtained from Cambridge Isotope Laboratories.
Monocyte isolation and cell culture
Primary human monocytes were obtained from healthy donors by leukapheresis and counterflow elutriation as described previously (16). For metabolic labeling studies, cells were incubated with 50 μg/ml of [13C3]serine, [D4]ethanolamine, and [D9]choline chloride.
Fatty acid analysis
Fatty acid analysis was performed as described previously with slight modifications (16). Briefly, fatty acid methyl esters (FAME) were generated with acetyl-chloride and methanol over night at room temperature and extracted with hexane. Total FA analysis was carried out using a Shimadzu 2010 GC-MS system. FAMEs were separated by a BPX70 column (10 m length, 0.10 mm diameter, 0.20 µm film thickness) from SGE using helium as carrier gas. The initial oven temperature was 50°C, which was programmed to increase with 40°C per min to 155°C, with 6°C per min to 210°C, and with 15°C per min to finally reach 250°C. The FA species and their positional and cis/trans isomers were characterized in scan mode and quantified by single-ion monitoring (SIM) mode detecting the specific fragments of saturated and unsaturated FAs (saturated: m/z 74; monounsaturated: m/z 55; diunsaturated: m/z 67; polyunsaturated: m/z 79). As an internal standard, nonnaturally occurring C13:0 was used.
Lipid extraction
Lipids were extracted according to the procedure described by Bligh and Dyer in the presence of not naturally occurring lipid species as internal standards (18). The chloroform phase was dried in a vacuum centrifuge and dissolved as described below for quantitative lipid analysis.
Lipid analysis
Lipids were quantified by electrospray ionization tandem mass spectrometry (ESI-MS/MS) in positive ion mode as described previously (19). In brief, samples were analyzed by direct flow injection using a HTS PAL autosampler, an Agilent 1100 binary pump, and triple quadrupole mass spectrometer (Quattro Ultima, Micromass). A precursor ion scan of m/z 184 specific for phosphocholine-containing lipids was used for PC (20). D9-choline labeled lipids were analyzed by precursor ion scan of m/z 193. Neutral loss scans of m/z 141 and m/z 185 were used for PE and PS, respectively. Analogous, neutral loss scans were used for stable isotope labeled D4-PE (m/z 145), 13C2-PE (m/z 143) and 13C3-PS (m/z 188). Free cholesterol (FC) and cholesteryl ester (CE) were quantified using a fragment ion of m/z 369 after selective derivatization of FC using acetyl chloride (21). Correction of isotopic overlap of lipid species and data analysis by self-programmed Excel macros were performed for all lipid classes. For all lipid classes, nonnaturally occurring lipid species were used as internal standards. Quantification was performed by standard addition calibration to cell homogenates using a number of naturally occurring lipid species for each lipid class.
Product ion spectra were generated in negative ion mode using a hybrid triple quadrupole linear ion trap mass spectrometer API 4000 Q-Trap (Applied Biosystems, Darmstadt, Germany) in the enhanced product ion spectrum mode at a scan speed of 1000 amu/s.
Thin-layer chromatography
PC, PE, triglycerides, and free fatty acids (FFA) were separated as described previously (22, 23). For GC-MS analysis of distinct lipid classes, the appropriate bands were scraped, homogenized in methanol, and subsequently used for FAME derivatization.
Statistical analysis
The level of significance for the difference between data sets was assessed using Student's independent t-test (*P < 0.01).
RESULTS
CLA metabolism: elongation and desaturation to conjugated fatty acids
First we characterized kinetics of cellular CLA uptake. Primary monocyte-derived macrophages isolated from healthy volunteers were treated with 10 µM and 30 µM CLA for 4 h and 24 h. Intracellular CLA concentrations were quantified by GC-MS. Both CLA isomers showed a time- and concentration-dependent uptake with a slightly faster uptake of t9,t11- compared with c9,t11-CLA (Table 1).
TABLE 1.
