Abstract
Lipids orchestrate biological processes by acting remotely as signaling molecules or locally as membrane components that modulate protein function. Detailed insight into lipid function requires knowledge of the subcellular localization of individual lipids. We report an analysis of the subcellular lipidome of the mammalian macrophage, a cell type that plays key roles in inflammation, immune responses, and phagocytosis. Nuclei, mitochondria, endoplasmic reticulum (ER), plasmalemma, and cytoplasm were isolated from RAW 264.7 macrophages in basal and activated states. Subsequent lipidomic analyses of major membrane lipid categories identified 229 individual/isobaric species, including 163 glycerophospholipids, 48 sphingolipids, 13 sterols, and 5 prenols. Major subcellular compartments exhibited substantially divergent glycerophospholipid profiles. Activation of macrophages by the Toll-like receptor 4-specific lipopolysaccharide Kdo2-lipid A caused significant remodeling of the subcellular lipidome. Some changes in lipid composition occurred in all compartments (e.g., increases in the levels of ceramides and the cholesterol precursors desmosterol and lanosterol). Other changes were manifest in specific organelles. For example, oxidized sterols increased and unsaturated cardiolipins decreased in mitochondria, whereas unsaturated ether-linked phosphatidylethanolamines decreased in the ER. We speculate that these changes may reflect mitochondrial oxidative stress and the release of arachidonic acid from the ER in response to cell activation.
Keywords: lipidome, membrane, lipopolysaccharide, mitochondria, endoplasmic reticulum, plasmalemma, nucleus
Lipids regulate and modify protein function and thus various biological processes by two distinct mechanisms. Signaling lipids, including free fatty acids, eicosanoids, sphingosine-1-phosphate, and lysophosphatidic acid, may be released from the sites of their generation in membranes and can subsequently affect receptors located remotely throughout tissues and cells [e.g., see (1–4) for reviews]. Structural lipids, which represent the bulk of the lipid in the organism, affect membrane-bound enzymes, transporters, and receptors in a local fashion by altering membrane properties or by specific binding to target proteins [reviewed in (5–8)].
A fundamentally accepted general concept of cell biology is the compartmentalization of biological processes within subcellular structures, termed organelles. Detailed information about the location of biochemical reactions is crucial for understanding their roles in cellular function and dysfunction. By the same token, knowing how different signaling and structural lipids affect cellular responses requires knowledge of their subcellular distribution. Traditional lipid analysis involves organic extraction of whole cells or tissues followed by separation and measurement of lipid classes and subclasses. When these approaches are taken, information regarding the subcellular distribution of lipid classes and the molecular identity of individual species of lipids is lost. Because the physical properties of individual membranes are determined not only by lipid head groups (lipid classes) but also by their acyl chain composition (6, 8), we sought to overcome these limitations by combining subcellular fractionation approaches we developed for the macrophage (9) with sophisticated mass spectrometric analysis of lipid species. In choosing a cell to apply these approaches to, we took advantage of the extraordinary biology of the macrophage. This cell type is ubiquitous throughout the mammalian kingdom and undergoes drastic transformation in response to activation (10–13). The LIPID MAPS Consortium has developed quantitative methods for comprehensively evaluating the composition, biosynthesis, and function of macrophage lipids (14) and has now applied these methods to subcellular fractions.
The classic pathway of macrophage activation is triggered by molecular components of pathogens, e.g., lipopolysaccharide (LPS), a complex structural lipid found in the outer membranes of most gram-negative bacteria (15, 16). Macrophages and other immune cell types detect submicromolar amounts of LPS (17–20) via the binding of this molecule to Toll-like receptor 4 (TLR-4) (15, 18, 21). It stimulates macrophages via its lipid anchor, which is termed lipid A. In our studies, we have employed Kdo2-lipid A (KLA), a homogenous subspecies of LPS that is specific for the TLR-4 receptor (17). Binding of LPS to TLR-4 and its associated coreceptors activates an intracellular signaling cascade that ultimately marshals cells of the adaptive immune system to resolve the infection. This process is accompanied by significant changes in the cellular levels of both signaling and structural lipids and in their metabolic pathways (Dennis et al., unpublished observations).
In the current study, we isolated the major subcellular organelles from resting and KLA-activated macrophages and determined the lipid composition of each organelle by LC-MS. We found that activation-induced lipid changes occurred on both global and subcellular levels and involved multiple lipid classes.
EXPERIMENTAL PROCEDURES
Materials
Immortalized mouse macrophage-like RAW264.7 cells were obtained from the ATCC (catalog no. TIB-71). DMEM (catalog no. 10-013) and PBS (catalog no. 21-031-CV) were from Mediatech. Fetal calf serum with low endotoxin content was from Hyclone (SH30071.03 ANG19242). KLA and lipid standards were from Avanti Polar Lipids. Iodixanol (OptiPrep from Axis-Shield) was obtained from Sigma-Aldrich. Solvents were chromatography-grade and purchased from OmniSolv. All other reagents/kits were from Sigma-Aldrich. Aqueous solutions were prepared using distilled-deionized water (catalog no. 25-055-CV) from Mediatech. Isolation media were prepared K+- and Na+-free; pH was adjusted by addition of Tris base (TRIZMA).
Tissue culture
Cells were maintained, treated, and fractionated as previously described (9). Briefly, RAW264.7 cells were maintained between passages 4 and 24 at 37°C and 10% CO2. The medium was composed of high glucose- and l-glutamine-containing DMEM supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For an experiment, five T-150 flasks of cells were plated at a density of 3.6 × 107 cells/flask in 24 ml of medium. At 24 h after plating, cells were treated with vehicle or 100 ng/ml KLA for another 24 h followed by subcellular fractionation.
Subcellular fractionation
Cells were harvested by scraping in 35 ml of PBS, pelleted at 200 g for 7 min, resuspended in 35 ml of isolation medium (250 mM sucrose, 10 mM HEPES-Tris, pH 7.4, 1 mM EGTA-Tris), and pelleted again to remove salts. For effective homogenization, cells were subjected to mild osmotic shock by resuspension in 35 ml of slightly hypotonic medium (isolation medium containing 100 mM sucrose) and thereafter pelleted. The supernatant was set aside; the cell pellet was carefully transferred to a 7 ml glass Dounce homogenizer, 10 ml of the supernatant was added, and the mixture homogenized by 40 strokes with a tight-fitting pestle. The resulting slurry was then combined with the remaining supernatant. Any alterations to this protocol resulted in ineffective cell lysis and organelle separation and decreased final yield.
