Abstract
The cytokines IL-1 and TNF induce expression of a series of genes that regulate inflammation through activation of NF-κB signal transduction pathways. TAK1, a MAPKKK, is critical for both IL-1- and TNF-induced activation of the NF-κB pathway. TAB2, a TAK1-binding protein, is involved in IL-1-induced NF-κB activation by physically linking TAK1 to TRAF6. However, IL-1-induced activation of NF-κB is not impaired in TAB2-deficient embryonic fibroblasts. Here we report the identification and characterization of a novel protein designated TAB3, a TAB2-like molecule that associates with TAK1 and can activate NF-κB similar to TAB2. Endogenous TAB3 interacts with TRAF6 and TRAF2 in an IL-1- and a TNF-dependent manner, respectively. Further more, IL-1 signaling leads to the ubiquitination of TAB2 and TAB3 through TRAF6. Cotransfection of siRNAs directed against both TAB2 and TAB3 inhibit both IL-1- and TNF-induced activation of TAK1 and NF-κB. These results suggest that TAB2 and TAB3 function redundantly as mediators of TAK1 activation in IL-1 and TNF signal transduction.
Keywords: IL-1/NF-κB/TAB2/TAB3/TAK1
Introduction
The pro-inflammatory cytokines IL-1 and TNF have several effects in the inflammation process. Stimulation of cells with IL-1 or TNF initiates a cascade of signaling events, including activation of NF-κB and mitogen-activated protein kinases (MAPKs) such as JNK and p38. These, in turn, upregulate the expression of many pro-inflammatory genes in the nucleus (Dinarello, 1996; Baud and Karin, 2001). NF-κB is normally sequestered in the cytoplasm of resting cells by association with inhibitory IκB proteins. This interaction masks the nuclear localization signal of NF-κB, preventing its nuclear translocation (Ghosh et al., 1998; Karin and Ben-Neriah, 2000; Li and Verma, 2002). Stimulation by IL-1 or TNF results in the phosphorylation of the IκB proteins, tagging them for ubiquitination and subsequent proteosome-mediated degradation. This results in the release of NF-κB, which translocates to the nucleus where it activates the transcription of specific target genes (Karin and Ben-Neriah, 2000; Li and Verma, 2002). Phosphorylation of IκB in response to extracellular stimuli is carried out by the IκB kinase (IKK) complex, which is comprised of two catalytic subunits, IKKα and IKKβ, as well as the modulator NEMO/IKKγ (Silverman and Maniatis, 2001; Ghosh and Karin 2002; Li and Verma, 2002).
Members of the TNF-receptor-associated factor (TRAF) family of adaptor proteins are involved in coupling stimulation of the TNF receptors (TNFRs) and the IL-1 receptor (IL-1R) to NF-κB activation and other downstream events (Silverman and Maniatis, 2001). TRAF2 plays a critical role in signal transduction mediated by both TNFR1 and TNFR2, and has been implicated in TNF-induced activation of NF-κB and MAPKs (Yeh et al., 1997). Similarly, TRAF6 is important for the transduction of IL-1-induced signals, including those resulting in NF-κB and MAPK activation (Cao et al., 1996; Lomaga et al., 1999; Naito et al., 1999). The TRAF proteins physically and functionally connect TNFRs and IL-1R to intracellular protein kinases, thereby linking these receptors to downstream signaling pathways.
TAK1, a member of the MAPKKK family, also participates in the IL-1-mediated signaling pathway (Yamaguchi et al., 1995; Ninomiya-Tsuji et al., 1999). Following exposure of cells to IL-1, endogenous TAK1 is recruited to the TRAF6 complex and activated, whereupon it stimulates NF-κB and MAPK activation. In previous studies, the yeast two-hybrid system was employed to isolate TAB1 and TAB2 proteins that interact with TAK1 (Shibuya et al., 1996; Takaesu et al., 2000). TAB1 was found to augment the kinase activity of TAK1 when coexpressed (Shibuya et al., 1996), indicating that it functions as an activator of TAK1. TAB2 was shown to be an intermediate in the IL-1 signaling pathway (Takaesu et al., 2000, 2001). TAB2 functions as an adaptor that links TAK1 and TRAF6 in response to IL-1 and thereby mediates TAK1 activation. These results suggest that IL-1 activation of the NF-κB and MAPK cascades involves the formation of a TRAF6–TAB2–TAK1 complex. In addition, a biochemical study has identified TRIKA1 and TRIKA2 as signaling components that are able to activate the IKK complex in a TRAF6-dependent manner (Deng et al., 2000; Wang et al., 2001). TRIKA1 consists of the ubiquitin-conjugating enzyme Ubc13 and the Ubc-like protein Uev1A, while TRIKA2 is a ternary complex composed of TAK1, TAB1 and TAB2. Thus, TRAF6-mediated ubiquitination appears to play an important role in TAK1 activation.
Previously, in order to explore the physiological importance of TAK1 in activating the NF-κB pathway in response to IL-1, we utilized small interfering RNA (siRNA) directed against TAK1 (Takaesu et al., 2003). Our previous studies confirmed that TAK1 is critical for IL-1-induced activation of the NF-κB pathway. Furthermore, our results indicated that TAK1 is also important for NF-κB activation in response to TNF via the inducible association of TAK1 with TRAF2 (Takaesu et al., 2003). On the other hand, recent studies have demonstrated that mouse embryo fibroblasts (MEFs) deficient in TAB2 exhibit normal IL-1- and TNF-induced activation of NF-κB (Sanjo et al., 2003). This result indicates that TAB2 is not essential for IL-1 or TNF signaling in MEFs. However, there remains the possibility that another TAB2-like molecule may compensate for the loss of TAB2 and support TAK1 kinase activity.
Here we describe the identification and characterization of TAB3, a novel TAK1-binding protein that is closely related to TAB2 in structure. TAB3 activates TAK1 and mediates its interaction with TRAF2 and TRAF6. TAB3 rapidly and transiently associates with TRAF6 and TRAF2 in an IL-1- and a TNF-dependent manner, respectively. IL-1 stimulation and TRAF6 overexpression induce ubiquitination of the TAB2 and TAB3 proteins. In this study, we utilized siRNA directed against TAB2 and TAB3 to further explore their roles in activation of the NF-κB and MAPK pathways following treatment of cells with IL-1 and TNF. These studies demonstrate that TAB2 and TAB3 are important for NF-κB and MAPK activation in response to IL-1 and TNF, and suggest that TAB2 and TAB3 have redundant functions as mediators of TAK1 activation in IL-1 and TNF signal transduction.
Results
Isolation of TAB3
In an attempt to identify new signal transducers involved in TAK1 activation in the IL-1 and TNF signaling pathways, we searched EST databases for sequences similar to TAB2. Multiple human and murine EST sequences were found which encode polypeptides sharing significant homology with TAB2. We cloned one full-length human TAB2 homolog by PCR from a human kidney cDNA library. This human TAB2-like gene, termed TAB3, was predicted to encode a 712 amino acid polypeptide (Figure 1A). Similar to TAB2, TAB3 contains a ubiquitin-binding motif (Shih et al., 2003) near its N-terminus and is predicted to encode an α-helical coiled-coil region in its C-terminus. Thus TAB3 is structurally related to TAB2. We also found two highly conserved homologs of TAB3 in Xenopus laevis. While this manuscript was in preparation, one of the homologs was reported by Munoz-Sanjuan and coworkers (Munoz-Sanjuan et al., 2002). Both of the Xenopus cDNAs resemble TAB3 more closely than TAB2. These workers (Munoz-Sanjuan et al., 2002) also described mouse and human TAB3. Furthermore, Drosophila contains a TAB2/3-like protein carrying the ubiquitin-binding motif and the α-helical coiled-coil region in its N- and C-termini, respectively (Figure 1A).
