Abstract
We previously described the use of a cell-based screening approach to identify small molecules that regulate adipocyte differentiation. Here we identify the amiloride derivative phenamil as an adipogenic compound. Phenamil acutely induces expression of the key transcription factor of adipogenesis, peroxisome proliferator-activated receptor γ (PPARγ) and, consequently, promotes the differentiation of multiple preadipocyte cell lines, including 3T3-L1 and F442A. Interestingly, the adipogenic action of phenamil is distinct from and additive with both PPARγ ligands and the previously identified adipogenic small molecule harmine. To identify signaling pathways mediating phenamil's effects, we performed transcriptional profiling of 3T3-F442A preadipocytes. ETS variant 4 (ETV4) was identified as a gene rapidly induced by phenamil but not by other adipogenic small molecules or PPARγ agonists. Transient expression of ETV4 in preadipocytes enhances the expression of PPARγ. Stable overexpression of ETV4 promotes expression of PPARγ and its downstream target genes and enhances morphological differentiation. Finally, knockdown of PPARγ expression by shRNA blocks the effects of phenamil on adipocyte differentiation and gene expression, but it does not block phenamil induction of ETV4, which suggests that ETV4 acts upstream of PPARγ in differentiation processes. These results identify a phenamil as new small molecule tool for the probing of adipocyte differentiation that acts, at least in part, through induction of ETV4 expression.
Keywords: ETS variant, peroxisome proliferator-activated receptor, diabetes, adipose tissue, fatty acid, nuclear receptor
Peroxisome proliferator-activated receptor γ (PPARγ) is a member of the nuclear receptor family that serves as the master transcriptional regulator of adipogenesis (1). PPARγ heterodimerizes with retinoid X receptor (RXR) and regulates adipogenic target gene expression. Downstream targets for PPARγ include a host of genes involved in lipid accumulation and metabolism, such as aP2, CD36, LPL, perilipin, and PEPCK. The transcriptional pathways that control adipocyte-specific expression of PPARγ remain to be fully elucidated. Transcription factors that have been reported to affect PPARγ expression include C/EBPβ and C/EBPδ (2); Kruppel-like zinc finger family of transcription factors KLF5 (3), KLF15 (4), and KLF2 (5); Krox-20 (6); GATA2/3 (7); TCF/Lef (8); E2F (9); and SMAD (10). The ability of multiple factors to influence PPARγ expression suggests a complex mode of regulation that requires the integration of multiple signaling pathways.
We have developed and validated cell-based screening methods for the identification of bioactive small molecules that affect lipid metabolism. We previously used this approach to characterize the small molecule harmine as an inducer of PPARγ expression and adipogenesis that has antidiabetic activity in mice (11). Interestingly, harmine's mechanism of action involves the inhibition of Wnt signaling in preadipocytes. By profiling the acute effects of harmine on preadipocyte gene expression, we subsequently identified the transcription factor Id2 as a mediator of harmine action in adipogenesis (12). We have now extended our screen for adipogenic small molecules in an effort to identify compounds with mechanisms of action distinct from that of harmine and PPARγ agonists.
We report here the identification of the small molecule diuretic phenamil as a stimulator of adipocyte differentiation. Interestingly, phenamil acts by inducing PPARγ mRNA expression in preadipocytes through a mechanism distinct from harmine. To gain insight into the mechanism of phenamil action, we performed transcriptional profiling of 3T3-F442A cells and identified ETV4 (ETS variant 4) as a putative mediator. Ectopic expression of ETV4 in preadipocytes promotes PPARγ expression and differentiation and appears to act upstream of PPARγ in the differentiation cascade. These results identify ETV4 as a modulator of adipocyte differentiation that is susceptible to small molecule regulation.
MATERIALS AND METHODS
Reagents and plasmids
Phenamil methanesulfonate was purchased from Sigma (#P203). GW7845 was kindly provided by T. Willson (Glaxo SmithKline). Insulin (#I0516), dexamethasone (#D2915), and 3-isobutyl-1-methylxanthine (IBMX, #I7018) were from Sigma. We used following primers for cloning ETV4: 5′-agtggatccgccgccatggagcggaggatgaaag-3′ (forward) and 5′-agtctcgagctactagtaagagtagccacc-3′ (reverse). Full-length cDNA was cloned into pBabe-puro and pCMV-SPORT. Oligonucleotides were from Integrated DNA Technologies.
Cell culture
3T3-L1 and 3T3-F442A preadipocyte cell lines were maintained and differentiated as previously described (11, 12). Retroviral overexpression of ETV4 was performed using pBabe-puro and the packaging cell line Phoenix E as described (13). 3T3-L1 cells were differentiated in 10% FBS supplemented with dexamethasone (1 μM), IBMX (0.5 mM), and insulin (5 μg/ml) for two days after confluence, followed by insulin alone. 3T3-F442A cells were differentiated in 10% FBS and insulin as described (12).
RNA and realtime PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). A 0.5 μg of total RNA was reverse-transcribed using MultiScribe (Applied Biosystems) and random hexamers according to the manufacturer's instructions. Real-time quantitative PCR (SYBR green) analysis was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Expression was normalized to 36B4. The following primers were used for ETV expression: ETV1, 5′-atggagaaaagtgcctgtacaat-3′ (forward) and 5′-ggtgtagtggggacactgga-3′ (reverse); ETV3, 5′-ctgggtggatgaggagga-3′ (forward) and 5′-acagcctgctttcattttcg-3′ (reverse); ETV4, 5′- cagcaggaagccaccact-3′ (forward) and 5′- ttgtctgggggagtcatagg-3′ (reverse); and ETV5, 5′- gcagtttgtcccagattttca-3′ (forward) and 5′- gcagctcccgtttgatctt-3′ (reverse).
