Abstract
Two high-density lipoprotein cholesterol quantitative trait loci (QTL), Hdlq1 at 125 Mb and Hdlq8 at 113 Mb, were previously identified on mouse distal chromosome 5. Our objective was to identify the underlying genes. We first used bioinformatics to narrow the Hdlq1 locus to 56 genes. The most likely candidate, Scarb1 (scavenger receptor B1), was supported by gene expression data consistent with knockout and transgenic mouse models. Then we confirmed Hdlq8 as an independent QTL by detecting it in an intercross between NZB and NZW (LOD = 12.7), two mouse strains that have identical genotypes for Scarb1. Haplotyping narrowed this QTL to 9 genes; the most likely candidate was Acads (acyl-coenzymeA dehydrogenase, short chain). Sequencing showed that Acads had an amino acid polymorphism, Gly94Asp, in a conserved region; Western blotting showed that protein levels were significantly different between parental strains. A previously known spontaneous deletion causes loss of ACADS activity in BALB/cBy mice. We showed that HDL levels were significantly elevated in BALB/cBy compared with BALB/c mice and that this HDL difference cosegregated with the Acads mutation. We confirmed that Hdlq1 and Hdlq8 are independent QTL on mouse chromosome 5 and demonstrated that Scarb1 and Acads are the underlying genes.
Keywords: high-density lipoprotein, QTL, mice
Because plasma high-density lipoprotein cholesterol (HDL) protects against coronary heart disease, considerable effort has been made to find the quantitative trait loci (QTL) and the genes that regulate HDL. Numerous QTL have been found in both inbred mouse strains and in human populations; these QTL often map to homologous locations in both species (1). Detecting QTL has been relatively easy. The next step of identifying causal QTL genes, however, has been more challenging, although the development of bioinformatic methods (2, 3) and improved genomic resources in the mouse (4) and genome-wide association studies in humans (5, 6) are improving the success of QTL gene identification.
QTL that are in close proximity on the same chromosome remain particularly challenging. In such situations, it is particularly difficult to identify the causal QTL genes. Although congenic strains are a powerful tool to separate these QTL, they are time consuming; therefore, we used a more efficient, genomic approach to decipher the presence of one or multiple QTL and identify the QTL genes. At least seven inbred mouse crosses (7–12) have identified HDL QTL on mouse distal chromosome (Chr) 5 (Table 1). However, the broad confidence intervals and shape of the logarithm of the odds ratio (LOD) score plots suggest that at least four of these crosses (B6 × 129, NZB × SM, B6 × NZB, and B6 × CAST) may have two closely linked QTL. We used advanced intercross lines between B6 and NZB at F11, which allows the accumulation of recombination events and thus narrows QTL (13) and confirmed that two QTL did exist between these two strains on distal Chr 5. These QTL were named Hdlq1 at 125 Mb and Hdlq8 at 113 Mb (11).
TABLE 1.
Mouse crosses that identified HDL QTL on distal Chr 5
| Cross | Peaks (Mb)a | 95% CI (Mb) | LOD | High Alleleb | Reference |
|---|---|---|---|---|---|
| (NZB × RF) F2 | 105 | 104–121 | 18 | NZB | 12 |
| (MRL × SJL)F2 | 107 | 4.9 | MRL | 12 | |
| NZO × (SJL × NZO) | 125 | 113–141 | 13 | NZO | 9 |
| (B6 × 129) F2 | 91, 125 | 79–127 | 3.3, 3.4 | 129 | 10 |
| (NZB × SM) F2 | 100, 120 | 80–130 | 10, 12 | NZB | 7 |
| (B6 × NZB) F11c | 113, 125 | 107–127 | 3.6, 5.8 | NZB | 11 |
| (B6 × CAST) F2 | 102, 130 | 65–144 | 6, 3.8 | CAST | 8 |
Chr, chromosome; QTL, quantitative trait loci.
Two locations and LOD scores were provided if two peaks were detected in the LOD score plot; each score corresponds to one peak.
The strain that carries the allele for high HDL levels at the QTL.
This cross was based on advanced intercross lines from a B6 × NZB intercross that was then randomly bred from F3-F11 to increase the number of recombination events.
In the present study, we first identified Scarb1 as the causal gene for Hdlq1. We then confirmed that Hdlq8 is an independent QTL by crossing two closely related strains that do not differ at the Scarb1 locus and thus were expected not to have a QTL at the Hdlq1 region. As expected, this cross detected Hdlq8 but had no QTL at Hdlq1. Subsequently, we used haplotype analysis, gene sequencing, expression studies, and a spontaneous mutation to demonstrate that Acads was the underlying gene for Hdlq8.
MATERIALS AND METHODS
Mice and diets
The details of the NZB/BlNJ (NZB) and NZW/LacJ (NZW) intercross were described previously (9, 13). For the BALB/cByJ [(BALB/cBy) × BALB/cJ (BALB/c)] intercross, female BALB/cBy and male BALB/c mice (The Jackson Laboratory, Bar Harbor, ME) were mated to generate F1 progeny, which were intercrossed to produce 97 F2 progeny. All mice were maintained in a temperature- and humidity-controlled environment with a 12 h light/12 h dark cycle and given unrestricted access to food and acidified water. Mice were fed standard chow containing 6% fat (5K52 LabDiet®, St. Louis, MO) until they were 8 weeks old, and then they were fed a high-fat atherogenic diet containing 15% dairy fat, 1% cholesterol, and 0.5% cholic acid as described previously (13) until they were euthanized at 16 weeks of age. Experiments were approved by the institutional Animal Care and Use Committee of The Jackson Laboratory.