Fig. 1. Structure of TAB3. (A) Comparison of amino acid sequences among hTAB3 (human), hTAB2 (human) and DTAB2 (Drosophila). They share the CUE domain (bold underline) and coiled-coil structure (box). Identical and conserved amino acids are indicated by black and gray boxes, respectively. DDBJ/EMBL/GenBank accession No. for hTAB3 is AY437560. (B) Schematic representation of various TAB3 constructs. Gray and black boxes indicate the CUE domain and coiled-coil structure, respectively.
TAB3 associates with TAK1
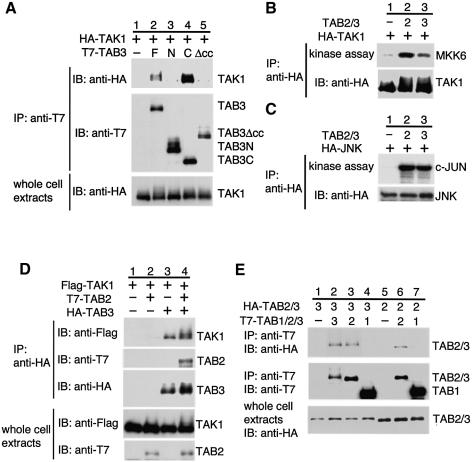
We have shown previously that TAB2 interacts with TAK1 through the C-terminal region of TAB2, and that overexpression of TAB2 can induce TAK1 kinase activity (Takaesu et al., 2000). To examine whether TAB3 functions similarly, the interaction of TAB3 with TAK1 was investigated in mammalian cells by in vivo coprecipitation. Human 293 embryonic kidney cells were cotransfected with T7-TAB3 and HA-TAK1. Cell extracts were immunoprecipitated with anti-T7 antibody, and coprecipitated HA-TAK1 was detected by immunoblotting with anti-HA antibody. TAB3 was found to associate with TAK1 (Figure 2A, lane 2). To verify that TAK1 associates with the C-terminal region of TAB3 in mammalian cells, we used two truncated proteins: T7-TAB3N, consisting of the N-terminus of TAB3 (amino acids 1–392), and T7-TAB3C, consisting of the C-terminus (amino acids 393–712) (Figure 1B). Immune complex assays showed that TAK1 coimmunoprecipitated with T7-TAB3C (Figure 2A, lane 4), but not with T7-TAB3N (lane 3). The C-terminal domain of TAB3 contains a coiled-coil structure (Figure 1A). To examine whether this coiled-coil region is involved in the interaction with TAK1, we constructed the mutant protein TAB3Δcc, which lacks this domain (amino acids 478–661) (Figure 1B). Coimmunoprecipitation analysis from cells coexpressing T7-TAB3Δcc and HA-TAK1 demonstrated that the TAB3Δcc protein failed to interact with TAK1 (Figure 2A, lane 5). These results confirm that the C-terminal coiled-coil region of TAB3 is responsible for its association with TAK1.
Fig. 2. TAB3 interacts with and activates TAK1. (A) Interaction of TAB3 with TAK1. The 293 cells were transfected with plasmids encoding T7-TAB3 full-length (F), T7-TAB3N (N), T7-TAB3C (C), T7-TAB3Δcc (Δcc) and HA-TAK1 as indicated. Complexes immunoprecipitated with anti-T7 antibody were immunoblotted with anti-HA or anti-T7 antibodies. Whole-cell extracts were immunoblotted with anti-HA antibody. (B and C) Activation of TAK1 and JNK by TAB3. The 293 cells were transfected with plasmids encoding HA-TAK1, HA-JNK, T7-TAB2 (2) and T7-TAB3 (3) as indicated. HA-TAK1 or HA-JNK was immunoprecipitated with anti-HA antibody. The immunoprecipitates were subjected to an in vitro phosphorylation assay using bacterially expressed MKK6 (B) or GST-c-Jun (C) as an exogenous substrate. The immunoprecipitates were analyzed by immunoblotting with anti-HA antibody. (D) Effect of TAB2 on the interaction between TAK1 and TAB3. The 293 cells were transfected with plasmids encoding HA-TAB3, T7-TAB2 and Flag–TAK1 as indicated. Complexes immunoprecipitated with anti-HA antibody were immunoblotted with anti-Flag, anti-T7 or anti-HA antibodies. Whole-cell extracts were immunoblotted with anti-Flag or anti-T7 antibodies. (E) Interaction among TAK1-binding proteins. The 293 cells were transfected with plasmids encoding HA-TAB2 (2), HA-TAB3 (3), T7-TAB1 (1), T7-TAB2 (2) and T7-TAB3 (3) as indicated. Complexes immunoprecipitated with anti-T7 antibody were immunoblotted with anti-HA or anti-T7 antibodies. Whole-cell extracts were immunoblotted with anti-HA antibody.
To examine whether TAB3 can induce TAK1 activation, we transfected 293 cells with HA-TAK1 in the presence or absence of the TAB3 expression vector. Transiently expressed TAK1 was immunoprecipitated using anti-HA antibody, and kinase activity was measured by in vitro kinase assay using MKK6 as a substrate. When expressed alone, TAK1 exhibited low basal kinase activity (Figure 2B, lane 1). However, coexpression of TAB3 led to a marked enhancement in TAK1 catalytic activity (lane 3), although the degree of activation was weaker than that induced by TAB2 (lane 2). Since activation of TAK1 induces JNK activation (Shirakabe et al., 1997), we analyzed the effect of ectopically expressed TAB3 on JNK activity. First, 293 cells were cotransfected with HA-JNK and TAB3 or TAB2. Then, JNK activity was determined by immunoprecipitation of JNK followed by in vitro kinase assay using GST-c-Jun protein as a substrate. We observed that TAB2 and TAB3 significantly induced activation of JNK (Figure 2C, lanes 2 and 3). Taken together, these results suggest that the function of TAB3 is similar to that of TAB2.
Since TAB2 and TAB3 each interact with TAK1 via their respective C-terminal coiled-coil domain, we examined whether the bindings of TAB2 and TAB3 to TAK1 are mutually exclusive. We cotransfected 293 cells with Flag-TAK1 and HA-TAB3 in the presence or absence of T7-TAB2. We confirmed that TAK1 and TAB3 could interact in the absence of TAB2 coexpression (Figure 2D, top panel, lane 3). Coexpression of T7-TAB2 did not block the interaction of TAB3 with TAK1 (lane 4). Furthermore, T7-TAB2 coimmunoprecipitated with HA-TAB3 (second panel, lane 4), suggesting that TAB2 and TAB3 form a complex. To confirm this possibility, T7-TAB2 and HA-TAB3 were coexpressed in 293 cells and coimmunoprecipitation analysis was performed. We found that TAB2 associated with TAB3 (Figure 2E, lane 3). Furthermore, when 293 cells were cotransfected with the combination of HA-TAB3 and T7-TAB3 or HA-TAB2 and T7-TAB2, we found that TAB3 and TAB2 each self-associated (lanes 2 and 6). These results suggest that homo- and hetero-oligomers of TAB2 and TAB3 form a complex with TAK1. We failed to detect any association of TAB1 with TAB3 or TAB2 when overexpressed (lanes 4 and 7).