c9,t11-CLA is elongated and desaturated, whereas t9,t11-CLA is mainly elongated in primary monocyte derived macrophages
| Treatment | Time | Concentration | c9,t11-CLA | SEM | t9,t11-CLA | SEM | CFA 20:2 | SEM | CFA 22:2 | SEM | CFA 24:2 | SEM | CFA 18:3 | SEM | CFA 20:3 | SEM | CFA 20:4 | SEM | % CLA Metabolism |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c9,t11-CLA | 4 h | 10 µM | 34.28 | 2.12 | 0.22 | 0.04 | 1.00 | 0.07 | 0.55 | 0.07 | 5.14 | ||||||||
| 30 µM | 98.62 | 3.15 | 0.83 | 0.03 | 1.17 | 0.04 | 0.63 | 0.02 | 2.67 | ||||||||||
| 24 h | 10 µM | 65.47 | 3.95 | 0.56 | 0.02 | 3.22 | 0.05 | 0.10 | 0.01 | 3.63 | 0.14 | 11.47 | |||||||
| 30 µM | 158.46 | 4.88 | 1.30 | 0.04 | 5.52 | 0.11 | 0.27 | 0.04 | 7.28 | 0.14 | 9.06 | ||||||||
| t9,t11-CLA | 4 h | 10 µM | 66.20 | 7.31 | 0.56 | 0.04 | 0.36 | 0.03 | 1.39 | ||||||||||
| 30 µM | 203.45 | 5.38 | 0.56 | 0.08 | 0.66 | 0.07 | 0.60 | ||||||||||||
| 24 h | 10 µM | 89.68 | 10.17 | 4.54 | 0.14 | 0.97 | 0.03 | 0.64 | 0.03 | 6.86 | |||||||||
| 30 µM | 210.93 | 21.49 | 7.13 | 0.43 | 2.29 | 0.18 | 0.94 | 0.01 | 4.91 |
Potential metabolites were determined with GC-MS. Boldface type: >1. Concentration: nmol/mg cell protein. CFA, conjugated fatty acid; CLA, conjugated linoleic acid; conc, concentration.
Next we asked whether c9,t11- and t9,t11-CLA are further metabolized to other fatty acids within the cells. Using GC-MS, we screened for new metabolites at different incubation and concentration times. Potential metabolites were identified by retention time analogies and their fragment patterns as standards for these metabolites are not available. In cells supplemented with c9,t11-CLA, we found metabolites fitting to conjugated fatty acids (CFA) 18:3 generated by desaturation and CFA 20:4 obtained through further elongation and desaturation (Table 1). Interestingly, we did not observe any desaturated CFAs originated from t9,t11-CLA; we found only the elongated metabolites CFA 20:2 and CFA 22:2. Of the incorporated c9,t11-CLA and t9,t11-CLA, 11% and 7%, respectively, were metabolized after 24 h to CFAs in macrophages.
c9,t11-CLA strongly affects cellular PC and PE species composition
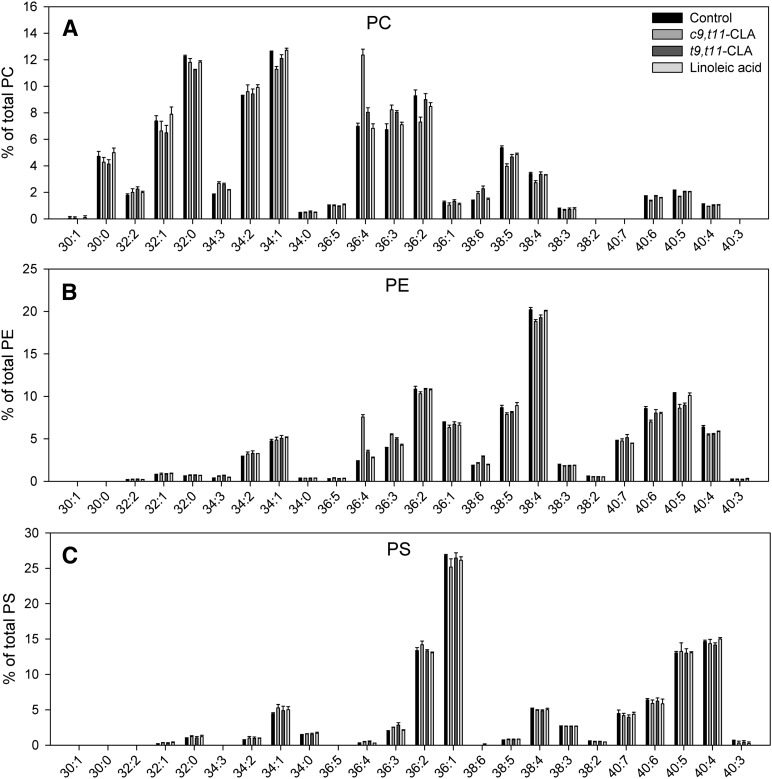
To test whether c9,t11-CLA or t9,t11-CLA modulate the species pattern of the individual glycerophospholipid classes, primary cells were supplied with single CLA isomers and PC, PE and PS species profiles were investigated by ESI-MS/MS. When cells were treated with c9,t11-CLA for 4 h we found a striking shift to PC 36:4, which was not observed when macrophages were treated with t9,t11-CLA or linoleic acid (Fig. 1A). As for PC, c9,t11-CLA incubation led to a shift to PE 36:4 (Fig. 1B), again t9,t11-CLA or linoleic acid had no effects on PE 36:4. The PS species profile was neither affected by c9,t11-CLA nor by t9,t11-CLA (Fig. 1C).