HDL measurement
We fasted mice for 4 h in the morning, and collected plasma and measured HDL concentrations as previously described at 8 and 16 weeks of age (13).
Genotyping F2 mice
We extracted DNA from (BALB/cBy × BALB/c) F2 mice tail tips using phenol-chloroform. Single-nucleotide polymorphims (SNPs) on Chr 5, including rs6226708 (33.7 Mb), rs13478257 (54.1 Mb), rs13478321 (72.1 Mb), rs3726313 (100.7 Mb), rs3668084 (116.2 Mb), and rs3685925 (139.8 Mb) were genotyped by the Allele Typing Service at The Jackson Laboratory. All positions are Build 36, NCBI.
QTL analysis
QTL mapping was carried out as previously described (10). Single loci on Chr 5 associated with HDL in NZB × NZW F2 progeny were analyzed using sex as an additive covariate. QTL confidence intervals were defined by a 1-unit decrease in the LOD score on either side of the peak marker.
Haplotype analysis
SNPs used in haplotype analysis were obtained from the most recent genotyping by the Center for Genome Dynamics (4), publicly available at http://cgd.jax.org/tools/diversityarray.shtml. For Hdlq1 we compared the haplotypes in the common interval from 121 to 132 Mb. We identified the regions where the haplotypes of the high and low allele strains differed from each other, with strains NZB and NZO/HILtJ (NZO) contributing a high allele, and strains C57BL/6J (B6), SJL/J (SJL) and SM/J (SM) contributing a low allele. For Hdlq8 we compared the haplotypes in the common interval from 107 to 117 Mb between all parental strains except CAST/EiJ (CAST), a strain recently derived from the wild, whose genome is unique. We identified the regions where strains B6, NZW, RF/J (RF), SJL, and SM, which contribute a low HDL allele, were identical; where strains NZB, MRL/J (MRL), and 129/SvImJ (129), which contribute a high HDL allele, were identical; and where the haplotypes of the high and low allele strains differed from each other. Genes that mapped to these regions were extracted from Ensembl (www.ensembl.org). Subsequently we compared the haplotypes of strains that did not detect a QTL on Chr 5.
Acads sequencing and genotyping
We amplified Acads using genomic DNAs from strains 129, B6, NZB, NZW, RF, SJL, MRL, C3H, PERA, I/Ln, and SM, and Scarb1 using genomic DNA from NZB and SM. Primers were designed to amplify each exon plus at least 50 nucleotides of the adjacent introns. Purified PCR products using ExoSAP-IT (USB, Cleveland, OH) were subjected to thermocycle sequencing, and the resulting fragments were analyzed on capillary-based machines by The Jackson Laboratory DNA Sequencing Service. Sequence analysis was done by aligning the sequence to the genomic B6 sequence using Sequencher 4.2 (GeneCodes Technology, Ann Arbor, MI). We amplified DNA fragments covering the deletion of Acads found in BALB/cBy using primers 5′-GCTGCCATGTTGAAAGACAA and 5′-AAGGCAAGTCCCTTTCTGGT in (BALB/cBy × BALB/c) F2 mice. The amplified fragments were genotyped by 2.5% NuSieve 3:1 agarose gel electrophoresis (Cambrex Corporation, Charles City, IA).
Real-time PCR of Scarb1
Using real-time PCR, we examined the expression of Scarb1 in liver samples from five males of each of the following strains: NZB, SM, B6, CAST, NZO, and SJL. cDNA samples were mixed with SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and gene-specific primers in a total volume of 25 µl. The primer pairs are as follows: Scarb1 forward 5′-TGCTCAAGAATGTCCGCATA and reverse 5′-ACGGTGTCGTTGTCATTGAA; β-actin forward 5′-CTTCTTGGGTATGGAATCC and reverse 5′-GCTCAGGAGGAGCGGTGAT. We performed PCR in 96-well optical reaction plates with an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Cycling parameters were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. PCR reactions were done in triplicate for each strain, and the expression of Scarb1 was normalized to the expression of β-actin. A comparative Ct (ΔΔCt) was applied to the raw Ct values to establish the relative gene expression between strains.
Western blot analysis of ACADS
We dissected liver tissue from three males of each strain, flash froze the samples in liquid nitrogen, and performed liver protein extraction and Western blotting as previously described (14). Rabbit anti-ACADS serum, kindly provided by Dr. Vockley of the Children's Hospital of Pittsburg, was used for the primary immunostain; goat anti-rabbit IgG horseradish peroxidase was used for the secondary immunostain. Rabbit anti-β-tubulin conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA) was used as the control.
Real-time PCR for Hdlq8
Using real-time PCR, we examined the expression of the nine genes in the narrowed Hdlq8 interval in liver samples from five females of NZB, NZW, and B6. cDNA samples were mixed with SYBR Green Master Mix (Applied Biosystems) and gene-specific primers in a total volume of 25 µl. We performed PCR in 96-well optical reaction plates with an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Cycling parameters were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. PCR reactions were done in triplicate for each strain, and the expression of Scarb1 was normalized to the expression of Gapdh.
Statistical analysis
We used one-way ANOVA to determine the allele effects on HDL levels. Data were analyzed using GraphPad Prism (Windows v5.00; GraphPad Software, San Diego, CA).