TAB3 is involved in NF-κB activation
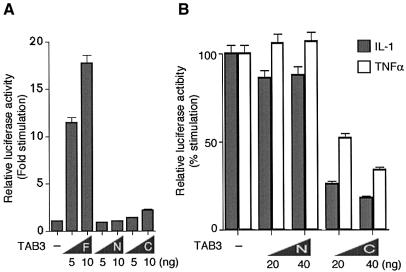
Given the sequence homology of TAB3 to TAB2, a known activator of NF-κB (Takaesu et al., 2000), we investigated whether TAB3 might play a role in NF-κB activation. When TAB3 and an NF-κB-dependent luciferase reporter were cotransfected into 293 cells, TAB3 was found to activate the reporter gene in a dose-dependent manner (Figure 3A). Intact TAB3 was required for this activity, as truncated derivatives of TAB3 failed to induce NF-κB activity. Wild-type and mutant proteins were expressed in comparable amounts, as shown by western blot analysis (data not shown).
Fig. 3. TAB3 is involved in NF-κB activation pathway. (A) Effects of TAB3 on NF-κB-dependent reporter gene activity. The 293 cells were transfected with luciferase reporter plasmid (0.1 µg) and the indicated amounts of plasmids encoding TAB3 full-length (F), TAB3N (N) and TAB3C (C). After 24 h incubation, cells were harvested and luciferase activity measured. The values shown are the average of one representative experiment in which each transfection was performed in duplicate. (B) Effects of TAB3 on IL-1- and TNFα-induced NF-κB-activation. 293IL-1RI cells were transfected with luciferase reporter plasmid (0.1 µg) and the indicated amounts of plasmids encoding TAB3N (N) and TAB3C (C). IL-1β (5 ng/ml) or TNFα (10 ng/ml) was added to each plate 3 h after transfection. Cells were harvested 24 h after transfection and luciferase activity was measured. The values shown are the average of one representative experiment in which each transfection was performed in duplicate.
We next tested whether TAB3 is involved in the IL-1 or TNF signaling pathway that leads to NF-κB activation. Since mutants of TAB2 lacking the C-terminus function as dominant negatives (Takaesu et al., 2000), we made a homologous truncated derivative of TAB3 (TAB3C) (Figure 1B). First, 293 cells were transfected with an NF-κB-dependent luciferase reporter and increasing concentrations of TAB3C. Later, cells were treated with IL-1 or TNF and luciferase activity was determined. Figure 3B shows that increasing concentrations of TAB3C potently inhibited induction of NF-κB by IL-1 and TNF. These results suggest that TAB3 may be a common downstream mediator of NF-κB activation by IL-1 and TNF.
TAB3 associates with TRAF6 and TRAF2
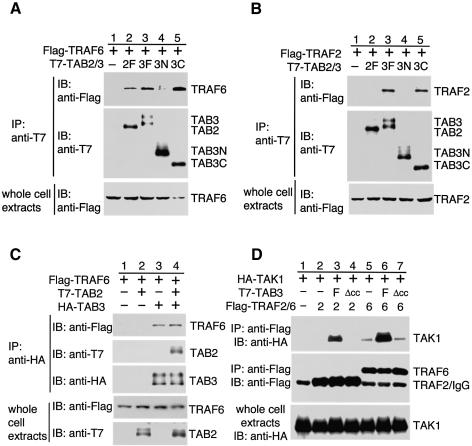
IL-1 and TNF activate their signal pathways via distinct families of cell-surface receptors. However, both pathways utilize members of the TRAF family of adaptor proteins as signal transducers (Silverman and Maniatis, 2001). The TRAF proteins share homology at their C-terminal domains, but their binding properties and activities differ. For example, whereas TRAF6 is essential for IL-1 signaling (Lomaga et al., 1999; Naito et al., 1999), TRAF2 is involved in TNF signaling (Yeh et al., 1997). Recently, it was shown that TAB2 interacts specifically with TRAF6 (Takaesu et al., 2000). Therefore we tested the ability of TAB3 to bind to TRAF6 and TRAF2. T7-TAB2 or T7-TAB3 was expressed together with Flag-TRAF6 or Flag-TRAF2 in 293 cells and immunoprecipitated with anti-T7 antibody. The immune complexes were subjected to immunoblotting with anti-Flag antibody. As observed previously, TAB2 was found to interact with TRAF6 (Figure 4A, lane 2), but not with TRAF2 (Figure 4B, lane 2). In contrast, TAB3 was found to coprecipitate both TRAF6 (Figure 4A, lane 3) and TRAF2 (Figure 4B, lane 3) efficiently. This is consistent with the notion that TAB3 is involved in both the IL-1 and TNF signaling pathways. To determine whether TAB2 and TAB3 competitively bind to TRAF6, we analyzed the interaction between TAB3 and TRAF6 in the presence or absence of TAB2. When Flag-TRAF6 and HA-TAB3 were expressed together with T7-TAB2 in 293 cells, TAB2 formed a complex with TAB3 (Figure 4C, second panel, lane 4) and did not interfere with the interaction between TAB3 and TRAF6 (top panel, lane 4). These results suggest that TAB2 and TAB3 bind cooperatively, but not competitively, to TRAF6.
Fig. 4. TAB3 mediates the interaction of TAK1 with TRAF6 and TRAF2. (A and B) Interaction of TAB3 with TRAF6 and TRAF2. The 293 cells were transfected with plasmids encoding T7-TAB2 full-length (2F), T7-TAB3 full-length (3F), T7-TAB3N (3N), T7-TAB3C (3C), Flag-TRAF6 and Flag-TRAF2 as indicated. Complexes immunoprecipitated with anti-T7 antibody were immunoblotted with anti-Flag or anti-T7 antibodies. Whole-cell extracts were immunoblotted with anti-Flag antibody. (C) Effect of TAB2 on the interaction between TRAF6 and TAB3. The 293 cells were transfected with plasmids encoding HA-TAB3, T7-TAB2 and Flag-TRAF6 as indicated. Complexes immunoprecipitated with anti-HA antibody were immunoblotted with anti-Flag, anti-T7 or anti-HA antibodies. Whole-cell extracts were immunoblotted with anti-Flag or anti-T7 antibodies. (D) Effect of TAB3 on the interaction of TAK1 with TRAF2 and TRAF6. The 293 cells were transfected with plasmids encoding HA-TAK1, T7-TAB3 full-length (F), T7-TAB3Δcc (Δcc), Flag-TRAF2 (2) and Flag-TRAF6 (6) as indicated. Complexes immunoprecipitated with anti-Flag antibody were immunoblotted with anti-HA or anti-Flag antibodies. Whole-cell extracts were immunoblotted with anti-HA antibody.
To determine the regions within TAB3 responsible for its interaction with TRAF6 and TRAF2, we performed immunoprecipitation assays using deletion mutants of TAB3. We found that TRAF6 and TRAF2 coprecipitated with TAB3C (Figure 4A and B, lane 5), but not with TAB3N (lane 4). Thus the C-terminal domain of TAB3 is required for binding to TRAF6 and TRAF2. This is similar to TAB2, which also requires its C-terminal domain for binding to TRAF6. We next tested whether the C-terminal coiled-coil region of TAB3 is essential for its interaction with TRAF6 and TRAF2. We found that the TAB3Δcc mutant (Figure 1B), which lacks the coiled-coil motif, was still able to associate with TRAF6 (see Figure 6A below) and TRAF2 (data not shown). Thus, the coiled-coil motif of TAB3 is required for its interaction with TAK1, but not with TRAF6 or TRAF2.