Microarrays
Total RNA from 24 h phenamil- and GW7845-treated F442A cells were prepared using TRIzol and further purified using RNAeasy columns (Qiagen). cDNA preparation and hybridization to Mouse-6 expression Beadchip were performed by the Southern California Genotyping Consortium (SCGC). Data were analyzed using Beadstring.
RESULTS
Identification of phenamil as a small molecule regulator of PPARgamma expression
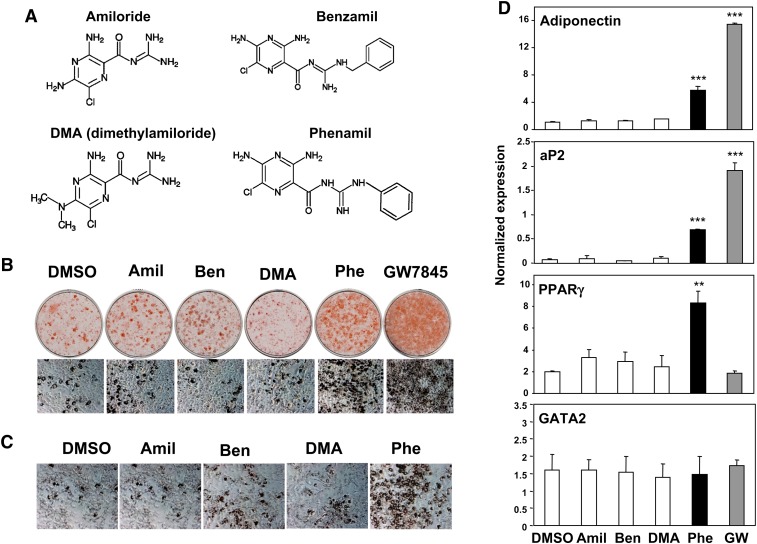
We identified the amiloride derivative phenamil (Fig. 1A) as an adipogenic compound by screening a BIOMOL library of bioactive compounds using a cell-based assay (11). Treatment of both 3T3-L1 and 3T3-F442A preadipocytes with phenamil followed by Oil Red O staining and morphological analysis confirmed the adipogenic effects of this compound (Fig. 1B, C). Consistent with their ability to stimulate morphologic differentiation, both phenamil and the PPARγ agonist GW7845 strongly promoted the expression of the adipogenic marker genes aP2, LPL, adiponectin CD36, and adipsin (Fig. 1D and data not shown).
Fig. 1.
Phenamil induces adipocyte differentiation and PPARγ expression in preadipocytes. A: Chemical structures of amiloride derivatives. Amiloride, benzamil, DMA, and phenamil are shown. B–D: Effects of amiloride derivatives on adipocyte differentiation. Phenamil (but not other derivatives) induces expression of adipogenic genes and adipocyte differentiation. B–C: Oil Red O staining of amiloride derivatives-treated cells. B: Differentiation of 3T3-L1 cells into adipocytes was induced by DMI and amiloride derivatives or GW7845 (GW; 20 nM). C: 3T3-F442A preadipocytes were treated for six days with the amiloride derivatives. D: 3T3-F442A cells were treated for six days with 10 μM amiloride derivatives or GW7845. Expression of adipogenic genes encoding PPARγ, aP2, and adiponectin were measured by real time PCR. *P < 0.05, **P < 0.005, ***P < 0.0005. amil, amiloride; ben, benzamil; DMA, dimethyl amiloride hydrochloride; phe, phenamil; PPARγ, peroxisome proliferator-activated receptor γ.
Since PPARγ is the central regulator of adipogenesis, we first tested whether this pro-adipogenic compound might act as an agonist for PPARγ. However, transient transfection assays for PPARγ activity using a chimeric GAL4-PPARγ receptor and a luciferase reporter containing GAL4 binding sites (14) showed that phenamil was inactive as a PPARγ agonist. As expected, the control PPARγ agonist GW7845 stimulated luciferase reporter activity in a dose-dependent manner (supplementary Fig. I-A). Furthermore, phenamil displayed no RXR ligand activity as determined by a similar chimeric GAL4-RXR reporter assay (supplementary Fig. I-B). These data suggest that phenamil promotes adipocyte differentiation through a mechanism distinct from that of PPARγ and RXR agonists.
Amilorides are a class of molecules widely used as diuretics in humans (15, 16), and they are known to inhibit epithelial sodium channel (ENaC) activity (17–21). To determine the correlation between the adipogenic effects of phenamil and ENaC activity, we assayed three additional amiloride derivatives also known to inhibit ENaC. The adipogenic activity of phenamil was not shared by amiloride itself or several closely related derivates (Fig. 1A–D). In addition, expression of the epithelial transporter targets of phenamil—ENaCα, ENaCβ, and ENaCγ—is not detectable in 3T3-L1 or 3T3-F442A preadipocytes (supplementary Fig. II).