The homogenate was brought to an isotonic state by the addition of 3.2 ml of the hypertonic medium (isolation medium containing 1.78 M sucrose) and then supplemented with 2 mM MgCl2 to preserve nuclei through subsequent steps. Differential centrifugation parameters were as follows: 200 g for 10 min to pellet nuclei/unbroken cells (crude “nuclear” pellet), 5,000 g for 10 min to pellet mitochondria, and 100,000 g for 1 h to pellet microsomes and plasmalemma fragments. Postnuclear and postmitochondrial supernatants were additionally purified by centrifugation at 300 g and 5,000 g for 10 min, respectively, to remove residual nuclei and mitochondria. The crude nuclear and mitochondrial pellets were additionally washed by resuspension and pelleting in Mg2+- containing and Mg2+-free media, respectively. The supernatant from the 100,000 g spin was retained as the cytosolic fraction.
The nuclear, mitochondrial, and microsomal pellets were subjected to centrifugation through step gradients of iodixanol in an SW-41 rotor. All gradient media were prepared according to the manufacturer's instructions based on the isolation medium above; the medium for nuclear preparation was supplemented with 5 mM MgCl2.
To purify nuclei, the crude nuclear pellet was brought to 25% iodixanol (final volume = 12 ml), iodixanol gradients were formed from the bottom up in three 12 ml tubes (4 ml 10%, 4 ml nuclei in 25%, 2.5 ml 30%, 1.5 ml 35%) and centrifuged at 10,000 g for 20 min. Nuclei banded at the 30/35% iodixanol interface.
The mitochondrial and microsomal pellets were resuspended in isolation medium and brought to 35% iodixanol (final volume = 6 ml) and fractionated by flotation for 2 h at 50,000 g in three 12 ml tubes each. The iodixanol gradient was formed with 2 ml 10% iodixanol, 4 ml 17.5% iodixanol, 4 ml 25% iodixanol, and 2 ml of the respective pellet resuspended in 35% iodixanol. Mitochondria banded at the 17.5/25% interface; plasmalemma and endoplasmic reticulum (ER) banded at the 10/17.5% and 17.5/25% interfaces, respectively. A 3rd fraction, originating from the microsomal pellet, banded at the most dense 25/35% interface, and was termed heavy microsomes (9).
All samples were frozen and stored at −80°C. Purity of fractions and composition of contaminants was determined as described previously (9). After completion of lipidomics analyses (see below), raw data were deconvoluted to determine organellar lipid levels as previously described (9) using proteomic marker ensembles derived from quadruplex iTRAQ analysis of fractions. Statistical analysis was performed using ANOVA.
Lipidomic analyses
Glycerophospholipids.
Glycerophospholipids from the majority of subclasses with the exception of cardiolipins (CL; see below) were extracted and analyzed as follows. Extraction was performed by modified Bligh and Dyer procedure using acidified methanol. Briefly, an equal volume of ice-cold methanolic HCl (∼0.05 N) and ice-cold CHCl3 was added to each of the fractions. Following 1 min of vortexing at 4°C, layers were separated by centrifugation (18,000 g for 5 min, 4°C) and the lower (organic) layer was collected. After the extraction and addition of standards, solvent was evaporated. The resulting lipid film was dissolved in 100 µl of isopropanol (IPA):hexane:100 mM NH4COOH(aq) 58:40:2 (mobile phase A). The mass spectrometric analysis and quantitation were performed essentially as previously described (22). The LC-MS technique was used with the utilization of synthetic odd-carbon phospholipid standards (four per each class). A MDS SCIEX 4000QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA) was used for the analyses, and coupled to it was a Shimadzu HPLC system (Shimadzu Scientific Instruments, Inc., Columbia, MD) consisting of a SCL 10 APV controller, two LC 10 ADVP pumps, and a CTC HTC PAL autosampler (Leap Technologies, Carrboro, NC). Phospholipids were separated on a Phenomenex Luna Silica column (Phenomenex, Torrance, CA) (2 × 250 mm, 5 µm particle size) using a 20 µl sample injection. A binary gradient consisting of IPA:hexane:100 mM NH4COOH(aq) 58:40:2 (mobile phase A) and IPA:hexane:100 mM NH4COOH(aq) 50:40:10 (mobile phase B) was used for the separation. The parameters of the mass spectrometer instrument and solvent gradient were as described in (22). Glycerophospholipid identification was achieved by comparison of fragmentation spectra of the samples to those of chemically defined phospholipid standards and literature, as described in (23).
CLs were quantified using normal phase LC coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems) equipped with an electrospray source. Normal phase LC using an Ascentis Si HPLC column (5 μm, 25 cm × 2.1 mm) was performed on an Agilent 1200 Quaternary LC system. Mobile phase A consisted of chloroform-methanol-aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform-methanol-water-aqueous ammonium hydroxide (600:340:50:5, v/v/v/v). Mobile phase C consisted of chloroform-methanol-water-aqueous ammonium hydroxide (450:450:95:5, v/v/v/v). The elution program consisted of a linear gradient begun at 100% mobile phase A to 100% mobile phase B over 14 min and held at 100% B for 11 min, followed by a 5 min linear gradient to 100% mobile phase C and held for 2 min. A 5 min gradient back to 100% B was then held for 2 min and returned to 100% A over 5 min and held for an additional 8 min. The LC flow rate was 300 μl/min. A postcolumn splitter diverted ∼10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer, with MS settings as follows: ion source (IS) = −4,200 V, curtain gas (CUR) = 20 psi, source gas 1 (GS1) = 20 psi, declustering potential (DP) = −55 V, and focusing potential (FP) = −265 V.
Four synthetic CL standards, CL(57:4), CL(61:1), CL(80:4) and CL(86:4) (from Avanti), were used as internal references. The CL(57:4) standard was 1′-[1,2-di-(9Z-tetradecenoyl)-sn-glycero-3-phospho], 3′-[1-(9Z-tetradecenoyl), 2-(10Z-pentadecenoyl)-sn-glycero-3-phospho]sn-glycerol. The 61:1 standard was 1′-[1,2-dipentadecanoyl-sn-glycero-3-phospho], 3′-[1-(pentadecanoyl), 2-(9Z-hexadecenoyl)-sn-glycero-3-phospho]sn-glycerol. The 80:4 standard was 1′-[1,2-di-(13Z-docosenoyl)-sn-glycero-3-phospho], 3′-[1-(13Z-docosenoyl), 2-(9Z-tetradecenoyl)-sn-glycero-3-phospho]sn-glycerol. The 86:4 standard was 1′-[1,2-di-(15Z-tetracosenoyl)-sn-glycero-3-phospho], 3′-[1-(15Z-tetracosenoyl), 2-(9Z-tetradecenoyl)-sn-glycero-3-phospho]sn-glycerol. The exact masses of these standards were 1,245.792, 1,307.902, 1,568.152 and 1,652.247 for CL(57:4), CL(61:1), CL(80:4), and CL(86:4), respectively. Data were analyzed as previously described (24).