RESULTS
Scarb1 is the gene underlying Hdlq1
We considered Scarb1 as a possible candidate gene for the QTL of four crosses (B6 × CAST, B6 × NZB, NZB × SM, and NZO × SJL), as the QTL was at the distal end of the chromosome, the confidence interval included the Scarb1 locus, and Scarb1 is well known to be involved in HDL metabolism. Among these crosses, the strains that carry the high alleles for HDL are CAST, NZB, and NZO; the strains that carry the low alleles are B6, SM, and SJL. We first performed haplotype analysis among the parental strains in the crosses that detected the QTL. We did not use the B6 × CAST cross because the wild-derived inbred strain CAST has a unique genotype that considerably reduces the value of haplotype analysis. Although 56 genes fit the expected haplotype pattern, Scarb1 was the most likely candidate gene. The haplotypes in strains NZB and NZO, which carry the high allele for HDL, were identical at the Scarb1 region, but they differed from the haplotypes shared by strains B6, SM, and SJL, which carry the low allele for HDL. The difference in haplotypes between QTL parental strains supported Scarb1 as the candidate for this QTL.
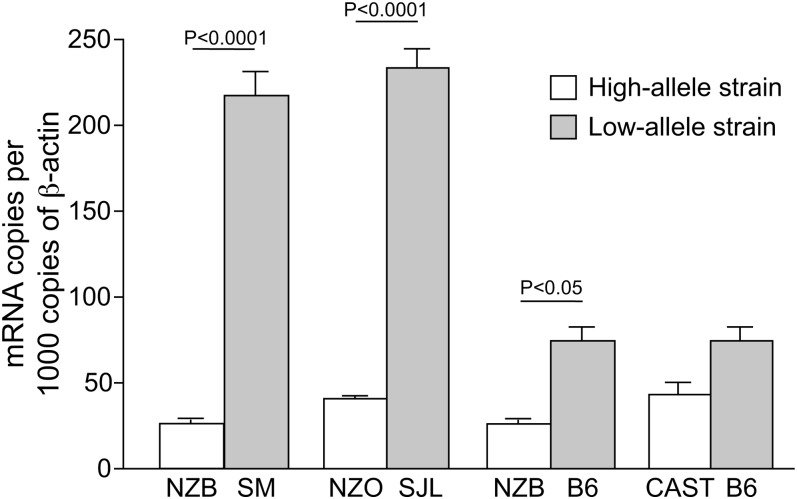
Next we looked at protein function and expression levels of Scarb1. We sequenced the entire coding region of Scarb1 in strains NZB and SM, compared the sequences with the B6 sequence in Ensembl, and found no amino acid change. We quantified Scarb1 expression using real-time PCR with mRNA extracted from liver of high allele (NZB, NZO, CAST) and low allele (SM, B6, SJL) strains (Fig. 1). The mRNA expression level of Scarb1 was significantly lower in strains that carry the high allele for HDL (NZB, NZO) compared with strains that carry the low allele for HDL (SM, B6, SJL) (P < 0.05). Although the difference was not significant, Scarb1 expression was somewhat higher in B6 mice than that in CAST mice. This observation is consistent with the scavenger receptor B1 (SR-B1) function, since Scarb1-deficient mice have increased HDL levels (15). Partial to complete SR-B1 deficiency results in an increase in plasma HDL (16), while hepatic overexpression of SR-BI in mice markedly reduces plasma HDL levels. Thus, we conclude that Scarb1 is a likely candidate for Hdlq1.
Fig. 1.
Real-time expression of Scarb1. Comparison between the parental strains from the crosses in which Hdlq1 was detected. The strains with the high HDL allele in the cross are indicated with the open bars, and the strains with the low HDL allele are indicated with the filled bars. In three strain combinations, the strain with the high HDL allele had a significantly lower expression of Scarb1 than the strain with the low HDL allele (*P < 0.0001, **P < 0.001).
Hdlq8 is distinct from Hdlq1 on distal Chr 5
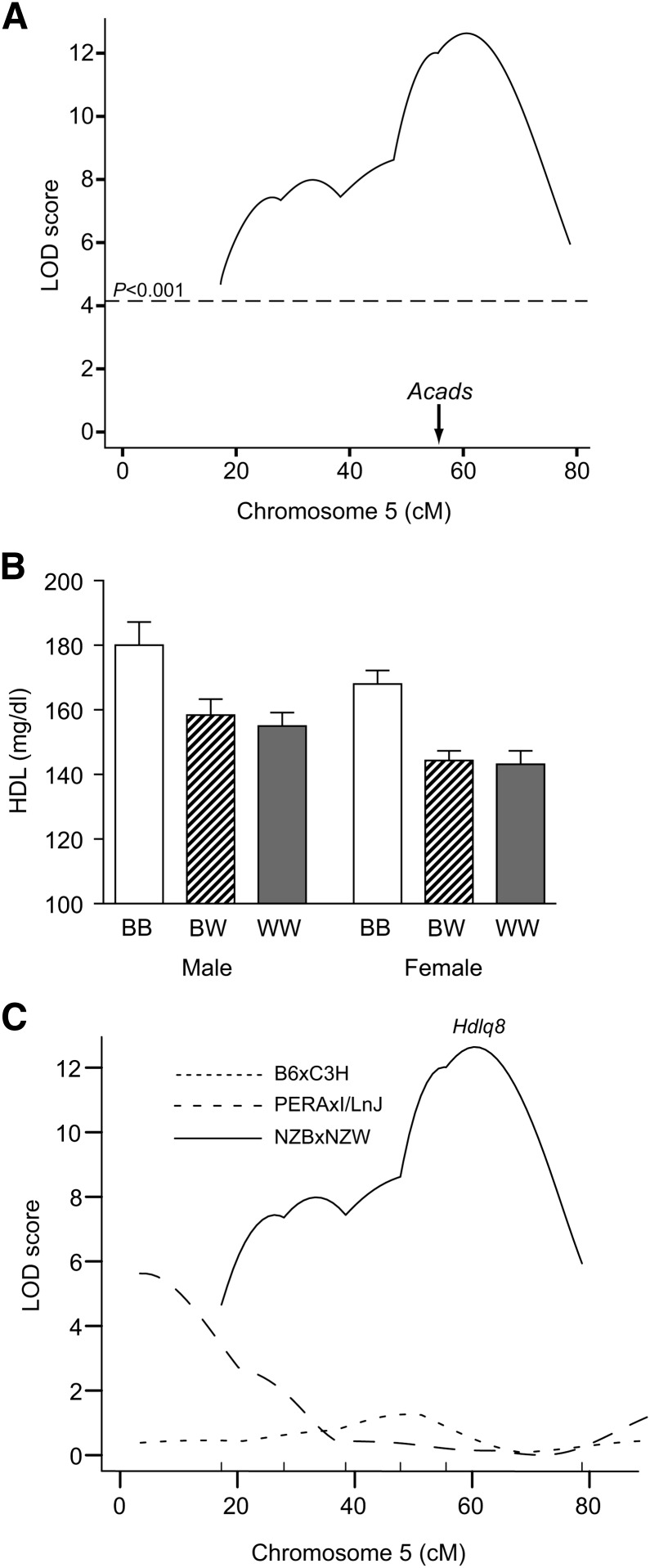