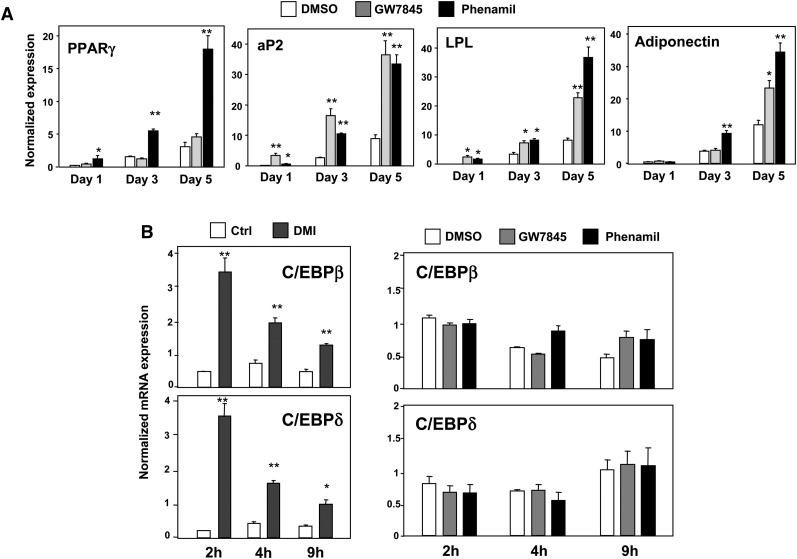
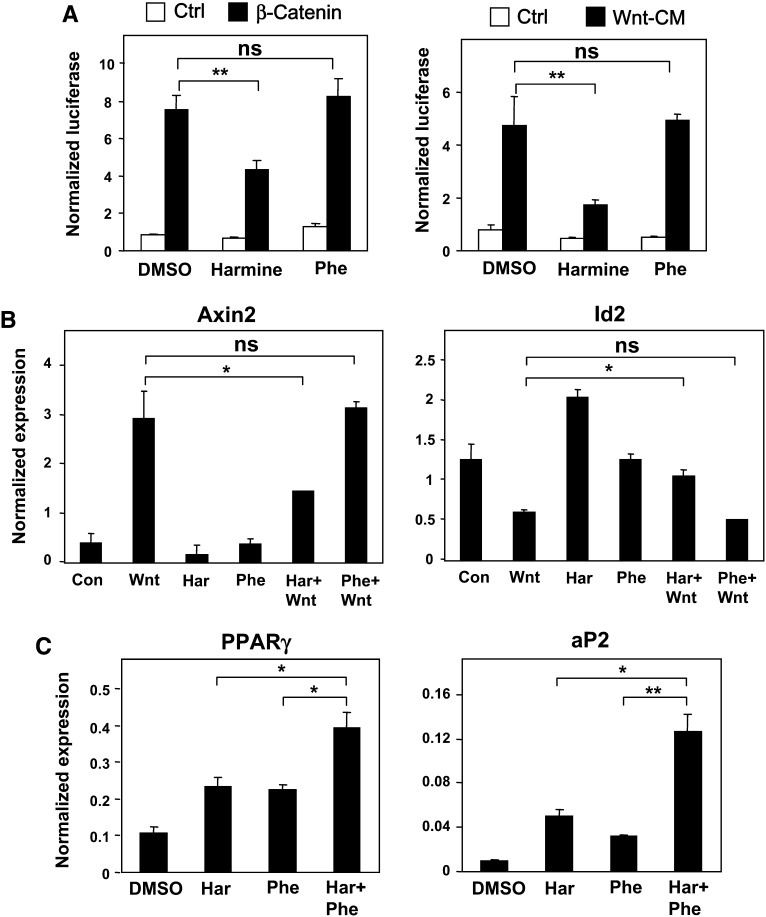
We next examined the time course of phenamil effects on adipocyte gene expression. Interestingly, in contrast to GW7845, phenamil induced the expression of PPARγ within 24 h, suggesting a distinct mode of action (Fig. 2A). Furthermore, cells treated with phenamil showed markedly elevated levels of PPARγ expression compared with GW7845-treated cells throughout the time course. The transcription factors C/EBPβ and C/EBPδ are postulated to induce PPARγ in response to IBMX and dexamethasone (2, 22); therefore, we considered these factors as potential mediators. However, phenamil did not affect the expression of C/EBPβ or C/EBPδ (Fig. 2B). The ability to acutely upregulate PPARγ expression in preadipocytes is also reminiscent of harmine, another adipogenic small molecule that we have recently characterized (11). Harmine promotes PPARγ expression through inhibition of Wnt signaling and induction of Id2 expression (11, 12). However, several lines of evidence suggest that the mechanism of action of phenamil is distinct from that of harmine. First, phenamil was unable to inhibit Wnt-dependent TOP-FLASH reporter activity in transient transfection assays (Fig. 3A). Second, harmine (but not phenamil) blocked the induction of the Wnt target genes Axin2 and Id2 by Wnt-3a in preadipocytes (Fig. 3B). Third, harmine and phenamil had additive effects on adipogenic gene expression and differentiation, consistent with different mechanisms of action (Fig. 3C). Together, these data indicate that phenamil acts via a mechanism distinct from that of the previously identified adipogenic chemicals—GW7845, harmine, and cAMP inducers—in adipogenesis.
Fig. 2.
Phenamil is a unique inducer of PPARγ expression in preadipocytes. A: Gene expression profile during differentiation of 3T3-F442A cells treated with GW7845 (20 nM) or phenamil (10 μM). PPARγ target genes aP2 and LPL were induced by GW7845 and phenamil; however, PPARγ expression was acutely induced by phenamil but not by GW7845. B: Induction of C/EBPβ and C/EBPδ by phenamil and standard differentiation cocktail (dexamethasone, IBMX, insulin) in 3T3-F442A cells. Standard differentiation cocktail induced the expression of C/EBPβ and C/EBPδ starting from 2 h treatment in 3T3-F442A cells, whereas phenamil and GW7845 did not alter expression of these two transcription factors. *P < 0.05, **P < 0.005, ***P < 0.0005. IBMX, 3-isobutyl-1-methylxanthine; PPARγ, peroxisome proliferator-activated receptor γ.
Fig. 3.