Sphingolipids.
Sphingolipids were extracted and analyzed according to the LIPID MAPS procedure (25). Briefly, 0.1 ml of sample was mixed with 0.5 ml of methanol and 0.25 ml of chloroform with internal standards added (0.5 nmol of each C12-SM, C12-Cer, C12-GlcCer, and C12-LacCer). The mixtures were sonicated, incubated overnight at 48°C in a heating block, and then cooled down. For alkaline extraction, 75 μl of 1 M KOH in methanol was added; samples were sonicated and incubated for 2 h at 37°C. After centrifugation, to remove insoluble material, supernatants were dried down using SpeedVac and neutralized with 3–5 μl of glacial acetic acid. Then 1 ml of chloroform and 2 ml of distilled deionized water were added; samples were mixed and centrifuged. The upper layer was carefully removed, leaving the interface (with some water). The bottom layer/interface was dried on a SpeedVac and used for LC-MS/MS analysis, which was performed using a 5 cm × 2.1 mm Supelcosil LC-NH2 3 μm particle size HPLC column and the following mobile phase: 98% 97:2:1 CH3CN:CH3OH: CH3COOH, 2% 64:15:20:1 CH3OH:H2O:CH3(CH2)3)OH:CH3 COOH containing 5 mM ammonium acetate.
Sterol lipids.
Sterols in subcellular samples were extracted and analyzed using methods described in McDonald et al. (26). Briefly, a mixture of deuterated sterols serving as internal standards was added to samples and extracted using a modified Bligh/Dyer procedure. Extracts were dried under nitrogen and reconstituted in methanol. Sterols were resolved by HPLC and detected by MS using an electrospray source in positive ion mode. The HPLC was equipped with a 250 × 2 mm Luna C18 column (3 μM particle size; Phenomenex, Torrance, CA). The A mobile phase was 85% methanol and the B mobile phase was methanol (both A and B contained 5 mM ammonium acetate). The mass spectrometer was operated in multiple reaction monitoring (MRM) mode for maximum sensitivity.
Prenol lipids.
Prenol lipids were extracted and analyzed using LIPID MAPS procedures (24) with modifications as outlined below. Coenzyme Q (CoQ) 9 and CoQ10 were quantified by LC-MRM experiments performed using a Shimadzu LC system (comprising a solvent degasser, two LC-10A pumps, and a SCL-10A system controller) coupled to a 4000 Q-Trap hybrid triple quadrupole linear ion-trap mass spectrometer equipped with a Turbo V ion source (Applied Biosystems). LC was operated at a flow rate of 200 μl/min with a linear gradient as follows: 100% of mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 4 min. Mobile phase A consisted of methanol-acetonitrile-aqueous 1 mM ammonium acetate (60/20/20, v/v/v). Mobile phase B consisted of 100% ethanol containing 1 mM ammonium acetate. A Zorbax SB-C8 reversed-phase column (5 μm, 2.1 × 50 mm) was obtained from Agilent.
MRM was performed in the positive ion mode with MS settings as follows: CUR = 10 psi, GS1 = 20 psi, GS2 = 30 psi, IS = +5000 V, source temperature (TEM) = 350°C, ihe = ON, DP = +100 V, entrance potential (EP) = +10 V, and collision cell exit potential (CXP) = +5 V. The voltage used for collision-induced dissociation was +55 V. To quantify CoQ9 and CoQ10 in the subcellular fractions of RAW 264.7 cells, a known quantity of CoQ6 (Sigma) was added as an internal reference. The MRM pairs for CoQ6, CoQ9, and CoQ10 are 608/197, 812/197, and 880/197, respectively. In these MRM pairs, the precursor ions are the [M+NH4]+ ions, and the m/z 197 is the major fragment ion corresponding to a proton adduct of the quinone ring of CoQ.
The MRM of dolichols was performed in the negative ion mode with MS settings as follows: CUR = 20 psi, GS1 = 20 psi, GS2 = 30 psi, IS = −4500 V, TEM = 350°C, ihe = ON, DP = −40 V, EP = −10 V, and CXP = −5 V. The voltage used for collision-induced dissociation was −40 V. The MRM pairs for dolichol-18, dolichol-19, and dolichol-20 were 1304.2/59, 1372.2/59, and 1440.2/59, respectively. The MRM pairs for internal references nor-dolichol-18, nor-dolichol-19, and nor-dolichol-20 (Avanti) were 1290.2/59, 1358.2/59, and 1426.2/59, respectively. In these MRM pairs, the precursor ions were the [M + acetate]− adduct ions, and the product ions were the acetate ions (m/z 59).
Databases and bioinformatics tools
Results of lipidomic analyses, as well as the respective search tools, are available online at the website of the LIPID MAPS/Nature Lipidomics Gateway (http://www.lipidmaps.org/resources/resources.html).
RESULTS
Lipidomic analyses were focused on the four major categories of mammalian lipids that were most likely to underlie lipid effects on protein function (i.e., glycerophospholipids, sphingolipids, sterols, and prenols). For our initial studies, we choose to not include the two lipid categories that contain relatively hydrophilic and extremely hydrophobic lipids (i.e., fatty acyls and glycerolipids, respectively). Relatively hydrophilic lipids such as eicosanoids, sphingoid bases, and their phosphates and fatty acids were excluded from analysis, because these lipids are expected to be very easily released from membranes of origin and are thus likely to redistribute during extensive subcellular fractionation. Additionally, these lipids typically function remotely as signaling molecules through receptor-mediated modes of action that are distinct from a local effect of membrane composition, and, thus, have no permanent in situ localization. Conversely, highly hydrophobic neutral lipids are mainly confined within specialized organelles, such as lipid droplets, and have little direct effect on membrane properties of the major organelles studied.
Lipid composition of subcellular compartments
A summary of the number of lipid species analyzed in the lipidomics analysis of the six major subcellular fractions and the whole cell homogenate is presented in Table 1 (complete quantitative data on each lipid species are available in supplementary Table I).
TABLE 1.