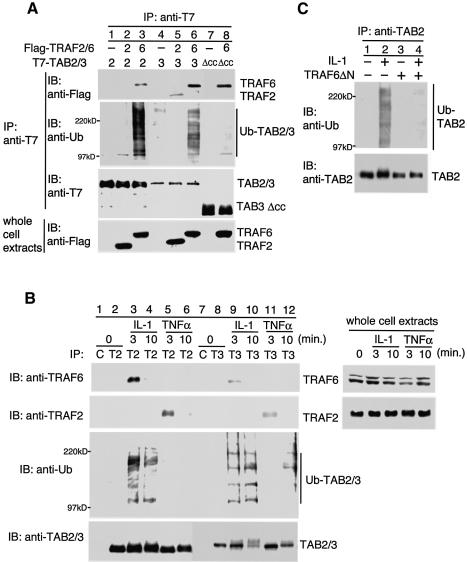
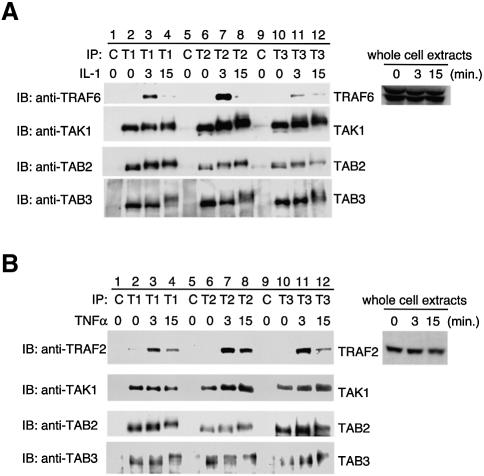
Fig. 6. Ubiquitination of TAB2 and TAB3. (A) Effect of TRAF2 and TRAF6 on ubiquitination of TAB2 and TAB3. 293IL-1RI cells were transfected with plasmids encoding T7-TAB2 (2), T7-TAB3 (3), T7-TAB3Δcc (Δcc), Flag-TRAF2 (2) and Flag-TRAF6 (6) as indicated. Complexes immunoprecipitated with anti-T7 antibody were immunoblotted with anti-Flag, anti-ubiquitin (anti-Ub) or anti-T7 antibodies. Whole-cell extracts were immunoblotted with anti-Flag antibody. (B) Effect of IL-1 and TNFα stimulation on ubiquitination of TAB2 and TAB3. 293IL-1RI cells were treated with IL-1 (10 ng/ml) or TNFα (20 ng/ml) for the indicated time periods. Cell extracts were subjected to immunoprecipitation with control IgG (C), anti-TAB2 (T2) or anti-TAB3 (T3) antibodies. Immunoprecipitated complexes were immunoblotted with anti-TRAF6, anti-TRAF2, anti-Ub, anti-TAB2 or anti-TAB3 antibodies. Whole-cell extracts were immunoblotted with anti-TRAF6 or anti-TRAF2 antibodies. (C) Effect of TRAF6ΔN on IL-1-induced ubiquitination of TAB2. 293IL-1RI cells were transfected with a plasmid encoding TRAF6ΔN. At 48 h post-transfection, cells were treated with IL-1 (10 ng/ml) for 10 min. Cell extracts were subjected to the immunoprecipitation with anti-TAB2 antibody. Immunoprecipitated complexes were immunoblotted with anti-Ub or anti-TAB2 antibodies.
We have previously shown that TAB2 functions as an adaptor protein mediating the association of TAK1 with TRAF6 (Takaesu et al., 2000). The ability of TAB3 to interact with both TAK1 and TRAF6 led us to hypothesize that TAB3 similarly acts as a link between TRAF6 and TAK1. To test this possibility, we analyzed the interaction between TAK1 and TRAF6 in both the presence and absence of TAB3. Although a small amount of TAK1 was found to associate with TRAF6 in the absence of exogenous TAB3 (Figure 4D, lane 5), overexpression of TAB3 strongly enhanced the association between TRAF6 and TAK1 (lane 6). This indicates that TAB3, at least when overexpressed, can link TRAF6 to TAK1. We next examined the effect of the TAB3Δcc mutant, lacking the coiled-coil motif, on TRAF6–TAK1 complex formation. As described above, we had observed that TAB3Δcc interacted with TRAF6 but not with TAK1. In the present experiment, we observed that overexpression of TAB3Δcc did not enhance the association between TRAF6 and TAK1 (lane 7). These results support the idea that TAB3 is an intermediate signaling molecule linking TAK1 and TRAF6.
We were interested to determine whether the presence of TAB3 could also affect the ability of TAK1 to interact with TRAF2. To address this point, Flag-TRAF2 and HA-TAK1 were cotransfected into 293 cells with or without T7-TAB3. In the absence of TAB3 expression, no association of TAK1 with TRAF2 could be determined (Figure 4D, lane 2). However, upon coexpression of TAB3, we were able to detect the association between TAK1 and TRAF2 (lane 3). Overexpression of TAB3Δcc failed to enhance the interaction of TAK1 with TRAF2 (lane 4). Taken together, these results suggest that TAB3 functions as an adaptor for the association of TAK1 with both TRAF6 and TRAF2.
Ligand-dependent endogenous interaction of TAB3 with TRAF6 and TRAF2
To evaluate the interaction of TAB3 with TRAF6 and TRAF2 under more physiological conditions, we examined the association of endogenous TAB3 with TRAF6 and TRAF2 in 293IL-1RI cells. A rabbit anti-TAB3 polyclonal antibody was generated to identify endogenous TAB3 protein. When lysates prepared from 293IL-1RI cells were subjected to immunoprecipitation followed by western blotting with anti-TAB3 antibody, one band at approximately 90 kDa was observed (Figure 5A, bottom panel, lane 10). This band was not observed when control IgG was used for immunoprecipitation (lane 9), indicating that it represents endogenous TAB3. Lysates from IL-1-treated cells were immunoprecipitated with anti-TAB3 antibody and then analyzed by immunoblotting with anti-TRAF6 antibody. We found that TRAF6 rapidly associated with TAB3 in an IL-1-dependent manner (top panel, lanes 10–12). This interaction was observed within 3 min after IL-1 treatment, and decreased thereafter. Thus the interaction between TAB3 and TRAF6 is physiologically induced by IL-1. The kinetics of IL-1-induced TRAF6–TAB3 association were similar to those for TRAF6–TAK1 (lanes 2–4) and TRAF6–TAB2 (lanes 6–8) association. TNF did not induce the interaction of TRAF6 with TAB2 or TAB3 (see Figure 6B). Thus the interaction of TRAF6 with TAB2 and TAB3 is signaled specifically and physiologically by IL-1. When TAK1 was immunoprecipitated with anti-TAK1 antibody, the TAK1 immunocomplexes were found to contain TAB2 and TAB3 even in the absence of IL-1 stimulation (Figure 5A, third and bottom panels, lane 2). In addition, we found that TAB2 and TAB3 associated constitutively (bottom panel, lane 6, and third panel, lane 10). Thus TAK1, TAB2 and TAB3 form a complex in the absence of stimulation. This is consistent with recent observations that the TRIKA2 complex containing TAK1, TAB1 and TAB2 is formed before IL-1 stimulation (Wang et al., 2001) and that TAK1, TAB1 and TAB2 are pre-associated on the membrane before stimulation (Jiang et al., 2002). Western blot analysis revealed that TAB3 proteins migrated more slowly on SDS–PAGE in cells treated with IL-1 (bottom panel, lanes 4, 8 and 12). These slowly migrating bands were eliminated by phosphatase treatment (data not shown), indicating that they may represent phosphorylated TAB3.