Phenamil and harmine act by different pathways. Phenamil does not modulate the Wnt signaling pathways. A-B: Phenamil effects on β-catenin-driven and Wnt-driven TOP FLASH activity in 293-T cells and endogenous Wnt target gene expression in 3T3-F442A cells. Wnt inhibitor harmine inhibited the activation of Wnt pathway in TOP FLASH (A) and induction of Wnt target genes Axin2 and Id2 (B). Phenamil does not regulate Wnt signaling pathways shown in TOP FLASH experiments (A) and expression of endogenous Wnt target genes (B). C: Phenamil and harmine showed an additive effect on the expression of PPARγ and its target gene aP2. 3T3-F442A cells were treated with harmine (1 μM), phenamil (1 μM), or both for 48 h. *P < 0.05, **P < 0.005; ns, not significant.
Identification of genes induced by phenamil
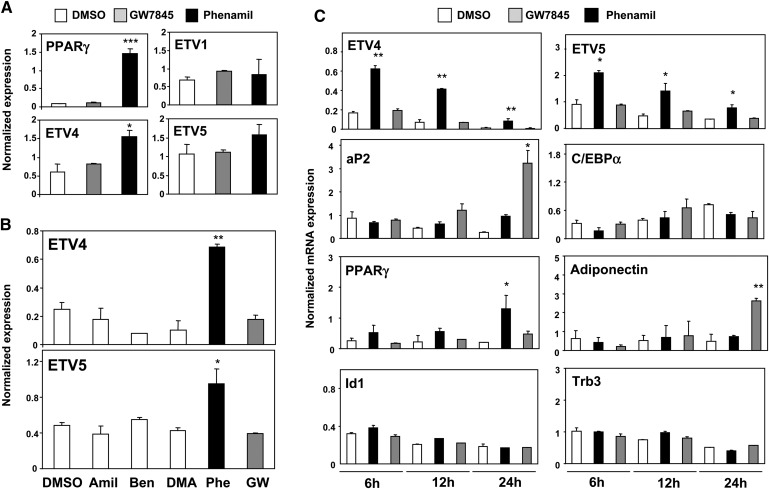
To gain insight into the biological pathway affected by this new adipogenic compound, we performed transcriptional profiling of 3T3-F442A cells treated with either phenamil or GW7845 for 24 h. We hypothesized that candidate mediators of phenamil action would be induced by phenamil but not by GW7845, because GW7845 does not induce PPARγ expression in 24 h (Fig. 4A). We focused in particular on transcriptional regulators. Only a limited number of genes, including ETV4, ETV5, EGR5, and PPARγ itself, met our criteria of being upregulated by phenamil (2-fold or more) but not regulated by GW7845 (less than 1.3-fold change) (Fig. 4A, B). Analysis of the time course of gene expression changes induced by phenamil showed that ETV4 and ETV5 were induced as early as 6 h, while PPARγ expression was induced later (Fig. 4C). These studies also confirmed that ETV4, ETV5, and PPARγ were upregulated by phenamil but not by GW7845. ETV3 and ETV1, another Pea3 family member, were not regulated by phenamil or GW7845, indicating that regulation of Pea3 members is not identical (Fig. 4A and data not shown). We also expected that candidate mediators of phenamil action would not be responsive to amiloride derivates that do not promote adipogenesis. Indeed, as shown in Fig. 4B, among amiloride derivatives tested, only phenamil induced ETV4 and ETV5 expression.
Fig. 4.
Identification of genes rapidly induced by phenamil in 3T3-F442A preadipocytes. A: 3T3-F442A cells were treated with either phenamil or GW7845 for 24 h. Comparison of effects by phenamil and GW7845 on ETV1, ETV4, ETV5, and PPARγ gene expression. Realtime PCR analysis confirmed that ETV4, ETV5, and PPARγ were upregulated by phenamil but not by GW7845. The other Pea3 member, ETV1, was not regulated by either phenamil or GW7845. B: Selective regulation of ETV4 and ETV5 by phenamil. ETV4 and ETV5 display a pattern of phenamil-specific induction. 3T3-F442A preadipocytes were treated with phenamil (10 μM) or GW7845 (20 nM) for 48 h at confluence, and mRNA levels were measured by real-time PCR. C: Time-course of regulation of selective genes by phenamil. ETV4 and ETV5 were induced by phenamil as early as 6 h, while PPARγ expression was induced later. Comparison of the effects by phenamil and GW7845 treatment on PPARγ and PPARγ target genes and Id1, trb3 expression are also shown. *P < 0.05, **P < 0.005, ***P < 0.0005. ETV1–5, EST variant 1-5; PPARγ, peroxisome proliferator-activated receptor γ.
The induction of ETV4 and ETV5 expression by phenamil was selective for preadipocyte cells and was not observed in osteogenic or mesenchymal cells (data not shown). We recently reported that phenamil promotes the osteoblastic differentiation of mesenchymal stem cells through a pathway involving the promotion of BMP signaling through Id1 and trb3 (23). This phenamil-stimulated pathway is appears to be selective for mesenchymal stem cells and does not operate in determined preadipocytes, suggesting that different downstream pathways operate in these cells (Fig. 4C). However, it remains possible that unidentified common upstream targets or mechanisms are regulated by the phenamil in both mesenchymal stem cells and preadipocytes.