Overview of the subcellular lipidome
| Nuclei |
Mitochondria |
ER |
Plasmalemma |
Dense Microsomes |
Cytosol |
Whole Cell |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid Categories | Control | KLA | Control | KLA | Control | KLA | Control | KLA | Control | KLA | Control | KLA | Control | KLA |
| Glycerophospholipids | 149 | 145 | 152 | 159 | 150 | 157 | 151 | 145 | 142 | 136 | 109 | 111 | 155 | 153 |
| Prenol lipids | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Sphingolipids | 48 | 48 | 47 | 47 | 48 | 47 | 48 | 45 | 48 | 47 | 47 | 46 | 48 | 48 |
| Sterol lipids | 13 | 12 | 12 | 12 | 12 | 12 | 13 | 12 | 11 | 9 | 5 | 9 | 12 | 11 |
| Total | 215 | 210 | 216 | 223 | 215 | 221 | 217 | 207 | 206 | 197 | 166 | 171 | 220 | 217 |
Each box shows the number of lipid molecular species that were detected and quantified in the specified subcellular fraction in the control or activated (KLA) states.
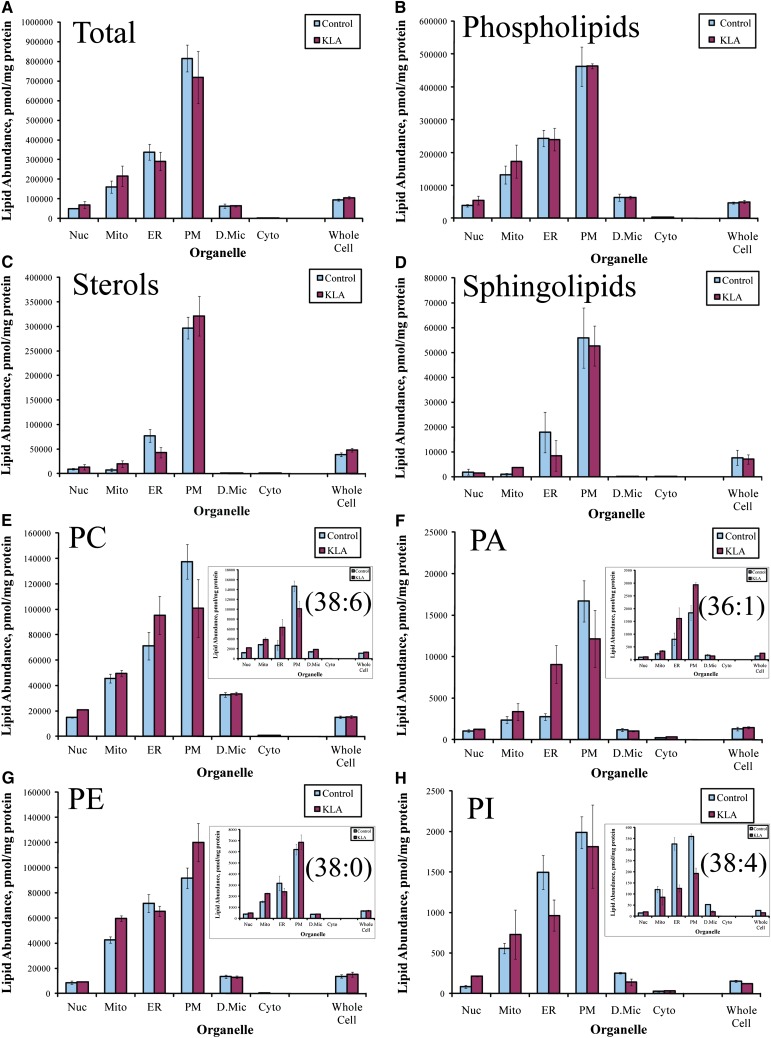
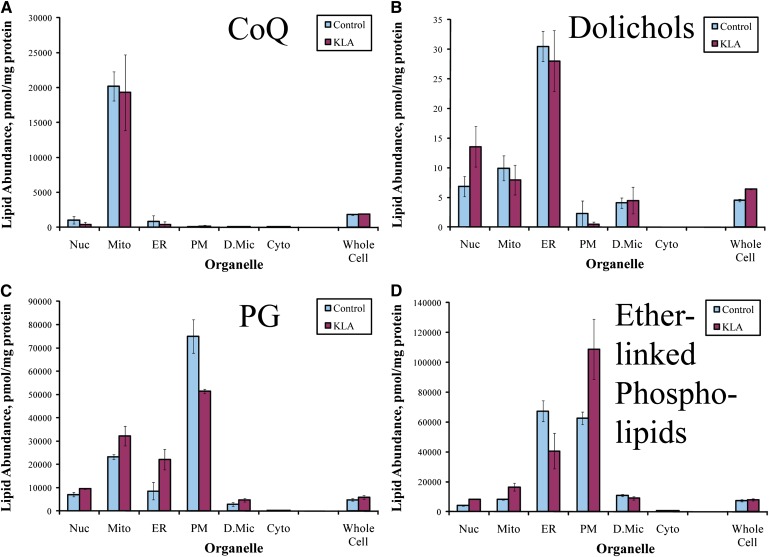
Lipid abundances for subcellular organelles/compartments were derived from the analyses of subcellular fractions using a deconvolution procedure (9). These data show an overall similarity for the subcellular distribution of different lipid species (maximal abundance in the plasmalemma and decreasing in the order ER > mitochondria > nuclei > dense microsomes >> cytoplasm) (Fig. 1). Lipid profiles (i.e., the relative abundance of various lipid species) are also similar among fractions and in general resemble the lipid profiles of the whole cell; however, detailed analyses reveal differences in both the subcellular distribution of individual lipids and classes and lipid profiles of organelles. For example, as expected, the subcellular localization of the ubiquinones was almost exclusively mitochondrial (Fig. 2A). Similarly, dolichols were remarkably absent from the plasma membrane and maximal in the ER (Fig. 2B). Phosphatidylglycerols and ether-linked phospholipids were present in all membranes but were relatively more abundant than other glycerophospholipid classes in mitochondria and ER, respectively (Fig. 2C, D).
Fig. 1.
Organelle lipid distribution profile (organellar factor). A: Total lipid. B–D: Total glycerophospholipids, sterol lipids, and sphingolipids, respectively. E–H: Total PC, PE, phosphatidic acid (PA), and phosphatidylinositol (PI), respectively (inserts: representative species). Data shown are sums of data for molecular species, mean ± SE, n = 3.
Fig. 2.
Subcellular distribution of lipids with specific localization. A: Total CoQ. B: Total dolichol. C: Total phosphatidylglycerol (PG). D: Total ether-linked phospholipids. Nuc, nuclei; Mito, mitochondria; PM, plasmalemma; D.Mic, dense microsomes; Cyto, cytoplasm. Data shown are sums of data for molecular species, mean ± SE, n = 3.