To confirm the existence of Hdlq8 without the confounding effect of the QTL caused by Scarb1, we carried out QTL analysis on Chr 5 using an intercross between strains NZB and NZW. These two strains are alike for 63.2% of their genome (17). At the Hdlq1 QTL, the SNP pattern surrounding the locus and the QTL gene Scarb1 is identical between NZB and NZW; at the Hdlq8 QTL, the SNP pattern is different. Therefore, this cross should not detect any QTL at Hdlq1. The genome scan for HDL identified a locus influencing plasma HDL on Chr 5 with a peak near SNP rs3668084 (116.2 Mb) and a significant LOD score of 12.7 (Fig. 2A). The LOD score at this locus did not differ when sex was used as an additive or interactive covariate, indicating that this QTL is not sex-specific. The QTL explained 13.8% of the total variance of HDL concentration. The 95% confidence interval extended from 104 to 120 Mb, which overlapped with the 107–117 Mb Hdlq8 region identified in the B6 × NZB advanced intercross lines (11). Because F2 mice that are NZB homozygous at rs3668084 (located on Chr 5 at 116,464,610 bp) had significantly higher HDL levels compared with mice with either the heterozygous or the homozygous NZW genotype at this locus (Fig. 2B), we determined that the allele for high HDL is recessive. These data confirm the existence of Hdlq8 separate from Hdlq1.
Fig. 2.
Hdlq8 was confirmed in the NZB × NZW intercross. A: LOD score plot for plasma HDL on Chr 5. B: The allele effect at peak marker SNP rs3668084 (116 Mb) on plasma HDL concentrations. The mice were divided into three groups: homozygous for the NZB allele (BB, n = 34 and 33 for males and females, respectively); heterozygous at the marker locus (BW, n = 56 and 63 for males and females, respectively); and homozygous for the NZW allele (WW, n = 26 and 32 for males and females, respectively). Values are expressed as mean HDL ± SEM of F2 mice with a particular genotype at the designated locus. C: The failure to detect Hdlq8 in crosses PERA × I (dashed line) and B6 × C3H (dotted line), while Hdlq8 is present in cross NZB × NZW (solid line).
Hdlq8 was narrowed by haplotype analysis
Haplotype analysis reduces the QTL regions by eliminating those regions that are identical by descent between the two strains as inferred by a shared SNP pattern. This strategy is particularly effective for the related strains NZB and NZW because they share so much of their genomes. However, haplotype analysis is based on the assumptions that the mutation causing the QTL is ancestral and that the genotyped SNPs are sufficiently dense to allow correct inference of the DNA regions that are identical by descent. These are fairly safe assumptions in this case. Not only are 97% of mutations ancestral, but this QTL was found in many different crosses (Table 1 and this cross), thus making it likely that the mutation is indeed ancestral. Furthermore, the density of SNPs is very high, as three of the parental strains, B6, NZW, and 129, are among those resequenced by Perlegen (http://mouse.perlegen.com/mouse/index.html), and all strains were genotyped recently by the Center for Genome Dynamics at The Jackson Laboratory (4).