ETV4 stimulates PPARγ expression and adipocyte differentiation
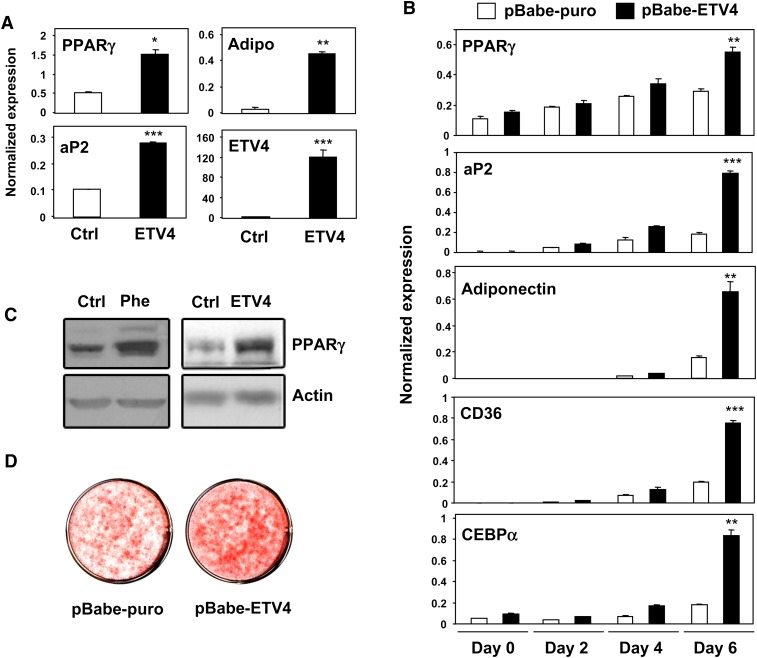
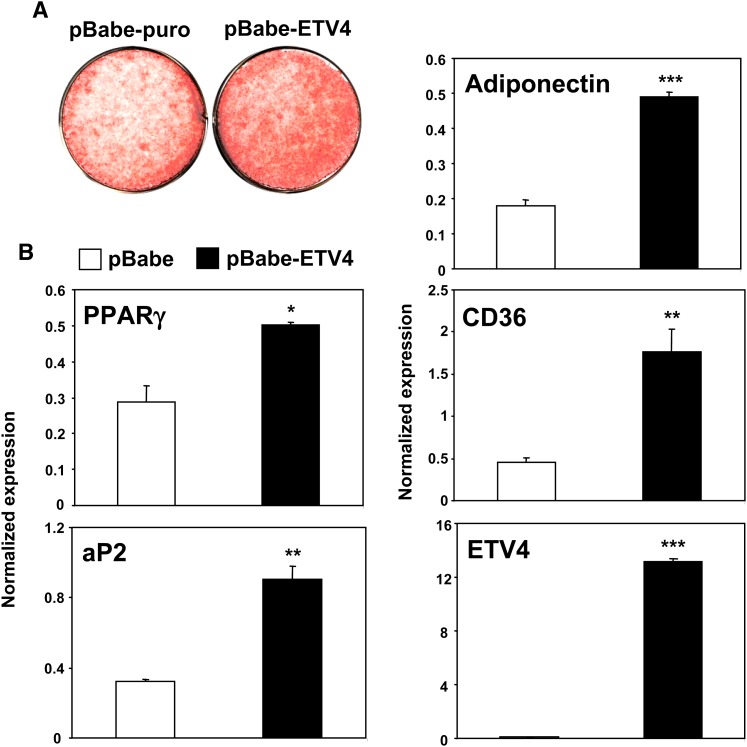
To address the hypothesis that ETV4 induction by phenamil may contribute to its ability promote PPARγ expression, we transiently overexpressed ETV4 in 3T3-L1 preadipocytes. Transient overexpression of ETV4 increased PPARγ mRNA expression and the mRNA of the target genes aP2 and adiponectin (Fig. 5A). This observation suggests that ETV4 can act upstream of PPARγ in the differentiation cascade. We then tested whether ETV4 could promote adipocyte differentiation when stably expressed. We generated preadipocytes stably expressing ETV4 using a retroviral vector (1). After stimulation of differentiation with adipogenic reagents, 3T3-L1 cells stably expressing ETV4 showed enhanced expression of PPARγ and its target genes aP2, CD36, adiponectin, and C/EBPα (Fig. 5B). Similar to the effects on PPARγ mRNA expression by phenamil, ETV4 also increased PPARγ protein expression in 3T3-L1 cells (Fig. 5C). Furthermore, ETV4-expressing cells showed increased capacity for morphological differentiation and lipid accumulation as determined by Oil Red O staining (Fig. 5D). To test whether the promotion of adipocyte differentiation by ETV4 was also observed in other preadipocytes cell lines, we generated ETV4-expressing 3T3-F442A cells. As shown in Fig. 6, stable ETV4 expression also induced adipogenic differentiation, PPARγ expression, and adipogenic markers in this cell line.
Fig. 5.
ETV4 stimulates PPARγ expression and adipocyte differentiation in 3T3-L1 cells. A: Transient overexpression of ETV4 induces expression of PPARγ and its target genes in 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were transiently transfected with an ETV4 expression vector or empty vector. Four days later, mRNA was collected, and gene expression was determined by real-time PCR. B-C: Promotion of adipocyte differentiation by stable overexpression of ETV4 in 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were infected with pBabe-based retrovirus harboring the ETV4 gene, and large stable pools were selected with puromycin (2 μg/ml). B: Expression of PPARγ and its target genes from day 0 to day 6. Differentiation of 3T3-L1 cells into adipocytes was induced by DI and GW7845 (GW; 30 nM). C: Oil red O staining of ETV4 overexpressing 3T3-L1 cells. *P < 0.05, **P < 0.005, ***P < 0.0005. ETV4, EST variant 1-4; PPARγ, peroxisome proliferator-activated receptor γ.
Fig. 6.