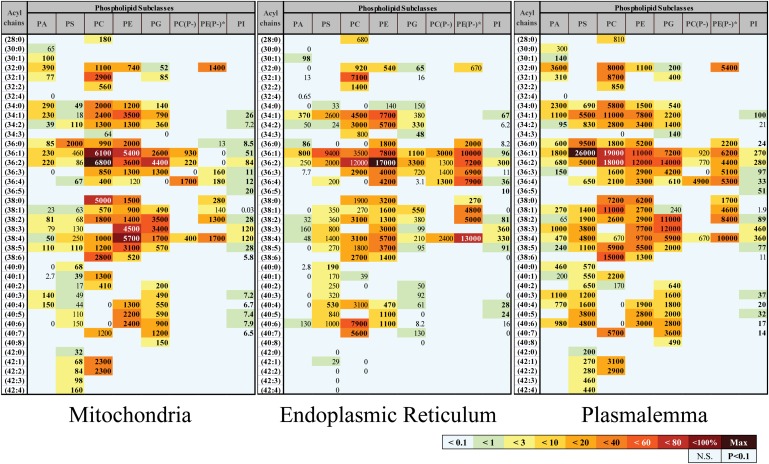
Distribution differences at the level of individual lipids were more subtle but could be observed by examining glycerophospholipid profiles of individual organelles (Fig. 3). For example, the two most abundant phospholipid species in mitochondria were phosphatidylcholines PC(36:2) and PC(36:1), whereas in ER they were phosphatidylethanolamines PE(36:2) and ether-linked (plasmalogen) PE(P-38:4) and in plasma membrane they were phosphatidylserine (36:1) and PC(36:1). The complete subcellular lipidome data are available for more detailed analysis as lipid abundance maps (supplementary Figs. I–XII).
Fig. 3.
Glycerophospholipidome of RAW264.7 macrophages is organelle specific. Each box represents an isobaric group; the values are expressed in pmol lipid/mg protein. Geographic map-like color-coding shows the abundance of a given lipid relative to the most abundant lipid species in the organelle; bold font denotes statistical significance. Acyl chains are abbreviated by the numbers of carbon atoms and double bonds per molecule. PA, phosphatidic acid; PG, phosphatidylglycerol; PI, phosphatidylinositol; PC(P-) and PE(P-), plasmenyl/plasmanyl analogs of PC and PE, respectively. *For simplicity, isobaric plasmenyl/plasmanyl species that differ by one double bond in their acyl chains are placed in a single box with the assumption of their plasmenyl nature [except the analyte with acyl chain composition (32:0), which is unequivocally plasmanyl PE (O-32:0)].
Factor-deconvolution of subcellular lipidome
The most important issue in the analysis of the subcellular lipidome is the spatial specificity of lipid distribution. Specific localization of particular lipid classes, subclasses, or individual lipids results from, and therefore contains information about, localization of their biosynthetic machinery and/or specificity of intracellular lipid transport systems (5, 27). However, in the absolute lipid abundance data (e.g., Fig. 3), this information is obscured by at least two other factors, specifically, organellar and species factors.
Indeed, an absolute amount of total lipid in an organelle is a function not only of various aspects of lipid metabolism but, in general, of the relative amount of membrane in the organelle, which we term “organellar factor”. If the amount of organellar protein is used as the measure of the organelle quantity, and, thus, total lipid content of the organelle is expressed as the lipid-to-protein ratio, then among membranous compartments, this ratio will be highest in the plasmalemma and lowest in mitochondria (the two most protein-poor and protein-rich organelles, respectively). It is reasonable to assume that these differences in the lipid-to-protein ratio are determined primarily by the protein rather than the lipid component of the organelle. In agreement with this assumption, all major lipid classes (and many of the phospholipid subclasses) show similar subcellular distributions that mirror total lipid distribution (Fig. 1). Therefore, we wished to eliminate this protein-driven organellar factor from the lipidomics analyses. The subcellular distribution of the total lipid, adjusted to exclude the organellar factor, must be a uniform distribution between organelles showing no difference between protein-rich (e.g., mitochondria) and protein-poor (e.g., plasmalemma) membranous compartments. Note, however, that some lipid subclasses show subcellular distributions markedly deviating from the general pattern (Fig. 2), and, therefore, elimination of the organellar factor will emphasize these specific deviations.
Similarly, the total amount of a particular lipid in an organelle is a function of such parameters as substrate availability and enzyme specificities that are independent of their localization. As an extreme example, the biosynthesis of polyunsaturated fatty acids (and corresponding phospholipids) is strictly dependent on the availability of essential fatty acids. These parameters are reflected in the total amount of the particular lipid in the whole cell (“species factor”) that may be expressed as a mole fraction of this lipid in the total lipid content of the cell. This factor is a bona fide target for lipidomic analysis of total cell, but it may be beneficial to eliminate it from the subcellular lipidomic analyses, because it is determined by factors that are localization independent.
To remove contributions of the organellar and species factors from the lipidomic data, we propose the following procedure. The total abundance of a particular lipid in an organelle may be presented as a product of the organellar factor (λ), the species factor (α), and, most importantly, a coefficient of the spatial specificity (distribution) δ (Eq. 1). More precisely, for each lipid k and organelle i:
| (Eq. 1) |
where Lorgi,k is abundance (pmol/mg protein) of lipid k in organelle i, λi is total lipid content in organelle i, organellar factor (pmol/mg protein), αk is relative lipid abundance, species factor (mole fraction), and δi,k is a coefficient of spatial specificity (specific lipid localization).
The λi and αk factors are easily determined from the comprehensive lipidome data (supplementary Figs. I–XII) using Eq. 2 and 3. For each organelle i, λi is derived by summing up all lipid abundances in the organelle (Eq. 2). For each lipid k, αk is derived by calculating mole fraction of the lipid in the whole cell (Eq. 3).
| (Eq. 2) |
| (Eq. 3) |
Therefore, the only unknown in Eq. 1 remains the factor of interest, the coefficient of spatial specificity δ, and this factor can be calculated. For this, Eq. 1 is converted to Eq. 4 after substitutions (Eq. 2 and Eq. 3).
For each lipid k:
| (Eq. 4) |
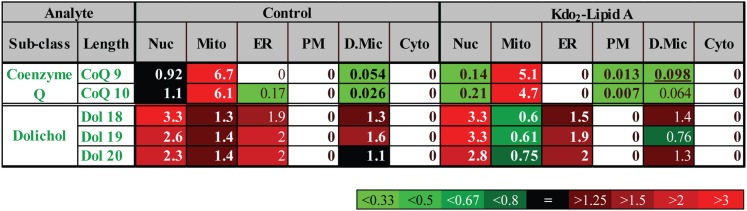
The coefficient of spatial specificity δi,k shows fold-difference between actual abundance of an analyte in an organelle and its predicted value from the whole cell level of the analyte (species factor) and the amount of total lipid in the organelle (organelle factor), or, in other words, a deviation from the general pattern of lipid distribution in the cell. δi,k equal to 1 indicates no deviation from the general pattern (lack of specificity of localization). δi,k > 1 indicates specific enrichment in a particular organelle; this is typically associated with δi,k < 1 in other organelles. An example of this analysis applied to the class of prenol lipids is shown in Fig. 4. As is evidenced by δ > 1, dolichol is specifically localized to the ER but, unexpectedly, the specific localization is even more pronounced for nuclei. As expected, CoQ shows highly specific mitochondrial localization. Note that, despite large (order of magnitude) differences in absolute abundances of different species of CoQ and dolichol (supplementary Fig. II), the specificity of their localization, as expressed by the coefficient of spatial specificity δ (Fig. 4), is the same.