Fig. 1.
c9,t11-CLA, but not t9,t11-CLA induces a shift to PC 36:4 and PE 36:4. A: PC species profile for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS (PC 36:4 upon c9,t11-CLA treatment: P < 0.001). B: PE species profile for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS (PE 36:4 upon c9,t11-CLA treatment: P < 0.001). C: PS species profile for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS. Values are mean ± SD of one representative experiment from three, each performed in triplicate. CLA, conjugated linoleic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
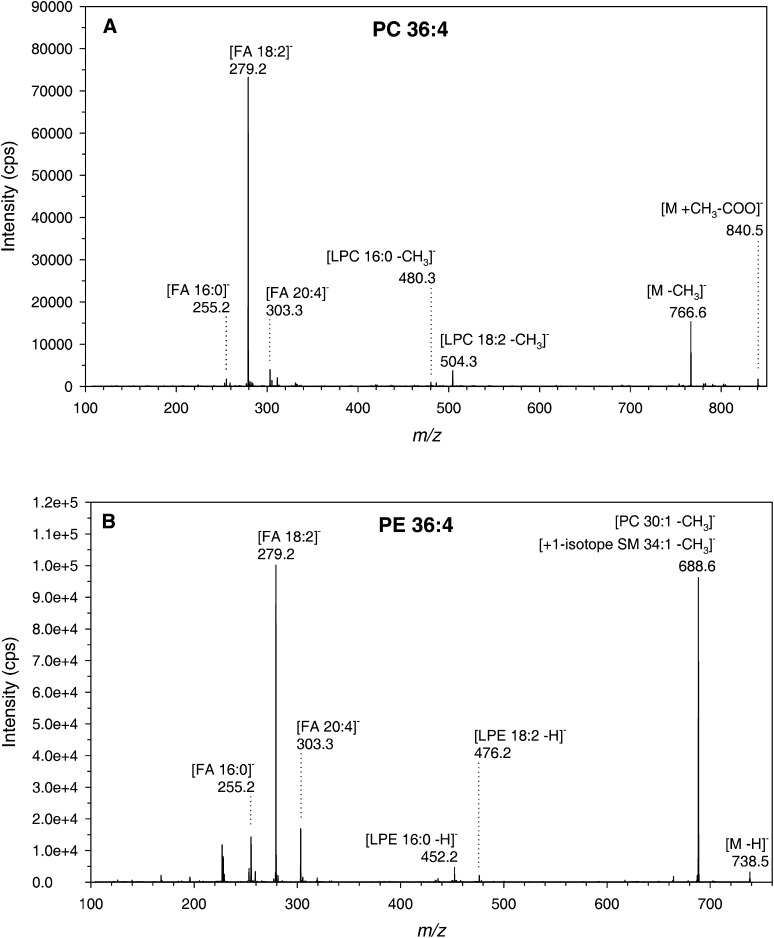
The fatty acyl compositions of PC/PE 36:4 were determined from product ion spectra in negative ion mode using fragmentation pattern described previously (24–27). We found that PC 18:2/18:2 and PE 18:2/18:2 primarily contribute to PC 36:4 and PE 36:4, because the major product ions detected were FA 18:2 (m/z 279), LPC 18:2 (m/z 504), and LPE 18:2 (m/z 476), respectively (Fig. 2A, B). However, we also observed a minor contribution of an acyl combination 16:0/20:4 (Fig. 2A, B).
Fig. 2.
PC 36:4 and PE 36:4 are primarily composed of FA 18:2. A: EPI spectrum of PC 36:4 of cells treated with 10 µM c9,t11-CLA for 4 h and analyzed by EPI spectra in negative ion mode. The product ions indicate a majority of PC 18:2/18:2 and a minor contribution of PC 16:0/20:4 to PC 36:4. B: EPI spectrum of PE 36:4 of cells treated with 10 µM c9,t11-CLA for 4 h and analyzed by EPI spectra in negative ion mode. The precursor ion of m/z 738 comprises, in addition to PE 18:2/18:2 and PE 16:0/20:4, fragment ions of chloride adduct ions of PC 30:1 and the +1 isotope peak of SM 34:1. CLA, conjugated linoleic acid; EPI, enhanced product ion; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin.
Taken together our data show that specifically c9,t11-CLA strongly affects cellular PC and PE species pattern by inducing a shift toward PC 18:2/18:2 and PE 18:2/18:2.