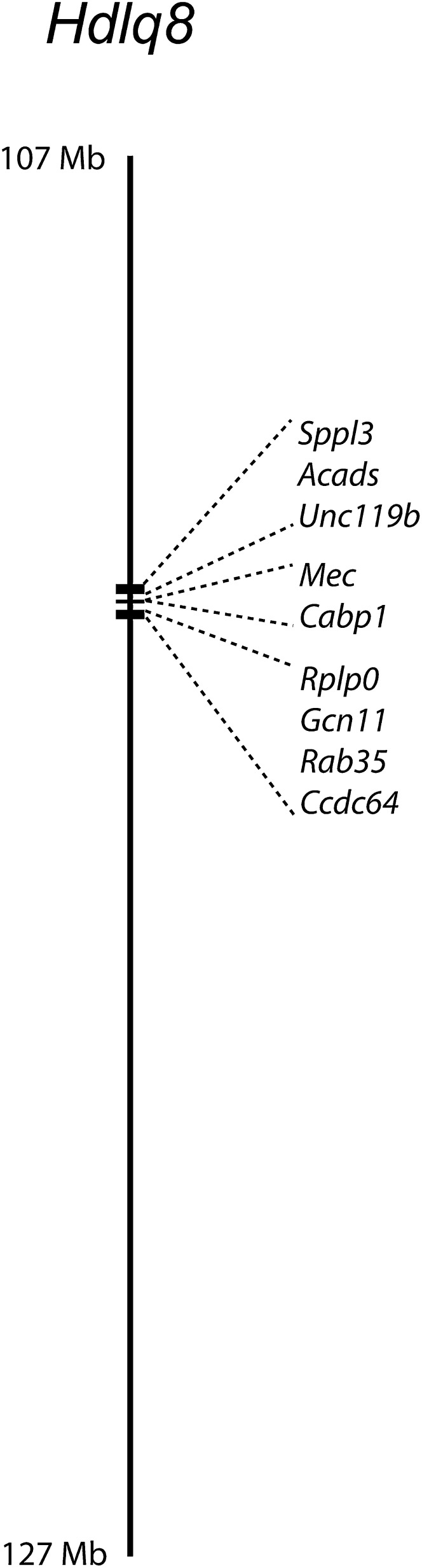
For the haplotype analysis, we analyzed six crosses that detected the QTL: B6 × 129, NZB × NZW, NZB × SM, NZB × B6, MRL × SJL, and NZB × RF. We did not use the NZO × (SJL × NZO) backcross in our haplotype analysis because this cross detected only a single QTL peak at 125 Mb and thus did not detect Hdlq8. We compared the haplotypes throughout the common Hdlq8 interval ranging from 107 to 117 Mb using a bioinformatic tool (available at http://cgd.jax.org/straincomparison/), searching for those regions where the SNPs were identical for NZB, 129, and MRL (all strains have alleles for high HDL); where the SNPs were identical for strains RF, SJL, B6, SM, and NZW (all strains have alleles for low HDL); and where the SNPs for the strains with high and low HDL alleles differed. This haplotype analysis narrowed Hdlq8 to a 0.6 Mb, nine-gene region (Fig. 3 and Table 2).
Fig. 3.
Haplotype analysis of the Hdlq8 region narrows the interval. For the haplotype analysis, we analyzed six crosses that detected the QTL: B6 × 129, NZB × NZW, NZB × SM, NZB × B6, MRL × SJL, and NZB × RF. We compared the haplotypes throughout the common Hdlq8 interval ranging from 107 to 117 Mb, searching for those regions where the SNPs were identical for NZB, 129, and MRL (all strains have alleles for high HDL); where the SNPs were identical for strains RF, SJL, B6, SM, and NZW (all strains have alleles for low HDL); and where the SNPs for the strains with high and low HDL alleles differed. This haplotype analysis narrowed Hdlq8 to a 0.6 Mb, nine-gene region.
TABLE 2.
Remaining candidate genes in Hdlq8 region analyzed for functional and expressional differences
| Gene | Coding SNP | Expression Difference | NZB versus B6 | NZB versus NZW |
|---|---|---|---|---|
| Sppl3 | No | No | 1.17 | −1.19 |
| Acads | Yes | No | −1.08 | −1.82 |
| Unc119b | No | No | 1.53 | −1.18 |
| Mlec | No | No | 1.18 | −1.40 |
| Cabp1 | No | No | −1.11 | −1.33 |
| Rplp0 | No | Yes | 1.38a | 1.05 |
| Gcn1l1 | No | No | 1.12 | −1.01 |
| Rab35 | No | No | 1.17 | −1.24 |
| Ccdc64 | No | NDb | NDb | NDb |
For measuring gene expression, cDNA from females was used for each strain and NZB was compared with B6 and NZW respectively. Gapdh was used as a control gene. SNP, single-nucleotide polymorphism.
P < 0.005.
ND = not determined. We were unable to design a specific primer pair.
We also analyzed the haplotypes of the parental strains for two crosses that failed to detect a QTL at this location, the PERA/EiJ (PERA) × I/LnJ (I/Ln) and the B6 × C3H/HeJ (C3H) crosses (9, 15). Haplotyping is often helpful with crosses that do not detect a QTL because the parental strains must share the identical haplotype for the QTL gene. Using the failure to find a QTL as evidence could be problematic, as a negative result could arise from an underpowered cross or the failure to reach significance. However, both of these crosses were carried out in our laboratory, and the LOD score plots for Chr 5 show no hint of a QTL in this region (Fig. 2C). Unfortunately, all nine candidate genes had identical SNPs for three of the four strains; the fourth strain PERA was recently derived from the wild, so although its SNPs were different from I/Ln, the comparison was not helpful.
Among the strains that led to the QTL, we then searched the SNP databases for genes with coding region SNPs that fit the strain pattern; only Acads had such SNPs. We also compared gene expression of the nine genes between NZB and B6 and between NZB and NZW (Table 2). Only Rplp0 showed a significantly higher expression in NZB compared with B6 (1.38-fold, P < 0.005), but this difference could not explain all the crosses (Table 2). Of these candidate genes, we selected Acads as the most likely because it codes for acyl-CoA dehydrogenase short chain, which is in the fatty acid pathway, and because it had a coding region difference predicted to change function in a highly conserved region.
Acads differs in sequence and encoded protein levels
We sequenced Acads in strains 129, NZB, B6, NZW, SM, MRL, SJL, and RF. Sequencing Acads revealed some splice site variants and one polymorphism changing the amino acid from aspartic acid to glycine at amino acid 94 (Table 3). As this polymorphism changes both the charge and polarity of the amino acid and is in a highly conserved region of the protein (Fig. 4), it is likely to change the function of its encoded protein. We also sequenced Acads in strains PERA, I, and C3H, which along with B6 are the parental strains that failed to detect a QTL. These four strains did not differ in the amino acid: all have aspartic acid at amino acid 94 (Table 3).
TABLE 3.