Fig. 4.
Specificity of localization of prenol lipids. Heat map of coefficient of spatial specificity δ. The numbers show fold-difference between actual lipid species abundance and its predicted value from the whole cell level of the lipid species (species factor) and the amount of total lipid in organelle (organellar factor). Nuc, nuclei; Mito, mitochondria; PM, plasmalemma; D.Mic, dense microsomes; Cyto, cytoplasm.
KLA-induced lipidome remodeling
Upon the interaction of KLA with TLR-4 receptors, the macrophages transition to an activated state that persists over the 24-h treatment period. This is evidenced by the release of eicosanoids, cytokines (e.g., tumor necrosis factor-α and interleukin-6) (17), and upregulation of characteristic enzymes (e.g., COX-2 and inducible nitric oxide synthase; data not shown). In agreement with the dramatic transformation of the macrophage, the subcellular lipidome similarly underwent profound remodeling upon activation, leading to alterations that affected all lipid categories studied and a majority of lipid classes/subclasses (supplementary Figs. XIII–XIX, which provide heat maps for the fold activation of the various lipid species examined). Some changes in lipid content were global, i.e., occurring to a similar degree in all (or most) subcellular membranes (e.g., Fig. 5); other changes were localized to a specific organelle (e.g., Figs. 6–9). Most changes were universal, i.e., occurring similarly in all structurally similar lipids (typically, lipids of the same subclass with similar acyl chains); however, in a minority of cases, higher chemical specificity was suggested. Although not all changes reached the level of statistical significance (significance is indicated by the presence of numbers in the heat maps in and supplementary Figs. XIII–XIX), we considered the trends (indicated by color alone on heat maps) as supporting information in establishing patterns of change.
Fig. 5.
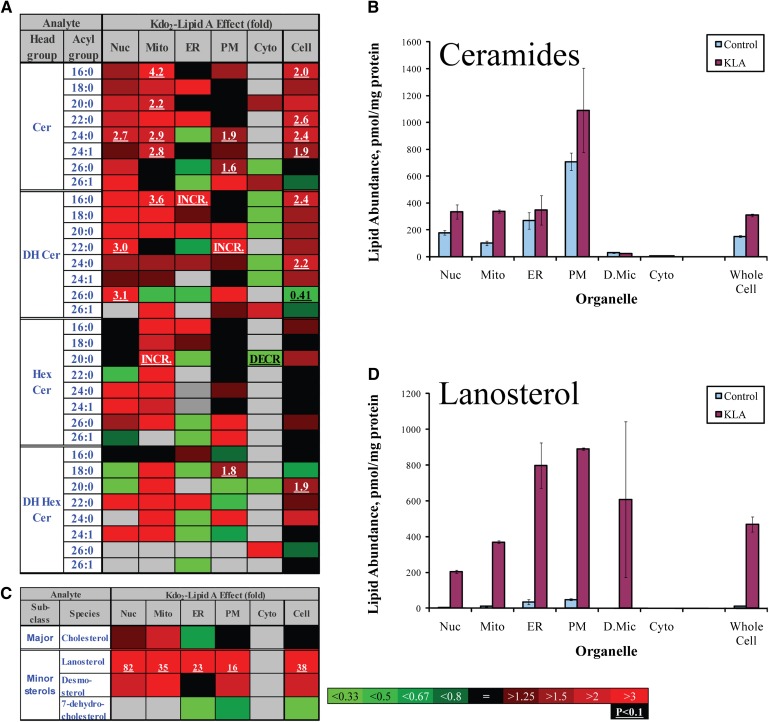
Global lipidome changes. A: Ceramide heat map showing fold-increase in response to KLA. Changes are color coded; numbers indicate statistically significant fold-changes. Cer, ceramide, DH, dihydro-, Hex, hexosyl (glucosyl or galactosyl). INCR., increase from zero; DECR., decrease to zero. Acyl groups are abbreviated by the numbers of carbon atoms and double bonds per molecule. B: Increase in total ceramide (sum of individual species is shown). C: Sterol lipids heat map showing fold-increase in response to KLA. D: Increase in lanosterol. Nuc, nuclei, Mito, mitochondria, PM, plasmalemma, D.Mic, dense microsomes, Cyto, cytoplasm. Data (B and D) are mean ± SE, n = 3.
Fig. 6.
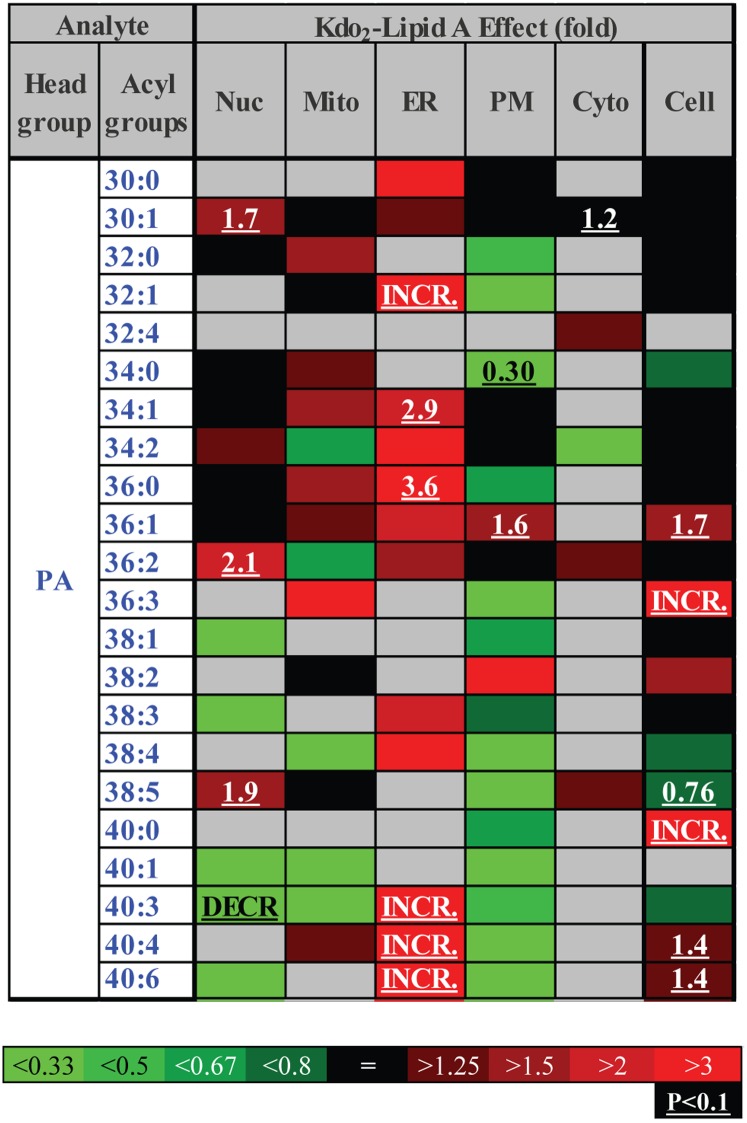
ER-specific increase in phosphatidic acid (heat map). Each box represents an isobaric group. Changes are color-coded; numbers indicate statistically significant fold-changes. INCR., increase from zero; DECR., decrease to zero; Nuc, nuclei; Mito, mitochondria; PM, plasmalemma; D.Mic, dense microsomes; Cyto, cytoplasm. Acyl groups are abbreviated by the numbers of carbon atoms and double bonds per molecule.