c9,t11-CLA activates cellular de novo PC and PE synthesis
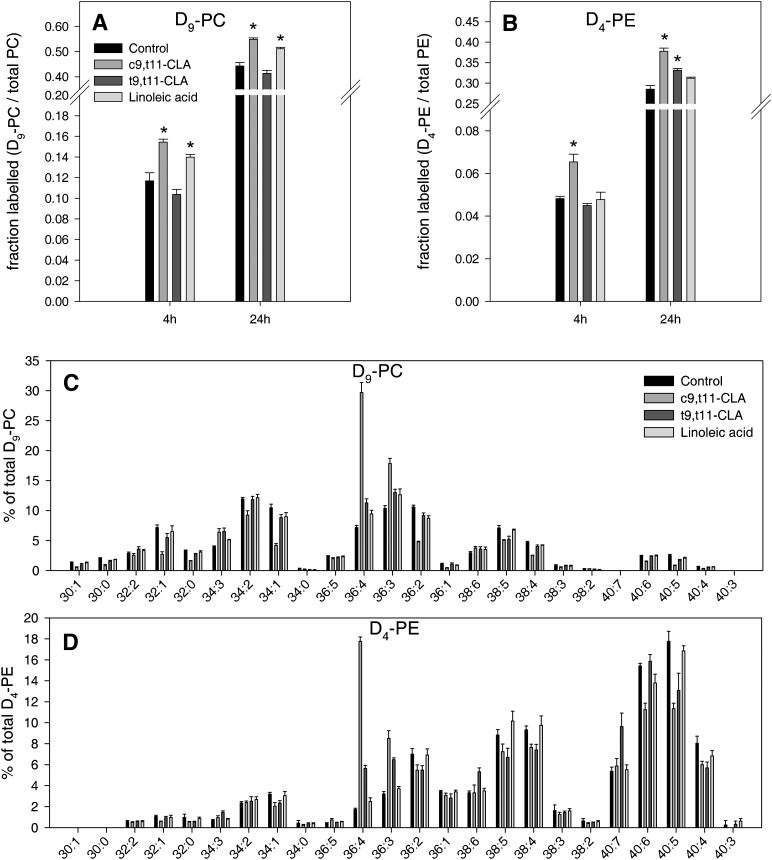
To analyze if the CLA isomers influence cellular glycerophospholipid biosynthesis, primary macrophages were supplied with stable isotope labeled choline (D9), ethanolamine (D4), and serine (13C3). Glycerophospholipid synthesis was quantified by ESI-MS/MS (supplementary ) (19). c9,t11-CLA, but not t9,t11-CLA, enhanced de novo PC (D9-PC) and PE (D4-PE) synthesis via the Kennedy pathway (Fig. 3A,B). PS synthesis (13C3-PS) and decarboxylation of PS to yield PE (13C2-PE) was not altered or reduced by c9,t11-, t9,t11-CLA, or linoleic acid (supplementary Fig. II-A, B).
Analysis of D9-PC and D4-PE profile showed that the species pattern of newly synthesized PC and PE changed in a similar way to the corresponding, unlabeled PC and PE species pattern (Fig. 3C, D). After 4 h c9,t11-CLA increased D9-PC 36:4 by 4-fold and D4-PE 36:4 by 10-fold compared with control and linoleic acid-treated cells. Calculation of the total PC 36:4 species shift comparing linoleic acid-treated with c9,t11-CLA-treated cells revealed an increased proportion of 20% for D9-PC 36:4 compared with 5.5% for PC 36:4. Taking into account that only 15% of total PC was labeled after 4 h, 39% of the PC 36:4 shift originated from de novo synthesis, and 61% was derived from FA remodeling. D4-PE 36:4 increased by 15.3% and PE 36:4 by 4.8% when comparing c9,t11-CLA with linoleic acid-treated cells. Because only 7% of total PE was labeled after 4 h, the majority of the total PE 36:4 shift was therefore derived from fatty acid remodeled PE (80%) compared with newly synthesized D4-PE (20%).
Fig. 3.
c9,t11-CLA increases de novo PC and PE synthesis via the Kennedy pathway and induces a shift toward newly synthesized PC 36:4 and PE 36:4. A: D9-PC synthesis in untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS. B: D4-PE synthesis in untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS. C: D9-PC species profile of untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS. D: D4-PE species profile of untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and quantified by ESI-MS/MS. Values are mean ± SD of one representative experiment from three, each performed in triplicate (*P < 0.01). CLA, conjugated linoleic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
In summary, c9,t11-CLA increases cellular PC and PE synthesis and induces shifts to PC 36:4 and PE 36:4, which are due to both de novo synthesis and fatty acid remodeling.