ETV4 stimulates adipocyte differentiation in 3T3-F442A cells. Overexpression of ETV4 induces expression of PPARγ and adipocyte differentiation in 3T3-F442A cells. A: Oil red O staining of ETV4 overexpressing 3T3-F442A cells. 3T3-F442A preadipocytes were infected with pBabe-based retrovirus harboring the ETV4 gene, and large stable pools were selected with puromycin (2 μg/ml). B: Expression of PPARγ and its target genes on day 6. *P < 0.05, **P < 0.005, ***P < 0.0005. ETV4, EST variant 1-4; PPARγ, peroxisome proliferator-activated receptor γ.
To further address the relative contribution of ETV4 to phenamil's effects, we employed both overexpression and knockdown approaches. First, we asked whether ectopic expression of ETV4 would eliminate the ability of phenamil to further induce PPARγ expression and differentiation. Control cells and ETV4-expressing 3T3-L1 cells were treated with DMSO or phenamil in the presence of hormonal cocktail. In both control and ETV4-expressing cells, phenamil enhanced the expression of PPARγ and its target genes. However, the fold induction by phenamil was clearly reduced in ETV4-expressing cells, suggesting that ETV4 accounts for some of phenamil's actions in adipocyte differentiation (supplementary Fig. III). However, the fact that the adipogenic effects were not completely blunted in ETV4-expressing cells implies that ETV4 is not the only mediator.
We also assessed the effects of phenamil in cells in which ETV4 was knocked down using an shRNA vector. These studies were inconclusive (data not shown) in that phenamil was still able to promote differentiation when ETV4 expression was knocked down (data not shown). Although it is possible that the degree of knockdown we achieved was not sufficient, we favor the interpretation that other factors induced by phenamil are contributing to its effects. In support of this hypothesis, we found that ETV5, which is also induced by phenamil (Fig. 4), is functionally redundant with ETV4 in preadipocytes. We generated 3T3-L1 preadipocytes stably expressing ETV5 using a retroviral vector. After stimulation of differentiation with adipogenic reagents, 3T3-L1 cells stably expressing ETV5 showed enhanced expression of PPARγ and adipocyte differentiation (supplementary Fig. IV). Taken together, these data are consistent with the hypothesis that ETV4 and ETV5 are functionally redundant and that their induction by phenamil likely contributes to the adipogenic effects of this compound.
ETV4 and ETV5 act upstream of PPARγ in adipogenesis
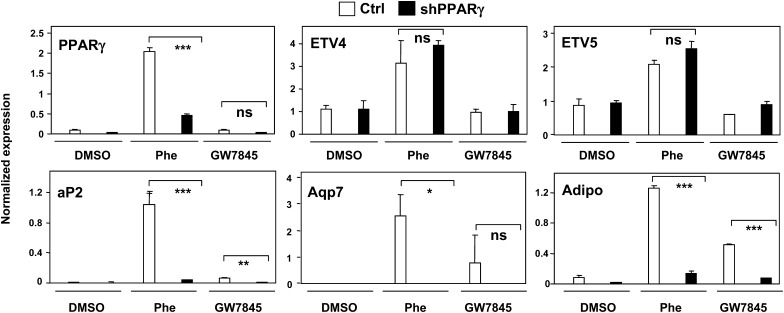
The above data strongly suggest that ETV4 and ETV5 act upstream of PPARγ to promote adipogenesis. To verify this idea, we generated stable 3T3-F442A cells expressing a shRNA directed against PPARγ. We and others have previously used this validated reagent to inhibit PPARγ mRNA and protein expression (12, 24). As expected, 3T3-F442A cells expressing PPARγ shRNA showed reduced PPARγ expression and markedly reduced differentiation and lipid accumulation (Fig. 7 and data not shown). Expression of the downstream target genes of PPARγ, including aP2, adiponectin and Aqp7, was greatly reduced in PPARγ shRNA-expressing cells, both in the presence and absence of phenamil (Fig. 7). These data indicate that phenamil's adipogenic effect is dependent on PPARγ expression. By contrast, the expression of both ETV4 and ETV5 was not affected by knockdown of PPARγ, suggesting that phenamil acts upstream of PPARγ (Fig. 7).
Fig. 7.
Knockdown of PPARγ blocks the adipogenic effects of phenamil but does not affect induction of ETV4. Stable 3T3-F442A cells expressing shRNAs against PPARγ were generated. ShRNAs effectively reduce PPARγ mRNA expression (upper left). Expression of ETV4 and ETV5 is not affected by knockdown of PPARγ, demonstrating that ETV4 is upstream of PPARγ (upper middle and right). Expression of downstream target genes of PPARγ, including aP2, adiponectin, and Aqp7, is reduced in PPARγ shRNA-expressing cells, both in the presence and absence of phenamil (bottom). Gene expression was determined by real-time PCR. *P < 0.05, **P < 0.005, ***P < 0.0005; ns, not significant. ETV4, EST variant 4; PPARγ, peroxisome proliferator-activated receptor γ.
DISCUSSION
We previously developed high-throughput screening approaches for the identification of small molecules that regulate differentiation of preadipocytes (11). In the present work, we have identified a new proadipogenic compound, phenamil, and utilized it as a chemical tool to identify transcriptional factors with the potential to affect preadipocyte differentiation.
Phenamil was previously characterized as an inhibitor of epithelial sodium channels (ENaC) and the Na+/H+ exchanger (17–21). Phenamil and benzamil were also reported to inhibit diamine oxidase (25). Interestingly, our data indicate that these targets are not responsible for the adipogenic effects of phenamil. First, other amiloride derivatives that target these proteins, such as benzamil, are not adipogenic (Fig. 1). Second, these putative protein targets of amiloride derivatives are not detectable in 3T3-L1 or 3T3-F442A preadipocytes (supplementary Fig. II). We also tested whether phenamil modulates previously recognized signaling pathways in adipogenesis, including the harmine-sensitive Wnt pathway. In contrast to harmine, phenamil did not alter Wnt signaling in preadipocytes (Fig. 3). Furthermore, harmine and phenamil had additive effects on adipogenic expression, consistent with distinct mechanisms of action (supplementary Fig. III-C).