Fig. 5. Ligand-dependent association of TRAF6 and TRAF2 with TAK1, TAB2 and TAB3. 293IL-1RI cells were treated with (A) IL-1 (10 ng/ml) or (B) TNFα (10 ng/ml) for the indicated time periods. Endogenous TAK1, TAB2 and TAB3 were immunoprecipitated with anti-TAK1 (T1), anti-TAB2 (T2) and anti-TAB3 (T3) antibodies, respectively. Complexes immunoprecipitated with control IgG (C) or each antibody was immunoblotted with anti-TRAF6, anti-TRAF2, anti-TAK1, anti-TAB2 or anti-TAB3 antibodies. Whole-cell extracts were immunoblotted with anti-TRAF6 or anti-TRAF2 antibodies.
We next examined the effect of TNF on the interaction of endogenous TAB3 with TRAF2. 293IL-1RI cells were either left untreated or were stimulated with TNF and the interaction of TRAF2 with TAK1, TAB2 and TAB3 was examined by immunoprecipitation. Stimulation with TNF resulted in a markedly greater interaction between TRAF2 and TAB3, whereas no significant interaction was observed in the absence of TNF (Figure 5B, top panel, lanes 10–12). In addition, TNF stimulation substantially increased the interaction of TRAF2 with TAK1 (lanes 2–4) and TAB2 (lanes 6–8). The kinetics of TNF-induced TRAF2–TAB3 association were similar to those for TRAF2–TAK1 and TRAF2–TAB2 association. The association was specific for TNF stimulation, because IL-1 treatment did not induce the interaction of TRAF2 with TAB2 or TAB3 (Figure 6B). Taken together, these results suggest that IL-1 and TNF stimulation transiently induces the formation of complexes composed of TRAF6–TAB2–TAB3–TAK1 and TRAF2–TAB2–TAB3–TAK1, respectively.
TAB2 and TAB3 are ubiquitinated in response to IL-1 stimulation
IL-1 signaling leads to TRAF6 polyubiquitination, thereby triggering activation of TAK1 (Deng et al., 2000; Wang et al., 2001). TRAF6 functions as an E3 ligase, facilitating the synthesis of polyubiquitination chains. Thus one of the targets of IL-1-induced ubiquitination is TRAF6 itself. The interaction of TRAF6 with TAB2 and TAB3 suggested that TAB2 and TAB3 might be substrates for TRAF6 ligase activity. To test this possibility, we transfected 293 cells with either T7-TAB2 or T7-TAB3 in the presence or absence of Flag-TRAF6. Cell extracts were immunoprecipitated with anti-T7 antibody and then immunoblotted for the presence of ubiquitin (Ub). TRAF6 association with TAB2 and TAB3 was detected in cells cotransfected with TRAF6 (Figure 6A, top panel, lanes 3 and 6). Immunoblot detection of Ub revealed a smear of material containing TAB2 and TAB3 between 100 kDa and 220 kDa (second panel, lanes 3 and 6). These results indicate that overexpression of TRAF6 can induce Ub modification in TAB2 and TAB3. When T7-TAB3Δcc, lacking the coiled-coil domain, was coexpressed with Flag-TRAF6, it was found to continue to associate with TRAF6 (top panel, lane 8) but was ubiquitinated very poorly (second panel, lane 8). Thus the coiled-coil domain of TAB3 is required for stimulation of TAB3 ubiquitination by TRAF6. TRAF2 has also been shown to act as an E3 ubiquitin ligase and to ubiquitinate itself (Shi et al., 2003). Coexpression of TRAF2 with TAB2 or TAB3 did not induce Ub modification of the TAB2 or TAB3 proteins (second panel, lanes 2 and 5).
We next tested whether IL-1 stimulation could trigger ubiquitination of TAB2 and TAB3. Endogenous TAB2 or TAB3 proteins were immunoprecipitated with anti-TAB2 or anti-TAB3 antibody, respectively, from 293IL-1RI cells, which had been treated with IL-1 or were untreated. Immunoprecipitates were immunoblotted with anti-TRAF6, anti-TRAF2 or anti-Ub antibody. We again observed that IL-1 stimulation induced association of TRAF6 with TAB2 and TAB3 (Figure 6B, top panel, lanes 3 and 9). Following IL-1 treatment, Ub modification of TAB2 or TAB3 was observed (third panel, lanes 3, 4, 9 and 10). Similarly, we examined the effect of TNF stimulation on Ub modification of TAB2 and TAB3 proteins. We observed that TNF stimulation induced association of TRAF2 with TAB2 and TAB3 (second panel, lanes 5 and 11). However, TNF induced weak ubiquitination of TAB3 (third panel, lane 12) but not TAB2 (lanes 5 and 6). Next, we investigated whether TRAF6 was required for the IL-1-induced Ub modification of TAB2. Since the E3 ligase activity of TRAF6 is dependent on its N-terminal RING finger motif (Deng et al., 2000; Wang et al., 2001), we examined the activity of a mutant TARF6 lacking the RING finger domain, TRAF6ΔN, on TAB2 modification in response to IL-1. We found that overexpression of TRAF6ΔN prevented IL-1-induced ubiquitination of TAB2 (Figure 6C, lane 4). Taken together, these results suggest that IL-1 signaling leads to ubiquitination of TAB2 and TAB3 through the action of TRAF6.
TAB2 and TAB3 have redundant functions in mediating IL-1 and TNF signaling
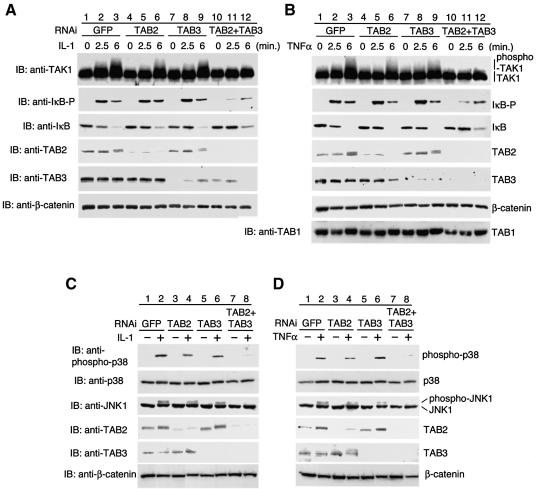
To investigate the functional roles of TAB2 and TAB3 in IL-1- and TNF-mediated signaling, we used siRNAs directed against their respective mRNAs. Western blot analysis showed that TAB2 siRNA transfection of HeLa cells caused a reduction in TAB2 protein levels (Figure 7A and B, fourth panels, lanes 4–6), but had little effect on TAB3 levels (fifth panels, lanes 4–6). Conversely, cells treated with TAB3 siRNA showed reduced levels of TAB3 protein (fifth panels, lanes 7–9) but had no effect on TAB2 (fourth panels, lanes 7–9). Furthermore, when cells were cotransfected with TAB2 and TAB3 siRNAs, levels of both proteins decreased (fourth and fifth panels, lanes 10–12). These results demonstrated that TAB2 and TAB3 siRNAs specifically blocked expression of their corresponding proteins. In the presence of both TAB2 and TAB3 siRNAs, TAK1 protein levels did not change (top panels, lanes 10–12), but TAB1 protein levels were slightly decreased (Figure 7B, bottom panel, lanes 10–12), suggesting that a decrease of TAB2/TAB3 protein levels may affect the stability of TAB1 protein.