c9,t11-CLA incorporates into glycerophospholipids, while t9,t11-CLA is preferentially bound to neutral lipids
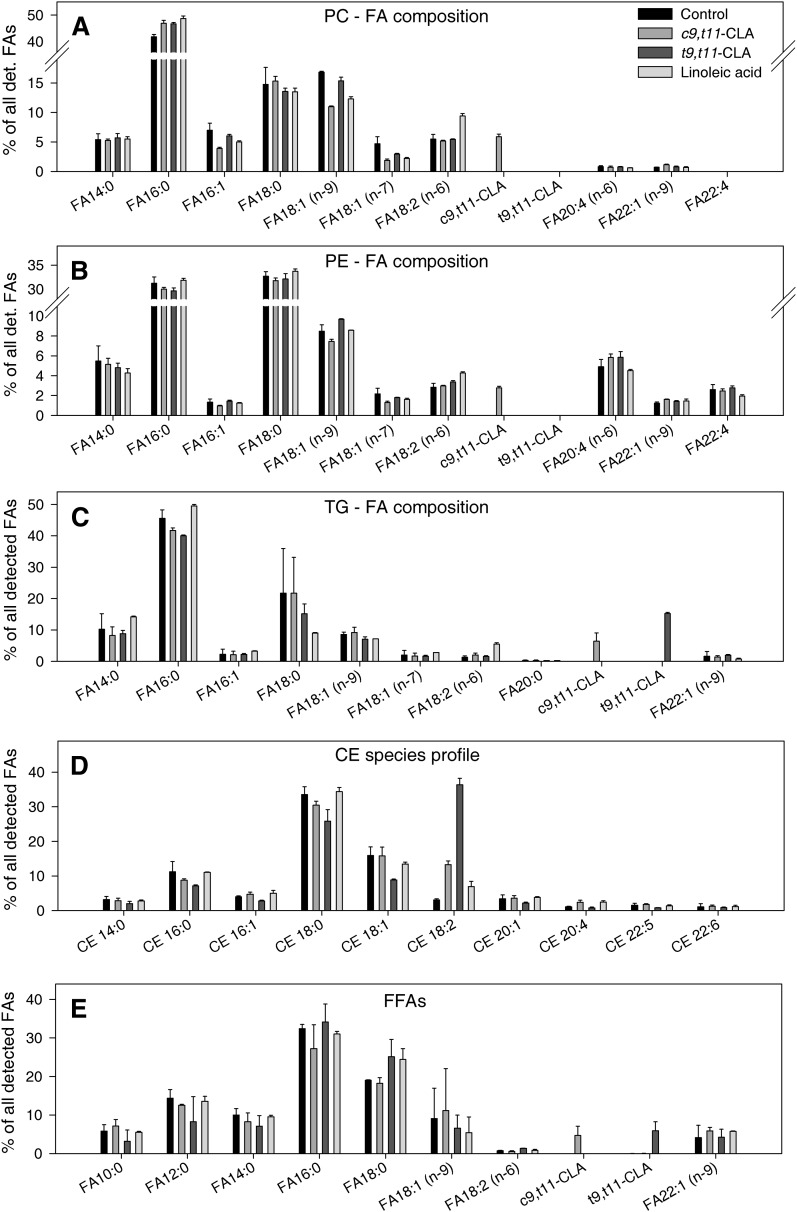
We hypothesized that particularly c9,t11-CLA incorporates in macrophage phospholipids and leads to an increase of PC 18:2/18:2 and PE 18:2/18:2. To test this hypothesis, lipid classes were separated by thin-layer chromatography (TLC) and analyzed regarding fatty acid composition by GC-MS. When cells were treated with CLA isomers, we detected c9,t11-CLA, but not t9,t11-CLA, in PC and PE fractions (Fig. 4A, B), which confirms our hypothesis that c9,t11-CLA incorporates into these cellular phospholipids. c9,t11-CLA contributed as equally as linoleic acid to the fatty acid composition of PC and PE. We did not detect c9,t11- or t9,t11-CLA in the PS fraction (data not shown) or any CFA (Table 1) in PC, PE, or PS.
Fig. 4.
Fatty acid composition of phospholipids (PC, PE) and neutral lipids (TG, CE), and cellular free fatty acids. A: FA composition of PC for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h, separated by TLC, methylated to generate FAMEs, and analyzed by GC-MS. B: FA composition of PE for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h, separated by TLC, methylated to generate FAMEs, and analyzed by GC-MS. C: FA composition of TGs for untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h, separated by TLC, methylated to generate FAMEs, and analyzed by GC-MS. D: CE species profile of untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h and analyzed by ESI-MS/MS. E: FFA in untreated cells and cells treated with 10 µM CLA or linoleic acid for 4 h, separated by TLC, methylated to generate FAMEs, and analyzed by GC-MS. Values are mean ± SD of one representative experiment from three, each performed in triplicate. CLA, conjugated linoleic acid; FA, fatty acid; FFA, free fatty acid; FAME, FA methyl ester; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
Because t9,t11-CLA was not found in the glycerophospholipid fraction, we asked whether this CLA isomer is bound to neutral lipids, including triglycerides and cholesterol esters. TLC and subsequent GC-MS analysis of cellular triglycerides demonstrated that t9,t11-CLA contributes 15%, whereas c9,t11-CLA contributes only 6% to the fatty acid composition of the triglycerides (Fig. 4C). Cellular cholesteryl ester (CE) species composition was analyzed by ESI-MS/MS, which revealed a 12-fold increase of CE 18:2 species for the t9,t11-CLA treatments and a 4-fold increase for the c9,t11-CLA-supplemented cells (Fig. 4D). Finally, we explored whether the CLA isomers are available as free fatty acids in the cells and found about equal proportions of unesterified c9,t11- and t9,t11-CLA (Fig. 4E).
In conclusion, our results show that c9,t11- and t9,t11-CLA differentially contribute to cellular lipid composition. c9,t11-CLA preferentially incorporates into PC and PE lipids, while the t9,t11-CLA isomer preferentially binds to neutral lipids.