Acads sequence variations among parents of crosses
| SNP | Build 37 | Position | Type | 129/NZB/ MRL/BALB/c | PERA | Othersa | CAST |
|---|---|---|---|---|---|---|---|
| rs33137118 | 115.563152 | exon 3 | Cn | G (Gly) | A (Asp) | A (Asp) | A (Asp) |
| rs33474157 | 115.562282 | exon 5 | Cs | G | A | A | G |
| rs6249609 | 115.561899 | exon 6 | Cs | C | C | T | C |
| rs6249067 | 115.561787 | intron 6 | splice site | G | A | A | |
| rs47957351 | intron 7 | intronic | G | A | A | ||
| rs6247384 | 115.561445 | intron 7 | intronic | A | G | G | G |
| rs6245762 | 115.561188 | intron 8 | intronic | G | A | A | A |
| rs6245197 | 115.561094 | intron 9 | intronic | C | T | T | T |
| intron 9 | splice site | del | C | C | del | ||
| rs33703028 | 115.561019 | intron 9 | splice site | C | T | T | T |
| rs33059464 | 115.560685 | exon 10 | 3′UTR | A | G | G | |
| rs33164602 | 115.560367 | exon 10 | 3′UTR | T | C | C |
SNP, single-nucleotide polymorphism.
Others includes NZW, B6, RF, SJL, SM, C3H, I/Ln.
Fig. 4.
Location of Acads amino acid changing polymorphism. Aspartic acid 94 (marked with *) in ACADS is in a conserved region of the protein sequence in mammals. Other strains include NZW, RF, SJL, B6, C3H, I/Ln, and PERA.
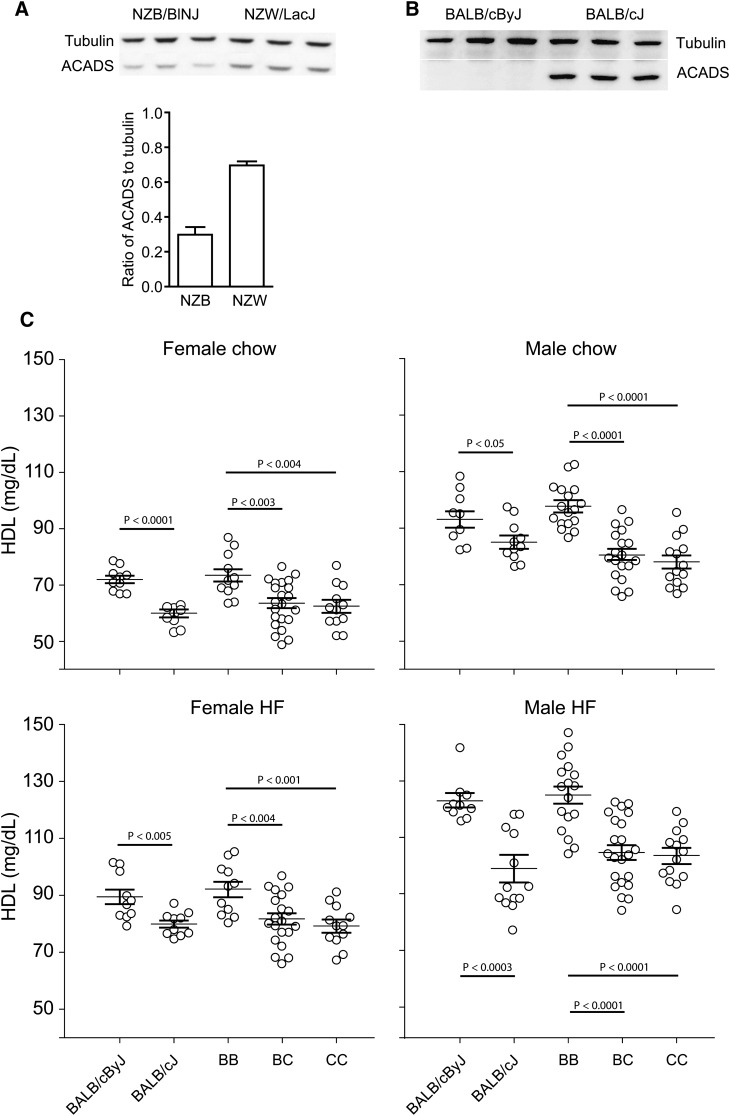
As previously reported, ACADS protein levels in liver are significantly higher in SM compared with NZB mice (14). To further investigate if this expression difference also exists between strains NZB and NZW, we measured ACADS levels in livers from NZB and NZW mice fed a high-fat diet for eight weeks. ACADS levels in NZW mice were increased significantly compared with NZB (Fig. 5A). Thus, a lower level of ACADS protein in the liver (strain NZB) results in higher plasma HDL; higher levels of ACADS protein (strains NZW and SM) results in lower plasma HDL.
Fig. 5.
ACADS expression and HDL levels in the cross of NZW × NZB and inbred strains BALB/cByJ and BALB/cJ mice. A: ACADS protein reduced in NZB compared with NZW mice as detected by Western blotting; densitometer readings provided under the gel (P < 0.0001). B: ACADS was totally deficient in BALB/cByJ mice compared with BALB/cJ. C: HDL concentrations in (BALB/cByJ × BALB/cJ) F2 mice and both parental strains on both chow (at 8 weeks of age) and after an 8-week high-fat (HF) diet (at 16 weeks of age). The F2 mice were divided into homozygous BALB/cByJ (BB), heterozygotes (BC), and homozygous BALB/cJ (CC) groups based on their genotypes in the ACADS-deficient region. Mice with BB genotype have significantly higher HDL levels compared with mice with either a BC or CC genotype (P < 0.01).