Fig. 7. Effects of TAB2 and TAB3 siRNAs on IL-1 and TNF signaling pathways. HeLa cells were transfected with annealed sense and antisense 21-mer siRNA oligonucleotides directed against Jellyfish GFP (GFP; as a control siRNA), TAB2 or TAB3 using Oligofectamine: 400 nM GFP siRNA; 200 nM TAB2 siRNA + 200 nM GFP siRNA; 200 nM TAB3 siRNA + 200 nM GFP siRNA; 200 nM TAB2 siRNA + 200 nM TAB3 siRNA. At 72 h post-transfection, the cells were treated with IL-1 (10 ng/ml) (A and C) or TNFα (10 ng/ml) (B and D) for the indicated times (A and B) or for 10 min (C and D). Western blot analysis was performed on extracts prepared from these cells using antibodies directed against TAK1, phospho-IκBα (IκB-P), IκBα, TAB2, TAB3, TAB1, phospho-p38, p38 or JNK1. Western blot analysis for β-catenin indicates that an equivalent amount of protein was present in each lane.
Previous studies have shown that activation of TAK1 coincides with TAK1 autophosphorylation, as evidenced by a shift in the mobility of TAK1 in SDS–PAGE (Kishimoto et al., 2000). To test the effect of TAB2 and TAB3 siRNAs on IL-1- and TNF-induced activation of TAK1, cells were transfected with the various siRNAs and treated with either IL-1 or TNF. Lysates were prepared and assayed for TAK1 activation. A shift in TAK1 mobility was observed in cells transfected with control GFP siRNA (Figure 7A and B, top panels, lane 3), or single transfection of TAB2 siRNA (lane 6) or TAB3 siRNA (lane 9). However, mobility shift following stimulation with IL-1 and TNF was significantly inhibited by the cotransfection of both TAB2 and TAB3 siRNAs (lane 12). These results suggest that TAB2 and TAB3 play a redundant but critical role in the IL-1- and TNF-induced activation of TAK1.
We next examined the ability of siRNAs directed against TAB2 and TAB3 to prevent IL-1- and TNF-induced NF-κB activation. Western blot analysis demonstrated increases in both phospho-IκBα and IκBα degradation in control cells following treatment with either IL-1 or TNF (Figure 7A and B, second and third panels, lanes 2 and 3). Transfection with either TAB2 or TAB3 siRNA alone had little effect on IκBα phosphorylation or degradation in response to IL-1 or TNF (lanes 5, 6, 8 and 9). However, when cells were transfected with TAB2 and TAB3 siRNAs together and then treated with either IL-1 or TNF, the amount of phospho-IκBα decreased and IκBα degradation was slightly inhibited (lanes 11 and 12). These findings were consistent with those obtained by gel retardation analysis, which demonstrated that IL-1 or TNF stimulation induced smaller increases in NF-κB binding in extracts prepared from cells transfected with both TAB2 and TAB3 siRNAs compared with control cells or cells transfected with each individual siRNA (Supplementary figure S1 available at The EMBO Journal Online). These data suggest that TAB2 and TAB3 have redundant functions in activating the IL-1- and TNF-induced NF-κB pathway.
In addition to NF-κB activation, IL-1 and TNF induce activation of p38 and JNK MAPKs (Baud and Karin, 2001). Similar experiments were performed to determine the effect of siRNAs directed against both TAB2 and TAB3 on IL-1- and TNF-induced activation of p38 and JNK. We observed no effect of transfecting individual TAB2 or TAB3 siRNA on activation of p38 and JNK induced by IL-1 or TNF (Figure 7C and D, top and third panels, lanes 4 and 6). However, the combination of TAB2 and TAB3 siRNAs reduced both IL-1- and TNF-induced activation of p38 and JNK (lane 8). Thus, TAB2 and TAB3 are important for activation of p38 and JNK in response to IL-1 and TNF. Taken together, these results suggest that TAB2 and TAB3 have redundant functions in mediating both IL-1 and TNF signal transduction pathways.
Discussion
Previous studies have demonstrated that the TAK1-associating protein TAB2 interacts with TRAF6 in an IL-1-dependent manner, resulting in the formation of a TRAF6–TAB2–TAK1 complex (Takaesu et al., 2000). Formation of this complex appears to be required for IL-1-mediated activation of NF-κB. TAB2 acts as an adaptor that links TAK1 and TRAF6, and thereby mediates the activation of TAK1 in the IL-1 signaling pathway. By this model, TAK1 and TAB2 are important components in the IL-1 pathway, but their physiological functions in this signal cascade have yet to be fully clarified. Recent studies using siRNA have demonstrated that TAK1 is critical not only for IL-1-mediated activation of the NF-κB pathway but also for TNF activation of this pathway (Takaesu et al., 2003). In addition, Vidal and coworkers have reported that dTAK1, a Drosophila melanogaster homolog of TAK1, controls the activity of the Rel/NF-κB-like transactivator Relish in the Imd pathway (Vidal et al., 2001). Taken together, these findings support the idea that TAK1 plays an essential role in the NF-κB activation pathway. Mice deficient for TAB2 exhibit embryonic lethal due to liver degeneration and apoptosis, yet embryonic fibroblasts deficient for TAB2 show no impairment in IL-1- or TNF-induced activation of NF-κB (Sanjo et al., 2003). This is in contrast with cells transfected with TAK1 siRNA, which do exhibit impaired IL-1 and TNF signaling. These findings demonstrate that the embryonic lethality caused by TAB2 deletion is not due to defective IL-1- or TNF-mediated signaling itself, but to some other defect leading to liver apoptosis. We and other workers (Munoz-Sanjuan et al., 2002) have now identified a novel TAK1-binding protein, termed TAB3, which is closely related to TAB2 in structure, in mouse, human and Xenopus laevis. In this study, we demonstrate that TAB2 and TAB3 have redundant functions in activating both IL-1 and TNF signaling pathways, and that TAB3 may compensate for the loss of TAB2.
We present several lines of evidence supporting the role of TAB3 as a signal transducer in both IL-1 and TNF signaling pathways. First, ectopic expression of TAB3 results in the activation of both NF-κB and JNK, and therefore mimics these two IL-1- and TNF-induced cellular responses. Secondly, the C-terminal half of TAB3 behaves as a dominant negative mutant that blocks both IL-1- and TNF-induced activation of NF-κB. Thirdly, endogenous TAB3 associates with TRAF6 and TRAF2 in an IL-1- and TNF-dependent manner, respectively. Fourthly, TAB3 interacts with TAK1 and thereby mediates its association with TRAF6 and TRAF2. Finally, IL-1 and TNF stimulation induces ubiquitination of TAB3. Collectively, these data are consistent with the idea that TAB3 participates in both IL-1 and TNF signaling by linking TAK1 with TRAF6 and TRAF2, which are upstream components of each pathway.