Fig. 7.
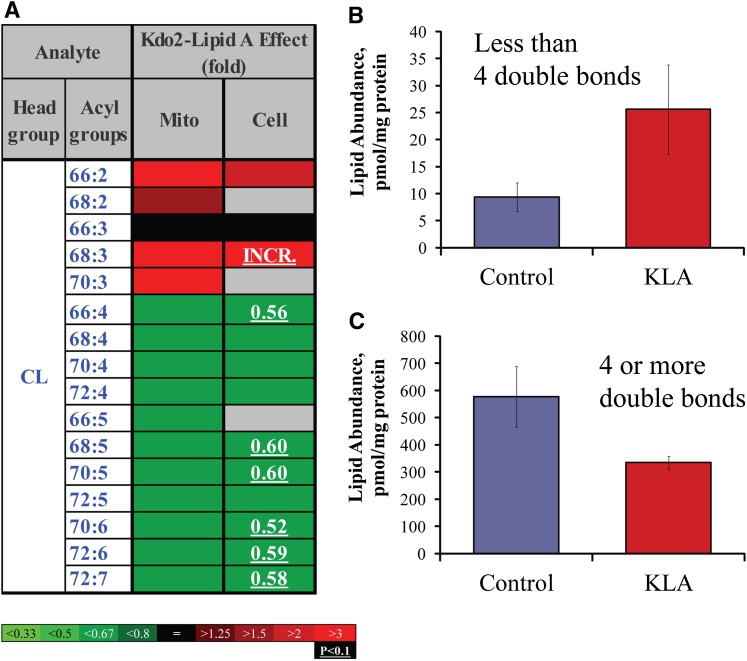
Shift of CL profile to less unsaturated species. A: Heat map. Each box represents an isobaric group. Changes are color-coded; numbers indicate statistically significant fold-changes. INCR., increase from zero. Acyl groups are abbreviated by the numbers of carbon atoms and double bonds per molecule. Mito, mitochondria. B: Highly unsaturated CLs (four or more double bonds). C: Moderately unsaturated CLs (two or three double bonds). Data (B and C) are mean ± SE, n = 3.
Fig. 8.
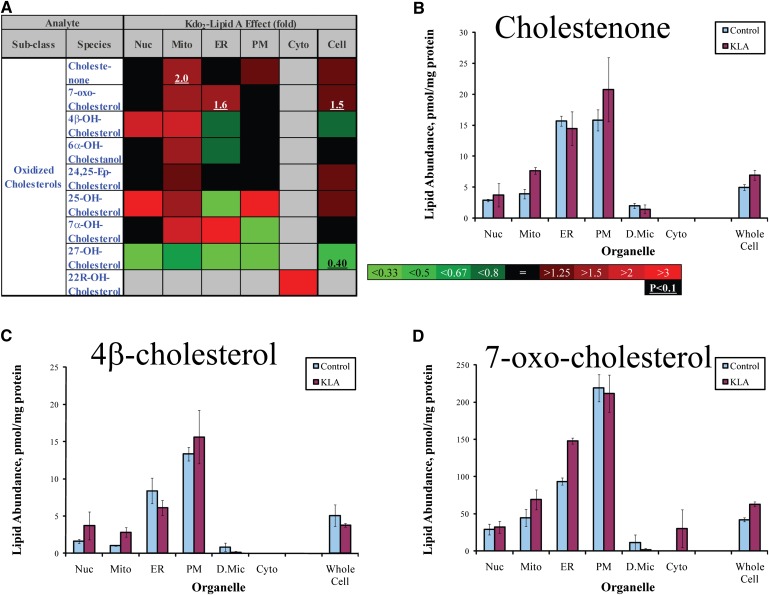
Location-specific oxysterol increases. A: Heat map. Changes are color-coded; numbers indicate statistically significant fold-changes. Nuc, nuclei; Mito, mitochondria; PM, plasmalemma; D.Mic, dense microsomes; Cyto, cytoplasm. B–E. Cholestenone (increase in mitochondria), 4-β-cholesterol (increase in mitochondria), 7-oxo-cholesterol (increase in ER). Data are mean ± SE, n = 3.
Fig. 9.
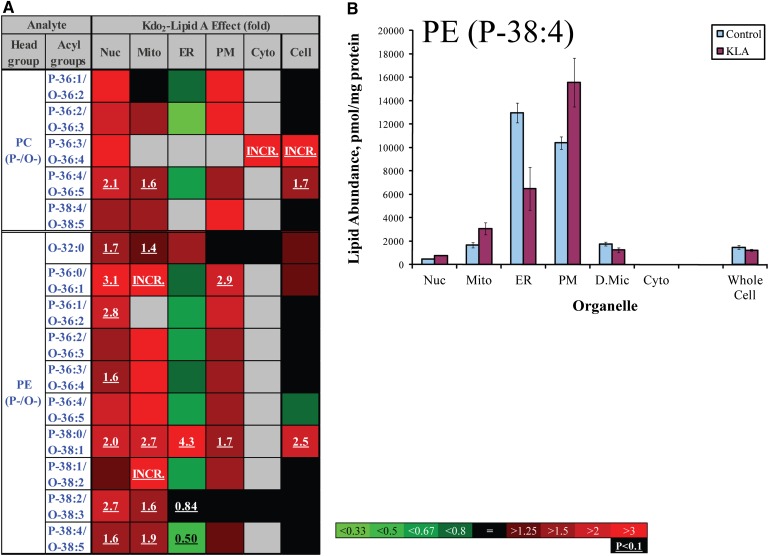
ER-specific decrease in unsaturated ether-linked lipids. A: Heat map. Each box represents an isobaric group. Changes are color-coded; numbers indicate statistically significant fold changes. INCR., increase from zero. PC (P-/O-), plasmenyl/plasmanyl analogs of PC; PE (P-/O-), plasmenyl/plasmanyl analogs of PE. Acyl groups are abbreviated by the numbers of carbon atoms and double bonds per molecule. Nuc, nuclei; Mito, mitochondria; PM, plasmalemma; D.Mic, dense microsomes; Cyto, cytoplasm. B: Plasmenyl/plasmanyl PE (P-38:4/O-38:5). Data are mean ± SE, n = 3.