BALB/cBy mice, which are deficient in ACADS, have higher HDL levels compared with BALB/c mice
The BALB/cBy strain is derived from the BALB/c strain. An Acads deletion appeared spontaneously in the 3′ end of the gene in BALB/cBy after the separation of this strain from BALB/c, causing a deficiency in ACADS activity in BALB/cBy that is not present in BALB/c (18). We examined ACADS protein levels in livers from BALB/cBy and BALB/c mice by Western blotting analysis and verified that ACADS was totally deficient in BALB/cBy mice (Fig. 5B). If Acads is the causal QTL gene, then this mutation in BALB/cBy should cause an increase in plasma HDL levels compared with its companion strain BALB/c. This prediction was true: HDL levels were significantly increased in BALB/cBy compared with BALB/c for both males and females fed a chow or high-fat diet (Fig. 5C).
To verify that the difference in HDL between BALB/cBy and BALB/c was due to the Acads mutation, we intercrossed these two strains, measured plasma HDL concentrations in F2 mice on a chow or high-fat diet at 16 weeks of age, and genotyped all F2 mice for the Acads deletion. Compared with mice homozygous for the BALB/c-like genotype, the HDL concentrations were significantly higher in mice homozygous for the BALB/cBy-like genotype (deficient in ACADS), regardless of sex or diet (Fig. 5C). This result proved that Acads deficiency caused an increase in HDL.
DISCUSSION
We used the genetic and genomic resources of the mouse to untangle a complex QTL found on distal Chr 5 in multiple crosses. Several of these crosses had a broad confidence interval and double peaks, suggesting the existence of more than one QTL in the region. Previously we used B6 × NZB advanced intercross lines, which accumulated large numbers of recombinations in the random breeding from generations F3-F11, to demonstrate that there were two QTL in the region: Hdlq8 at 113 Mb and Hdlq1 at 125 Mb (11). We narrowed the Hdlq1 region using haplotype mapping and tested an obvious candidate gene, Scarb1. We found several lines of evidence that it was the QTL gene for Hdlq1. We then focused on identifying the candidate gene for Hdlq8. Using haplotype analysis, we first confirmed the presence of Hdlq8 by selecting and crossing two strains (NZB and NZW) that differ in the Hdlq8 region but not in the Scarb1 region. The F2 mice from this cross did show a significant QTL at 115 Mb, confirming the existence of a QTL gene separate from Scarb1. We recognized the possibility that this cross might not have detected a QTL because NZB and NZW are alike for a short region from 107 to 109 Mb. If this had been the result, we would have then tested a cross comprising two other strains that were different in this 3 Mb region but were alike at Scarb1.
After confirming the QTL, we used haplotype analysis on all the strains to narrow the region. This reduced the region to nine genes. We sequenced the most likely candidate, Acads, and found a polymorphism that changed the charge and polarity of an amino acid located in a highly conserved region. Because the protein level was also decreased, this amino acid change may have destabilized the protein. Searching the literature, we found a previously identified mutation in Acads and showed that the mutation did alter the HDL phenotype. Taken together, this compelling evidence supports Acads as the causal gene for Hdlq8.
How a deficiency of ACADS protein functions to increase plasma HDL is not clearly known, but the literature does suggest a mechanism. Acads encodes short-chain acyl-CoA dehydrogenase, which participates in the β-oxidation of short-chain fatty acids. Acads-deficient BALB/cByJ mice accumulate and secrete fatty acid metabolites in the urine and develop fatty liver with hypoglycemia after fasting for 18 h (18, 19). Unsaturated fatty acids destabilize ABCA1, increase its degradation, and inhibit cholesterol efflux from macrophages (20, 21). Future studies are required to determine whether the increase in fatty acids caused by Acads deficiency affects this key step in HDL metabolism and whether the increase in HDL caused by Acads deficiency is atheroprotective.
The question of how many genes on distal Chr 5 can cause an HDL QTL is not completely resolved. Although there is a homologous human QTL at 12q24.23 (22, 23), two recent genome-wide association studies showed that SNPs within a cluster of five genes, including MVK and MMAB, showed significant association with HDL levels (5, 6). There is a 10 Mb gap between ACADS and the other gene cluster (ACADS at 119.65 Mb; MMAB at 108.47 Mb; and MVK at 108.49 Mb). In mice, however, all three genes are very close together (Acads at 115.37 Mb; Mvk at 114.69 Mb; and Mmab at 114.70 Mb). In the case of BALB/cBy and BALB/c, the sequence of Mvk is identical in both strains, so Acads must be the gene responsible for the difference in HDL expression observed between these strains. However, Mvk does carry polymorphisms that change amino acids that differ among the other strains, giving rise to these QTL in the mouse, although no single polymorphism could explain all the crosses. We cannot exclude the possibility that Mvk might also play a role in HDL metabolism in mice. Mvk encodes mevalonate kinase, which catalyzes an early step in cholesterol biosynthesis. It may be that the HDL gene in human and mouse is different: MVK might be the HDL gene in human, and Acads might be the HDL gene in mouse. Future studies may elucidate the relationship of Mvk or Mmab and Acads in HDL metabolism. However, we think the evidence presented in this article, particularly finding the responsible polymorphism and the spontaneous mutation in BALB/cBy, demonstrates clearly that Acads can alter HDL levels.
Acknowledgments
The authors are most grateful to Harry Whitmore and Fred Rumill for their invaluable help in mouse husbandry, Cynthia McFarland for the excellent technical assistance, and Joanne Currer for editorial assistance.