As compared with transgenic knockout mice, siRNA does not totally prevent the translation of its target mRNA. However, siRNA has the advantage that it can be introduced either individually or in combination into the same cells to decrease the level of specific proteins. Here, siRNA has been utilized to specifically decrease the expression of TAB2 and TAB3 in order to better define their respective roles in regulating the IL-1 and TNF signaling pathways. Neither TAB2 nor TAB3 siRNA alone had any significant effect on IL-1- or TNF-induced activation of TAK1 and NF-κB, even though each of these individual siRNAs specifically decreased expression of their respective target protein. However, cotransfection of the TAB2 and TAB3 siRNAs resulted in inhibition of both IL-1- and TNF-induced activation of TAK1 and NF-κB. Thus, at least in HeLa cells, it seems likely that TAB2 and TAB3 have redundant roles in IL-1- and TNF-mediated signaling. Although our previous studies demonstrated that TAB2 functions mainly in the IL-1 signaling pathway (Takaesu et al., 2000, 2001), it appears that TAB2 is able to compensate for the loss of TAB3 in the TNF signaling pathway. The generation and analysis of TAB3-deficient mice will reveal whether mammalian TAB3 is in fact involved in IL-1- and TNF-mediated activation of NF-κB. Furthermore, it would be interesting to determine whether a Drosophila TAB2/3 homolog would be involved in dTAK1-mediated Imd pathway.
Our previous and present studies suggest a model in which IL-1 and TNF stimulation facilitates the formation of TRAF6–TAB2/TAB3–TAK1 and TRAF2–TAB2/TAB3–TAK1 complexes, respectively, leading to the activation of TAK1. Our result demonstrating that TAB2, TAB3 and TAK1 preform a complex in the absence of stimulation is consistent with a recent observation that the TRIKA2 complex containing TAK1, TAB1 and TAB2 is formed constitutively (Wang et al., 2001), and further suggests that TAB3 may be a component of the TRIKA2 complex. In the case of the IL-1 signaling pathway, TRAF6 acts as a mediator between the IL-1 receptor complex and TAK1, and TAB2/TAB3 function as adaptors that link TAK1 and TRAF6. In addition, these authors have demonstrated that ubiquitination of TRAF6 is involved in TAK1 activation by IL-1, leading to phosphorylation and activation of the IKK complex (Wang et al., 2001). How TRAF6 activates TAK1 remains unknown, although it apparently depends upon the E3 ligase activity of TRAF6, which promotes or perhaps stabilizes the oligomerization of TRAF6 (Deng et al., 2000; Wang et al., 2001). Interestingly, TAB2 and TAB3 contain a ubiquitin-binding domain, raising the possibility that TAB2 and TAB3 mediate TAK1 activation through a ubiquitin-mediated step. Consistent with this possibility, we found that TAB2 and TAB3 were ubiquitinated by either IL-1 stimulation or TRAF6 overexpression. These results suggest that stimulation of cells with IL-1 leads to the ubiquitination of TAB2 and TAB3 by stimulating the E3 ligase activity of TRAF6. Other studies have shown that TAK1 is activated by autophosphorylation (Kishimoto et al., 2000). Based on these results, we speculate that ubiquitination of the TAB2–TAB3 complex may alter the conformation of TAK1, which induces its kinase activity leading to autophosphorylation. Although TRAF2 is another E3 that plays an important role in TNF signaling (Yeh et al., 1997; Shi and Kehrl, 2003), it remains to be determined whether TRAF2 mediates ubiquitination of TAB2 and TAB3. Further study is required to clarify the exact mechanisms by which ubiquitination of TAB2 and TAB3 activates TAK1.
Materials and methods
Cloning of human TAB3
Full-length human TAB3 gene was amplified by PCR from a human kidney cDNA library (Clontech) with the following primers: 5′-GCC GGTTAACATCCATTTCC-3′ (5′-primer), 5′-TTTCACTTTCAACC TGGCGC-3′ (3′-primer) for the first reaction, and 5′-ATTTGCTCTGG CATGGCGCAAAG-3′ (5′-primer), 5′-ATTCAGGTGTACCGTGGCA TCTC-3′ (3′-primer) for the second reaction. The amplified cDNAs were subcloned into pCR2.1-TOPO (Invitrogen) and completely sequenced.
Expression vectors and antibodies
To overexpress T7-TAB3, we constructed pCMV-T7-TAB3, a vector expressing T7-TAB3 under the control of the cytomegalovirus (CMV) promoter. Mammalian expression vectors encoding TAK1, TAB2, TRAF6, TRAF2 and JNK have been described (Ninomiya-Tsuji et al., 1999; Takaesu et al., 2000). The TAB3 mutants, TAB3N, TAB3C and TAB3Δcc, were generated by PCR. Polyclonal rabbit antibody against TAB3 was produced using peptides corresponding to amino acids 635–648 of TAB3. This antibody reacted with TAB3 but not TAB2. Polyclonal rabbit antibodies against TAK1 and TRAF6 have been described (Ninomiya-Tsuji et al., 1999; Takaesu et al., 2000). Anti-TRAF2 and anti-β-catenin were from Pharmingen. Anti-ubiquitin, anti-p65 and anti-JNK were from Santa Cruz. Anti-phospho-IκBα, anti-IκBα, anti-phospho-p38 and anti-p38 were from Cell Signaling. Anti-T7 was from Novagen. Anti-HA (HA.11) was from Babco. Anti-Flag (M2) and control immunoglobulin G (IgG) were from Sigma.
Cell culture and transfection
The 293IL-1RI cells were described previously (Ninomiya-Tsuji et al., 1999). The HEK293, 293IL-1RI and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. The 293 cells in 100 mm plates were transfected with the expression plasmids (10 µg) by calcium phosphate precipitation.
Reporter gene assays
The 293 and 293IL-1RI cells (1.6 × 105 cells/well) were seeded into six-well (35 mm) plates. Cells were transfected by the calcium phosphate precipitate method 24 h after seeding with the NF-κB-Luc reporter gene plasmid, along with each expression vector as indicated. The total DNA concentration (1.7 µg) was kept constant by supplementing with empty vector DNAs. IL-1β (5 ng/ml) or TNFα (10 ng/ml) was added to each plate 3 h after transfection. At 24 h after transfection, luciferase activity was determined with the Luciferase Assay System (Promega). β-Gal vector (0.1 µg), under the control of the β-actin promoter, was used for normalizing transfection efficiencies. The values shown are the averages of one representative experiment in which each transfection was performed in duplicate.
Immunoprecipitation and immunoblotting
Cells were either left untreated or treated with IL-1β (5 ng/ml) or TNFα (10 ng/ml) for the indicated times. Cells were washed once with ice-cold phosphate-buffered saline and lysed in 0.3 ml 0.5% Triton X-100 lysis buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 12.5 mM glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM dithiothreitol (DTT), 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 20 µM aprotinin. Proteins from cell extracts were immunoprecipitated with various antibodies and protein G-Sepharose (Pharmacia). For immunoblotting, immunoprecipitates or whole-cell extracts were resolved by SDS–PAGE and transferred to Hybond-P membranes (Amersham). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized with horseradish-peroxidase-conjugated antibodies against rabbit or mouse IgG by using the Enhanced Chemiluminescence (ECL) Western Blotting System (Amersham).