The ETS genes encode a family of transcriptional regulators mediating tumorigenesis, proliferation, and differentiation. The Pea3 group of ETS-family transcription factors consists of three members: ETV1 (Er81), ETV4 (E1AF), and ETV5 (Erm) (26, 27). Pea3 members share the conserved ETS-domain and transactivating domains and are involved in wide range of cellular processes, including development, cell cycle control, proliferation, differentiation, and tumor metastasis. Previous studies have shown that ETVs can regulate the expression of target genes such as P21 (Cip), cyclooxygenase-2, and metalloproteases (MMP) (26, 28). Of note, P21 has been linked to adipogenesis through analysis of P21 null and knockdown cells (29). P21-null mice fed a high-fat diet showed reduced adipose tissue mass and improved insulin sensitivity.
We have identified ETV4 as a phenamil-responsive gene and shown that this factor can influence PPARγ expression and the differentiation potential of preadipocyte cell lines. Stable or transient expression of ETV4 stimulated expression of PPARγ. Similarly, retroviral expression of ETV4 enhanced the morphological differentiation of multiple preadipocyte cell lines. Knockdown of PPARγ expression by shRNA blocked the effects of phenamil on PPARγ target genes but did not alter expression of ETV4 itself, indicating that ETV4 acts upstream of PPARγ in the differentiation processes. Thus, ETV4 is a phenamil-inducible factor that modulates adipogenic gene expression and differentiation. Although our data suggest that ETV4 is a contributor to phenamil's adipogenic effects, the involvement of other factors cannot be excluded. In fact, we have identified other phenamil-responsive genes, including ETV5 that may also contribute to its adipogenic effects. Stable overexpression of ETV5 also stimulated adipogenesis, suggesting that common factor(s) might regulate expression of ETV genes (supplementary Fig IV). Furthermore, ETV1, another Pea3 family member, also stimulated the expression of PPARγ and adipogenesis, indicating that all three Pea3 members share similar activities in adipocyte differentiation (supplementary Fig. IV). Identification of the direct molecular target of phenamil in preadipocytes will be an interesting avenue for future studies. The precise targets on which ETV acts to affect adipocyte differentiation and PPARγ expression also remain to be determined.
The antidiabetic thiazolidinedione (TZD) drugs are known to exert their beneficial effects on insulin sensitivity by acting as direct ligands for PPARγ (30). TZDs activate PPAR target genes, resulting in increase in lipid flux and partitioning into adipocytes (31). Although effects of TZDs on many cell types have been documented, genetic studies support the hypothesis that adipose tissue is the primary target for the glucose-lowering effects of TZDs (32, 33). The clinical use of TZDs is limited by unfavorable side effects, including fluid retention and increased risk of congestive heart failure, indicating a need for additional therapeutic strategies. We previously identified the small molecule harmine as an inducer of PPARγ expression that has antidiabetic activity in mice (11). Harmine does not share the side effects associated with TZDs, suggesting that the induction of PPARγ expression could be an alternative approach in the treatment of insulin resistance. In the present work, we have identified phenamil as a second small molecule that induces PPARγ expression by a distinct mechanism. Characterization of the molecular pathways that mediate induction of PPARγ expression in response to such small molecules may provide insights into therapeutic strategies.
Supplementary Material
Acknowledgments
The authors thank Dr. Lim HJ for providing ETV1 and ETV5 constructs.
Footnotes
Abbreviations:
- ETV
- ETS variant
- IBMX
- 3-isobutyl-1-methylxanthine
- PPARγ
- peroxisome proliferator-activated receptor γ
- RXR
- retinoid X receptor
This work was supported by the Samsung Research Fund, Sungkyunkwan University (K.W.P.); by the Howard Hughes Medical Institute (P.T.); and by National Institutes of Health Grants HL-090553 and DK-063491. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Tontonoz P., Hu E., Spiegelman B. M. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid- activated transcription factor. Cell. 79: 1147–1156. [DOI] [PubMed] [Google Scholar]
- 2.Wu Z., Xie Y., Bucher N. L., Farmer S. R. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9: 2350–2363. [DOI] [PubMed] [Google Scholar]
- 3.Oishi Y., Manabe I., Tobe K., Tsushima K., Shindo T., Fujiu K., Nishimura G., Maemura K., Yamauchi T., Kubota N., et al. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1: 27–39. [DOI] [PubMed] [Google Scholar]
- 4.Mori T., Sakaue H., Iguchi H., Gomi H., Okada Y., Takashima Y., Nakamura K., Nakamura T., Yamauchi T., Kubota N., et al. 2005. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 280: 12867–12875. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee S. S., Feinberg M. W., Watanabe M., Gray S., Haspel R. L., Denkinger D. J., Kawahara R., Hauner H., Jain M. K. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator- activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 278: 2581–2584. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z., Torrens J. I., Anand A., Spiegelman B. M., Friedman J. M. 2005. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 1: 93–106. [DOI] [PubMed] [Google Scholar]