DISCUSSION
Although several groups have reported that c9,t11-CLA is metabolized in cells and incorporated into plasma and tissue lipids (15, 28), only a few studies investigated its effects on cellular lipids. Metabolism of t9,t11-CLA and its contribution to cellular lipids is completely unexplored.
Here we demonstrate that c9,t11-CLA and t9,t11-CLA are differentially metabolized to CFAs. We found that c9,t11-CLA is desaturated and elongated to CFA 20:4 (supplementary Fig. III), which is supported by results from other groups showing that this CLA isomer in particular is well metabolized up to CFA 20:3 and 20:4 (14, 29, 30). In contrast, the t9,t11-CLA isomer is only elongated to CFA 20:2 and CFA 22:2, but it is not desaturated. Because Agatha et al. showed that c9,c11-CLA is delta-5 and delta-6 desaturated in leukemia cells (14), we hypothesize that t9,t11-CLA is resistant for further desaturation due to the trans-double bond at position 9. This conclusion is supported by experiments with human skin fibroblasts demonstrating that the trans-fatty acids elaidic acid (t9-FA 18:1) and linoelaidic acid (t9,t12-FA 18:2) in contrast to the cis-fatty acids oleic acid (c9-FA 18:1) and linoleic acid (c9,c12-FA 18:2) inhibit delta-5 and delta-6 desaturation (31, 32).
Our results further show that c9,t11-CLA leads to a shift toward PC and PE 18:2/18:2 (Figs. 1, 2). As we could detect c9,t11-CLA in the PC and PE fractions (Fig. 4A, B), we assume that at least one acyl chain of PC/PE 18:2/18:2 is c9,t11-CLA. This finding might also contribute to the reported anti-inflammatory effects of c9,t11-CLA, such as inhibition of eicosanoid release (5). Due to its incorporation, c9,t11-CLA might displace n-6 fatty acids like arachidonic acid in membrane phospholipids as precursor of eicosanoid production (1). Interestingly, our results further demonstrate that t9,t11-CLA is not incorporated into cellular phospholipids. However, an un-expected finding is that t9,t11-CLA leads to a shift to newly synthesized PE 40:7 (Fig. 3D). Because analysis of a product ion spectrum reveals that primarily PE 18:1/22:6 contributes to PE 40:7 (data not shown) and as we could not detect t9,t11-CLA in the PE fraction (Fig. 4B), we conclude that this shift might not be due to incorporation of this CLA isomer in PE 40:7. Our results further demonstrate that t9,t11-CLA is preferentially bound to neutral lipids, including triglycerides (TG) and cholesterol esters, proposing that a trans-double bond at position 9 favors incorporation into neutral lipids rather than into phospholipids.
Detailed metabolic profiling demonstrated that c9,t11-CLA activates cellular PC and PE de novo biosynthesis, we did not find an increased mRNA expression of genes related to PC and PE synthesis upon CLA treatment (supplementary Fig. IV). A major reason for this finding might be that c9,t11-CLA, due to its incorporation, fuels and, therefore, drives PC and PE synthesis. However, another explanation for this finding might be a metabolic activation of enzymes necessary for phospholipid (PL) synthesis. Upon induction the key enzyme of PC synthesis, choline-phosphate cytidyltransferase (CCT) is rapidly translocated from a soluble, inactive form to a membrane-associated, active form (33). Translocation of this enzyme is triggered by membrane curvature stress (34).
PUFA acyl chains and double bonds dramatically alter physical properties of cellular membranes (35). While acyl chains of unconjugated PUFAs can freely rotate after the double bonds, the conjugated double-bond system characteristic for all CLAs leads to a planar, rigid structure. t9,t11-CLA is a relatively straight molecule, whereas c9,t11-CLA structure is bended due to the cis-double bond (supplementary Fig. V-A, B). These differential structural features of the supplied CLA isomers may very well explain the observed PC synthesis rates (Fig. 2A) using the above- described model for CCT activation by membrane curvature stress. It is also supported by data from Yin et al. showing that incorporation of c9,t11-CLA into phospholipids substantially alters physical membrane properties and function (36, 37). However, another possible explanation for the enhanced PL synthesis rates might be that c9,t11-CLA treatment enhances PC and PE synthesis by increasing the diacylglycerol (DAG) content necessary for the last step of the Kennedy pathway (supplementary Fig. I).
To summarize, our results show that geometrical isomers of CLA have different effects on macrophage lipid metabolism. c9,t11-CLA activates cellular PC and PE synthesis and incorporates into these lipids, which might contribute to the reported biological effects of this isomer. Our data are a good example to demonstrate that phospholipid species composition strongly depends on exogenous fatty acid supply. Moreover, these findings are a valuable contribution to better understand fundamentals of cellular membrane biology, such as the relation between fatty acid structure and its effects on glycerophospholipid metabolism.