Footnotes
Abbreviations:
- Chr
- chromosome
- LOD
- logarithm of the odds ratio
- QTL
- quantitative trait loci
- SNP
- single-nucleotide polymorphism
- SR-B1
- scavenger receptor B1
This work was supported by National Institutes of Health Grants HL-081162 and HL-077796 (B.P.), American Heart Association Grant 0725905T (Z.S.), an American Heart Association fellowship (M.S.L.), and National Cancer Institute Cancer Core Grant CA-034196 (The Jackson Laboratory). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Rollins J., Chen Y., Paigen B., Wang X. 2006. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc. Med. 16: 220–234. [DOI] [PubMed] [Google Scholar]
- 2.DiPetrillo K., Wang X., Stylianou I. M., Paigen B. 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21: 683–692. [DOI] [PubMed] [Google Scholar]
- 3.Burgess-Herbert S. L., Cox A., Tsaih S. W., Paigen B. 2008. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 180: 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., Paigen B. J., Graber J. H., de Villena F. P., Churchill G. A. 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods. 6: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korstanje R., Li R., Howard T., Kelmenson P., Marshall J., Paigen B., Churchill G. 2004. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J. Lipid Res. 45: 881–888. [DOI] [PubMed] [Google Scholar]
- 8.Mehrabian M., Castellani L. W., Wen P. Z., Wong J., Rithaporn T., Hama S. Y., Hough G. P., Johnson D., Albers J. J., Mottino G. A., et al. 2000. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei × C57BL/6J). J. Lipid Res. 41: 1936–1946. [PubMed] [Google Scholar]
- 9.Su Z., Ishimori N., Chen Y., Leiter E. H., Churchill G. A., Paigen B., Stylianou I. M. 2009. Four additional mouse crosses improve the lipid QTL landscape and identify Lipg as a QTL gene. J Lipid Res. 50: 2083–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Z., Wang X., Tsaih S. W., Zhang A., Cox A., Sheehan S., Paigen B. 2009. Genetic basis of HDL variation in 129/SvImJ and C57BL/6J mice: importance of testing candidate genes in targeted mutant mice. J. Lipid Res. 50: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Le Roy I., Nicodeme E., Li R., Wagner R., Petros C., Churchill G. A., Harris S., Darvasi A., Kirilovsky J., et al. 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13: 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wergedal J. E., Ackert-Bicknell C. L., Beamer W. G., Mohan S., Baylink D. J., Srivastava A. K. 2007. Mapping genetic loci that regulate lipid levels in a NZB/B1NJxRF/J intercross and a combined intercross involving NZB/B1NJ, RF/J, MRL/MpJ, and SJL/J mouse strains. J. Lipid Res. 48: 1724–1734. [DOI] [PubMed] [Google Scholar]
- 13.Su Z., Cox A., Shen Y., Stylianou I. M., Paigen B. 2009. Farp2 and Stk25 are candidate genes for the HDL cholesterol locus on mouse chromosome 1. Arterioscler. Thromb. Vasc. Biol. 29: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stylianou I. M., Affourtit J. P., Shockley K. R., Wilpan R. Y., Abdi F. A., Bhardwaj S., Rollins J., Churchill G. A., Paigen B. 2008. Applying gene expression, proteomics and SNP analysis for complex trait gene identification. Genetics. 178: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittenburg H., Lyons M. A., Li R., Kurtz U., Wang X., Mossner J., Churchill G. A., Carey M. C., Paigen B. 2006. QTL mapping for genetic determinants of lipoprotein cholesterol levels in combined crosses of inbred mouse strains. J. Lipid Res. 47: 1780–1790. [DOI] [PubMed] [Google Scholar]
- 16.Stylianou I. M., Svenson K. L., VanOrman S. K., Langle Y., Millar J. S., Paigen B., Rader D. J. 2009. Novel ENU-induced point mutation in scavenger receptor class B, member 1, results in liver specific loss of SCARB1 protein. PLoS ONE. 4: e6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Z., Tsaih S. W., Szatkiewicz J., Shen Y., Paigen B. 2008. Candidate genes for plasma triglyceride, FFA, and glucose revealed from an intercross between inbred mouse strains NZB/B1NJ and NZW/LacJ. J. Lipid Res. 49: 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiffer S. P., Prochazka M., Jezyk P. F., Roderick T. H., Yudkoff M., Patterson D. F. 1989. Organic aciduria and butyryl CoA dehydrogenase deficiency in BALB/cByJ mice. Biochem. Genet. 27: 47–58. [DOI] [PubMed] [Google Scholar]
- 19.Wood P. A., Amendt B. A., Rhead W. J., Millington D. S., Inoue F., Armstrong D. 1989. Short-chain acyl-coenzyme A dehydrogenase deficiency in mice. Pediatr. Res. 25: 38–43. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Kurdi-Haidar B., Oram J. F. 2004. LXR-mediated activation of macrophage stearoyl-CoA desaturase generates unsaturated fatty acids that destabilize ABCA1. J. Lipid Res. 45: 972–980. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Oram J. F. 2002. Unsaturated fatty acids inhibit cholesterol efflux from macrophages by increasing degradation of ATP-binding cassette transporter A1. J. Biol. Chem. 277: 5692–5697. [DOI] [PubMed] [Google Scholar]
- 22.Bielinski S. J., Tang W., Pankow J. S., Miller M. B., Mosley T. H., Boerwinkle E., Olshen R. A., Curb J. D., Jaquish C. E., Rao D. C., et al. 2006. Genome-wide linkage scans for loci affecting total cholesterol, HDL-C, and triglycerides: the Family Blood Pressure Program. Hum. Genet. 120: 371–380. [DOI] [PubMed] [Google Scholar]
- 23.Ng M. C., So W. Y., Lam V. K., Cockram C. S., Bell G. I., Cox N. J., Chan J. C. 2004. Genome-wide scan for metabolic syndrome and related quantitative traits in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21-q25. Diabetes. 53: 2676–2683. [DOI] [PubMed] [Google Scholar]