- 7.Tong Q., Dalgin G., Xu H., Ting C. N., Leiden J. M., Hotamisligil G. S. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 290: 134–138. [DOI] [PubMed] [Google Scholar]
- 8.Kennell J. A., O'Leary E. E., Gummow B. M., Hammer G. D., MacDougald O. A. 2003. T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol. Cell. Biol. 23: 5366–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fajas L., Landsberg R. L., Huss-Garcia Y., Sardet C., Lees J. A., Auwerx J. 2002. E2Fs regulate adipocyte differentiation. Dev. Cell. 3: 39–49. [DOI] [PubMed] [Google Scholar]
- 10.Choy L., Derynck R. 2003. Transforming growth factor-beta inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. J. Biol. Chem. 278: 9609–9619. [DOI] [PubMed] [Google Scholar]
- 11.Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. 2007. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 5: 357–370. [DOI] [PubMed] [Google Scholar]
- 12.Park K. W., Waki H., Villanueva C. J., Monticelli L. A., Hong C., Kang S., MacDougald O. A., Goldrath A. W., Tontonoz P. 2008. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-gamma expression and adipocyte differentiation. Mol. Endocrinol. 22: 2038–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8: 1224–1234. [DOI] [PubMed] [Google Scholar]
- 14.Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. 1995. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 83: 803–812. [DOI] [PubMed] [Google Scholar]
- 15.Padilla M. C., Armas-Hernandez M. J., Hernandez R. H., Israili Z. H., Valasco M. 2007. Update of diuretics in the treatment of hypertension. Am. J. Ther. 14: 154–160. [DOI] [PubMed] [Google Scholar]
- 16.Lant A. F., Smith A. J., Wilson G. M. 1969. Clinical evaluation of amiloride, a potassium-sparing diuretic. Clin. Pharmacol. Ther. 10: 50–63. [DOI] [PubMed] [Google Scholar]
- 17.Benos D. J. 1982. Amiloride: a molecular probe of sodium transport in tissues and cells. Am. J. Physiol. 242: C131–C145. [DOI] [PubMed] [Google Scholar]
- 18.Frelin C., Barbry P., Chassande O., Vigne P., Lazdunski M. 1988. The structure of the amiloride sensitive apical Na+ channel from pig kidney. Kidney Int. 26 (Suppl.): S12–S13. [PubMed] [Google Scholar]
- 19.Kleyman T. R., Cragoe E. J., Jr 1988. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 105: 1–21. [DOI] [PubMed] [Google Scholar]
- 20.Blazer-Yost B. L., Helman S. I. 1997. The amiloride-sensitive epithelial Na+ channel: binding sites and channel densities. Am. J. Physiol. 272: C761–C769. [DOI] [PubMed] [Google Scholar]
- 21.Bobkov Y. V., Ache B. W. 2007. Block by amiloride derivatives of odor-evoked discharge in lobster olfactory receptor neurons through action on a presumptive TRP channel. Chem. Senses. 32: 149–159. [DOI] [PubMed] [Google Scholar]
- 22.Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., Spiegelman B. M. 1999. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 3: 151–158. [DOI] [PubMed] [Google Scholar]
- 23.Park K. W., Waki H., Kim W. K., Davies B. S., Young S. G., Parhami F., Tontonoz P. 2009. The small molecule phenamil induces osteoblast differentiation and mineralization. Mol. Cell. Biol. 29: 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S., Bennett C. N., Gerin I., Rapp L. A., Hankenson K. D., Macdougald O. A. 2007. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer- binding protein alpha and peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 282: 14515–14524. [DOI] [PubMed] [Google Scholar]
- 25.Chassande O., Renard S., Barbry P., Lazdunski M. 1994. The human gene for diamine oxidase, an amiloride binding protein. Molecular cloning, sequencing, and characterization of the promoter. J. Biol. Chem. 269: 14484–14489. [PubMed] [Google Scholar]
- 26.Shindoh M., Higashino F., Kohgo T. 2004. E1AF, an ets-oncogene family transcription factor. Cancer Lett. 216: 1–8. [DOI] [PubMed] [Google Scholar]
- 27.de Launoit Y., Baert J. L., Chotteau-Lelievre A., Monte D., Coutte L., Mauen S., Firlej V., Degerny C., Verreman K. 2006. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta. 1766: 79–87. [DOI] [PubMed] [Google Scholar]
- 28.Funaoka K., Shindoh M., Yoshida K., Hanzawa M., Hida K., Nishikata S., Totsuka Y., Fujinaga K. 1997. Activation of the p21(Waf1/Cip1) promoter by the ets oncogene family transcription factor E1AF. Biochem. Biophys. Res. Commun. 236: 79–82. [DOI] [PubMed] [Google Scholar]
- 29.Inoue N., Yahagi N., Yamamoto T., Ishikawa M., Watanabe K., Matsuzaka T., Nakagawa Y., Takeuchi Y., Kobayashi K., Takahashi A., et al. 2008. Cyclin-dependent kinase inhibitor, p21WAF1/CIP1, is involved in adipocyte differentiation and hypertrophy, linking to obesity, and insulin resistance. J. Biol. Chem. 283: 21220–21229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann J. M., Moore L. B., Smith-Oliver T. A., Wilkison W. O., Willson T. M., Kliewer S. A. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J. Biol. Chem. 270: 12953–12956. [DOI] [PubMed] [Google Scholar]
- 31.Lehrke M., Lazar M. A. 2005. The many faces of PPARgamma. Cell. 123: 993–999. [DOI] [PubMed] [Google Scholar]
- 32.Khoursheed M., Miles P. D., Gao K. M., Lee M. K., Moossa A. R., Olefsky J. M. 1995. Metabolic effects of troglitazone on fat-induced insulin resistance in the rat. Metabolism. 44: 1489–1494. [DOI] [PubMed] [Google Scholar]
- 33.Olefsky J. M., Saltiel A. R. 2000. PPAR gamma and the treatment of insulin resistance. Trends Endocrinol. Metab. 11: 362–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.