Supplementary Material
Acknowledgments
The authors thank Jolante Aiwanger, Doreen Müller, Simone Peschel, and Barbara Tille for outstanding technical assistance.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- CFA
- conjugated fatty acid
- CLA
- conjugated linoleic acid
- FC
- free cholesterol
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- TG
- triglyceride
This work was supported by Deutsche Forschungsgemeinschaft Grants SCHM 654/9-1 and SCHM 654/9-2) and by the Seventh Framework Programme of the EU-funded “LipidomicNet” (proposal 202272).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Schmitz G., Ecker J. 2008. The opposing effects of n-3 and n-6 fatty acids. Prog. Lipid Res. 47: 147–155. [DOI] [PubMed] [Google Scholar]
- 2.Kramer J. K., Cruz-Hernandez C., Deng Z., Zhou J., Jahreis G., Dugan M. E. 2004. Analysis of conjugated linoleic acid and trans 18:1 isomers in synthetic and animal products. Am. J. Clin. Nutr. 79: 1137S–1145S. [DOI] [PubMed] [Google Scholar]
- 3.Aldai N., Osoro K., Barron L. J., Najera A. I. 2006. Gas-liquid chromatographic method for analysing complex mixtures of fatty acids including conjugated linoleic acids (cis9trans11 and trans10cis12 isomers) and long-chain (n-3 or n-6) polyunsaturated fatty acids. Application to the intramuscular fat of beef meat. J. Chromatogr. A. 1110: 133–139. [DOI] [PubMed] [Google Scholar]
- 4.Kraft J., Kramer J. K., Schoene F., Chambers J. R., Jahreis G. 2008. Extensive analysis of long-chain polyunsaturated fatty acids, CLA, trans-18:1 isomers, and plasmalogenic lipids in different retail beef types. J. Agric. Food Chem. 56: 4775–4782. [DOI] [PubMed] [Google Scholar]
- 5.Eder K., Ringseis R. 2010. Metabolism and actions of conjugated linoleic acids on atherosclerosis-related events in vascular endothelial cells and smooth muscle cells. Mol. Nutr. Food Res. 54: 17–36. [DOI] [PubMed] [Google Scholar]
- 6.Azizian H., Kramer J. K. 2005. A rapid method for the quantification of fatty acids in fats and oils with emphasis on trans fatty acids using Fourier Transform near infrared spectroscopy (FT-NIR). Lipids. 40: 855–867. [DOI] [PubMed] [Google Scholar]
- 7.Saba A., Mazzini F., Raffaelli A., Mattei A., Salvadori P. 2005. Identification of 9(E),11(E)-18:2 fatty acid methyl ester at trace level in thermal stressed olive oils by GC coupled to acetonitrile CI-MS and CI-MS/MS, a possible marker for adulteration by addition of deodorized olive oil. J. Agric. Food Chem. 53: 4867–4872. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell P. L., McLeod R. S. 2008. Conjugated linoleic acid and atherosclerosis: studies in animal models. Biochem. Cell Biol. 86: 293–301. [DOI] [PubMed] [Google Scholar]
- 9.Bassaganya-Riera J., Hontecillas R. 2006. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin. Nutr. 25: 454–465. [DOI] [PubMed] [Google Scholar]
- 10.Kelley N. S., Hubbard N. E., Erickson K. L. 2007. Conjugated linoleic acid isomers and cancer. J. Nutr. 137: 2599–2607. [DOI] [PubMed] [Google Scholar]
- 11.Ringseis R., Eder K. 2009. Influence of conjugated linoleic acids on functional properties of vascular cells. Br. J. Nutr. 102: 1099–1116. [DOI] [PubMed] [Google Scholar]
- 12.Ecker J., Langmann T., Moehle C., Schmitz G. 2007. Isomer specific effects of conjugated linoleic acid on macrophage ABCG1 transcription by a SREBP-1c dependent mechanism. Biochem. Biophys. Res. Commun. 352: 805–811. [DOI] [PubMed] [Google Scholar]
- 13.Ecker J., Liebisch G., Patsch W., Schmitz G. 2009. The conjugated linoleic acid isomer trans-9,trans-11 is a dietary occurring agonist of liver X receptor alpha. Biochem. Biophys. Res. Commun. 388: 660–666. [DOI] [PubMed] [Google Scholar]
- 14.Agatha G., Voigt A., Kauf E., Zintl F. 2004. Conjugated linoleic acid modulation of cell membrane in leukemia cells. Cancer Lett. 209: 87–103. [DOI] [PubMed] [Google Scholar]
- 15.Burdge G. C., Lupoli B., Russell J. J., Tricon S., Kew S., Banerjee T., Shingfield K. J., Beever D. E., Grimble R. F., Williams C. M., et al. 2004. Incorporation of cis-9,trans-11 or trans-10,cis-12 conjugated linoleic acid into plasma and cellular lipids in healthy men. J. Lipid Res. 45: 736–741. [DOI] [PubMed] [Google Scholar]