In vitro kinase assays
Bacterially expressed GST-c-Jun or MKK6 were described previously (Shirakabe et al., 1997). Aliquots of immunoprecipitates were incubated with substrates (1 µg) in 10 µl kinase buffer containing 10 mM HEPES pH 7.4, 1 mM DTT, 5 mM MgCl2 and 5 µCi [γ-32P]ATP at 25°C for 2 min. Samples were resolved by SDS–PAGE and phosphorylated proteins were visualized by autoradiography.
RNA oligonucleotides
siRNAs with two thymidine residues (dTdT) at the 3′ end of the sequence were designed against the TAB3 (sense 5′-CCACCUCAACAGCCA UCUU-3′) and TAB2 (sense 5′-CCUCCAGCACUUCCUCUUC-3′) mRNAs along with their corresponding antisense RNA oligonucleotides (Japan Bio Service). These RNAs were dissolved in DEPC-treated water to 50 µM, heated to 90°C in buffer (30 mM HEPES–KOH pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate) and annealed at 37°C. As a control, we used siRNA directed against Jellyfish GFP (Nippon Gene).
Transfection of RNA oligonucleotides
HeLa cells (1 × 105 cells/well) were seeded into six-well (35 mm) plates to 20–30% confluency, and transfection of the RNA oligonucleotides was performed using Oligofectamine (Invitrogen) to a final RNA concentration of 400 nM. The cells were harvested at different time points post-treatment with IL-1β (10 ng/ml) or TNFα (10 ng/ml) and lysed for use in western blot analysis.
Gel retardation assays
HeLa cells were harvested 30 min after treatment with IL-1β (10 ng/ml) or TNFα (10 ng/ml) and lysed for use in gel retardation assays. NF-κB oligonucleotides were from Promega. Binding reactions were performed at room temperature for 10 min by incubating 7 µl of total lysates and 0.035 pmol of labeled oligonucleotides in 15 µl of binding buffer (10 mM Tris pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 4% glycerol, 0.75 µg poly(dI-dC), 1.5 µg salmon sperm DNA, 1.5 µg BSA).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank M.Lamphier for critical reading of the manuscript. This work was supported by special grants from CREST, Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan and Yamanouchi Foundation for Research on Metabolic Disorders (K.M.).
References
- Baud V. and Karin,M. (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Cao Z., Xiong,J., Takeuchi,M., Kurama,T. and Goeddel,D.V. (1996) TRAF6 is a signal transducer for interleukin-1. Nature, 383, 443–446. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang,C., Spencer,E., Yang,L., Braun,A., You,J., Slaughter,C., Pickart,C. and Chen,Z.J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell, 103, 351–361. [DOI] [PubMed] [Google Scholar]
- Dinarello C.A. (1996) Biologic basis for interleukin-1 in disease. Blood, 87, 2095–2147. [PubMed] [Google Scholar]
- Ghosh S. and Karin,M. (2002) Missing pieces in the NF-κB puzzle. Cell, 109, S81–S96. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Ninomiya-Tsuji,J., Qina,Y., Matsumoto,K. and Li,X. (2002) Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phopsphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol. Cell. Biol., 22, 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. and Ben-Neriah,Y. (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol., 18, 621–663. [DOI] [PubMed] [Google Scholar]
- Kishimoto K., Matsumoto,K. and Ninomiya-Tsuji,J. (2000) TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem., 275, 7359–7364. [DOI] [PubMed] [Google Scholar]
- Li Q. and Verma,I.M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol., 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Lomaga M.A., Yeh,W.C., Sarosi,I., Duncan,G.S., Furlonger,C., Ho,A., Morony,S.,.Capparelli,C., Van,G. et al. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40 and LPS signaling. Genes Dev., 13, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Sanjuan I., Bell,E., Altmann,C.R., Vonica,A. and Brivanlou,A.H. (2002) Gene profiling during neural induction in Xenopus laevis: regulation of BMP signaling by post-transcriptional mechanisms and TAB3, a novel TAK1-binding protein. Development, 129, 5529–5540. [DOI] [PubMed] [Google Scholar]
- Naito A., Azuma,S., Tanaka,S., Miyazaki,T., Takaki,S., Takatsu,K., Nakao,K., Nakamura,K., Katsuki,M. et al. (1999) Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells, 4, 353–362. [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto,K., Hiyama,A., Inoue,J., Cao,Z. and Matsumoto,K. (1999) The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature, 398, 252–256. [DOI] [PubMed] [Google Scholar]
- Sanjo H., Takeda,K., Tsujimura,T., Ninomiya-Tsuji,J., Matsumoto,K. and Akira,S. (2003) TAB2 is essential for prevention of apoptosis in fetal liver but not for interleukin-1 signaling. Mol. Cell. Biol., 23, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S. and Kehrl,J.H. (2003) Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J. Biol. Chem., 278, 15429–15434. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Yamaguchi,K., Shirakabe,K., Tonegawa,A., Gotoh,Y., Ueno,N., Irie,K., Nishida,E. and Matsumoto,K. (1996) TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science, 272, 1179–1182. [DOI] [PubMed] [Google Scholar]
- Shih S.C., Prag,G., Francis,S.A., Sutanto,M.A., Hurley,J.H. and Hicke,L. (2003) A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J., 22, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakabe K., Yamaguchi,K., Shibuya,H., Irie,K., Matsuda,S., Moriguchi,T., Gotoh,Y., Matsumoto,K. and Nishida,E. (1997) TAK1 mediates the ceramide signaling to stress-activated protein kinase/c-Jun N-terminal kinase. J. Biol. Chem., 272, 8141–8144. [DOI] [PubMed] [Google Scholar]
- Silverman N. and Maniatis,T. (2001) NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev., 15, 2321–2342. [DOI] [PubMed] [Google Scholar]
- Takaesu G., Kishida,S., Hiyama,A., Yamaguchi,K., Shibuya,H., Irie,K., Ninomiya-Tsuji,J. and Matsumoto,K. (2000) TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell, 5, 649–658. [DOI] [PubMed] [Google Scholar]
- Takaesu G., Ninomiya-Tsuji,J., Kishida,S., Li,X., Stark,G.R. and Matsumoto,K. (2001) Interleukin-1 (IL-1) receptor-associated kinase leads to activation of TAK1 by inducing TAB2 translocation in the IL-1 signaling pathway. Mol. Cell. Biol., 21, 2475–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G., Surabhi,R.M., Park,K.J., Ninomiya-Tsuji,J., Matsumoto,K. and Gaynor,R.B. (2003) TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol., 326, 105–115. [DOI] [PubMed] [Google Scholar]
- Vidal S., Khush,R.S., Leulier,F., Tzou,P., Nakamura,M. and Lemaitre,B. (2001) Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NFκB-dependent innate immune responses. Genes Dev., 15, 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Deng,L., Hong,M., Akkaraju,G.R., Inoue,J. and Chen,Z.J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature, 412, 346–351. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Shirakabe,K., Shibuya,H., Irie,K., Oishi,I, Ueno,N., Taniguchi,T., Nishida,E. and Matsumoto,K. (1995) Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science, 270, 2008–2011. [DOI] [PubMed] [Google Scholar]
- Yeh W.C., Shahinian,A., Speiser,D., Kraunus,J., Billia,F., Wakeham,A., de la Pompa,J.L., Ferrick,D., Hum,B. et al. (1997) Early lethality, functional NF-κB activation and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity, 7, 715–725. [DOI] [PubMed] [Google Scholar]