- 16.Ecker J., Liebisch G., Englmaier M., Grandl M., Robenek H., Schmitz G. 2010. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. Proc. Natl. Acad. Sci. USA. 107: 7817–7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard J. W. 2009. Trophic macrophages in development and disease. Nat. Rev. Immunol. 9: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 19.Binder M., Liebisch G., Langmann T., Schmitz G. 2006. Metabolic profiling of glycerophospholipid synthesis in fibroblasts loaded with free cholesterol and modified low density lipoproteins. J. Biol. Chem. 281: 21869–21877. [DOI] [PubMed] [Google Scholar]
- 20.Liebisch G., Lieser B., Rathenberg J., Drobnik W., Schmitz G. 2004. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim. Biophys. Acta. 1686: 108–117. [DOI] [PubMed] [Google Scholar]
- 21.Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. 2006. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta. 1761: 121–128. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz G., Assmann G., Bowyer D. E. 1984. A quantitative densitometric method for the rapid separation and quantitation of the major tissue and lipoprotein lipids by high-performance thin-layer chromatography. I. Sample preparation, chromatography, and densitometry. J. Chromatogr. 307: 65–79. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz G., Lenczyk M., Ord D., Bowyer D. E., Assmann G. 1984. A quantitative densitometric method for the rapid separation and quantitation of the major lipids of tissues and lipoproteins by high-performance thin-layer chromatography. II. Reduction of the densitometric data. J. Chromatogr. 307: 81–89. [DOI] [PubMed] [Google Scholar]
- 24.Ekroos K., Ejsing C. S., Bahr U., Karas M., Simons K., Shevchenko A. 2003. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J. Lipid Res. 44: 2181–2192. [DOI] [PubMed] [Google Scholar]
- 25.Han X., Yang J., Cheng H., Ye H., Gross R. W. 2004. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal. Biochem. 330: 317–331. [DOI] [PubMed] [Google Scholar]
- 26.Hsu F. F., Turk J. 2005. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J. Am. Soc. Mass Spectrom. 16: 1510–1522. [DOI] [PubMed] [Google Scholar]
- 27.Hsu F. F., Turk J. 2000. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 11: 797–803. [DOI] [PubMed] [Google Scholar]
- 28.Goedecke J. H., Rae D. E., Smuts C. M., Lambert E. V., O'Shea M. 2009. Conjugated linoleic acid isomers, t10c12 and c9t11, are differentially incorporated into adipose tissue and skeletal muscle in humans. Lipids. 44: 983–988. [DOI] [PubMed] [Google Scholar]
- 29.Sebedio J. L., Juaneda P., Dobson G., Ramilison I., Martin J. C., Chardigny J. M., Christie W. W. 1997. Metabolites of conjugated isomers of linoleic acid (CLA) in the rat. Biochim. Biophys. Acta. 1345: 5–10. [DOI] [PubMed] [Google Scholar]
- 30.Banni S., Petroni A., Blasevich M., Carta G., Cordeddu L., Murru E., Melis M. P., Mahon A., Belury M. A. 2004. Conjugated linoleic acids (CLA) as precursors of a distinct family of PUFA. Lipids. 39: 1143–1146. [DOI] [PubMed] [Google Scholar]
- 31.Rosenthal M. D., Whitehurst M. C. 1983. Selective effects of isomeric cis and trans fatty acids on fatty acyl delta 9 and delta 6 desaturation by human skin fibroblasts. Biochim. Biophys. Acta. 753: 450–459. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal M. D., Doloresco M. A. 1984. The effects of trans fatty acids on fatty acyl delta 5 desaturation by human skin fibroblasts. Lipids. 19: 869–874. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto H., Banchio C., Vance D. E. 2008. Transcriptional regulation of phosphatidylcholine biosynthesis. Prog. Lipid Res. 47: 204–220. [DOI] [PubMed] [Google Scholar]
- 34.Attard G. S., Templer R. H., Smith W. S., Hunt A. N., Jackowski S. 2000. Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc. Natl. Acad. Sci. USA. 97: 9032–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaikh S. R., Edidin M. 2006. Polyunsaturated fatty acids, membrane organization, T cells, and antigen presentation. Am. J. Clin. Nutr. 84: 1277–1289. [DOI] [PubMed] [Google Scholar]
- 36.Yin J. J., Mossoba M. M., Kramer J. K., Yurawecz M. P., Eulitz K., Morehouse K. M., Ku Y. 1999. Effects of conjugated linoleic acid on oxygen diffusion-concentration product and depletion in membranes by using electron spin resonance spin-label oximetry. Lipids. 34: 1017–1023. [DOI] [PubMed] [Google Scholar]
- 37.Yin J. J., Kramer J. K., Yurawecz M. P., Eynard A. R., Mossoba M. M., Yu L. 2006. Effects of conjugated linoleic acid (CLA) isomers on oxygen diffusion-concentration products in liposomes and phospholipid solutions. J. Agric. Food Chem. 54: 7287–7293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.