An example of global lipidome changes was the KLA-induced production of ceramides (Fig. 5A, B). This phenomenon was suggested previously (28) and was observed at the whole cell level (Fig. 5); however, mapping of the subcellular lipidome demonstrated directly that ceramides generated during activation reached all major cellular membranes (Fig. 5A, B) and, therefore, may exert biological effects throughout the entire cell. Unexpectedly, a global change pattern was also observed for two cholesterol precursors, lanosterol and desmosterol (Fig. 5C, D).
An example of a local but structurally universal lipidome change is shown in Fig. 6. All detected species of phosphatidic acid demonstrated marked increases in the ER, whereas the changes were mixed in the other organelles and in the whole cell. The mitochondrial lipid CL demonstrated a more complex response to cell activation (Fig. 7). Unsaturated CL species with four or more double bonds significantly decreased, whereas less unsaturated species tended to increase. Oxidized sterols did not show an unequivocal pattern of change; however, the majority of these lipids tended to increase in mitochondria (with the exception of 27-hydroxycholesterol, which decreased throughout the cell) (Fig. 8). Similarly ether-linked phospholipids (Fig. 9) showed a complex pattern of change: although these lipids increased in all major cellular membranes, most of members of this class tended to decrease in the ER, with the most pronounced decrease shown by highly unsaturated PE (P-38:4/O-38:5). Note that for ether-linked phospholipids, unlike other phospholipid subclasses, the ER is the main subcellular location (Fig. 2D and 3); thus, these decreases may have biological significance.
DISCUSSION
The present study reports a comprehensive analysis of the subcellular lipidome of a mammalian cell, a murine macrophage (RAW264.7). The abundances of the major species of membrane lipids were determined in all major cellular organelles and compartments in both resting and activated cells. To fully consider the lipidome remodeling that takes place upon macrophage activation, we note that it is insufficient to consider in isolation the degree of change induced by KLA treatment in a given lipid as is done in the heat maps ( and supplementary Figs. XIII–XIX). This is because a low abundance lipid is more likely to undergo large fold-changes, whereas an abundant lipid is unlikely to undergo a substantial change in amount but is more likely to determine the properties of a given membrane. An exception to this consideration is found in signaling lipids such as ceramides, which may act via high affinity interactions with specific proteins and thus confer biological effects despite a low abundance. Because there is no a priori way to predict signaling properties of a lipid, we have found it insightful to virtually superimpose two types of maps, the heat map ( and supplementary Figs. XIII–XIX) and the lipid abundance map (supplementary Figs. I ).
The observed changes characterize the activated state of macrophages; however, it may be possible that early transient changes associated with TLR-4 signaling were undetected at the 24 h time point. It should also be noted that, although acyl chain composition may be influenced by fatty acid availability (in the cell culture medium or in the in vivo extracellular milieu), our data indicate that the lipid composition is a function of additional organelle-specific factors as the fatty acyl profiles differ between lipid classes/subclasses and subcellular organelles (Fig. 3 and supplementary Figs. I, III–IX, and XI, XII).
As is true for any “omics” study, the dynamic lipidome that we mapped is no more, but also no less, than a powerful hypothesis-building engine. Even a superficial analysis allows us to propose several testable hypotheses that may in the future provide further understanding of various aspects of lipid metabolism and macrophage physiology.
Mitochondrial oxidative stress
Oxidative stress (sometimes referred to as the respiratory burst) is a part of the host defense response mounted by macrophages in response to infection and inflammation. A major element of this response is the release of reactive oxygen species (ROS) arising from the activation of NADPH oxidase, an enzyme of the plasmalemma (29). The released ROS assist in the destruction of the pathogen in part by oxidizing membrane lipid components. In macrophage and nonmacrophage cell types, mitochondria are also capable of generating large quantities of ROS (30). A prerequisite of mitochondrial ROS production is severe respiratory inhibition (31), a condition that may also be met in activated macrophages. In the experiments reported here, respiration was inhibited by up to 90% (data not shown), which suggests that mitochondria may have contributed to the total oxidative stress generated by the activated macrophages. In most cases, whether mitochondrial ROS production makes a contribution to the oxidative stress is difficult to determine; however, analysis of the subcellular lipidome provides circumstantial support for the potential role of mitochondria. The data of Fig. 8 show mitochondria-specific 2-fold increases in cholestenone and 4β-hydroxycholesterol and a somewhat smaller fold-change in 7-oxocholesterol. These oxidized sterols are thought to be products of nonenzymatic reactions and as such are a readout of oxidative stress in their vicinity. In contrast, the amounts of these oxidized sterols did not change in plasmalemma upon activation, suggesting little contribution from the NADPH oxidase.
Changes in CL levels are also consistent with local oxidative stress. The selective loss of more unsaturated species (Fig. 7) may be due to oxidation. Polyunsaturated fatty acyls are highly susceptible to oxidation because of the presence of vulnerable bis-allylic carbons. It is likely that in CLs that have three double bonds per four acyl chains (which we define here as “less unsaturated”), few of the acyl chains are polyunsaturated; therefore, these CLs will be resistant to oxidation. With an increasing number of double bonds per molecule, the likelihood of occurrence of polyunsaturated fatty acyls increases, leading to oxidative stress susceptibility and loss of native CL as was observed in the data of Fig. 7.
Thus, based on the lipidomics analysis, the hypothesis of mitochondrial oxidative stress in activated macrophages gains some credibility and additional experiments may be designed to further test it. Two main caveats become evident from these considerations. First, our initial subcellular lipidome has focused only on major lipid classes and lacks, e.g., information about oxidized CLs and other phospholipids. Second, the species resolution at the level of isobaric groups does not allow direct conclusions about precise fatty acyl composition of the lipids. However, more advanced levels of lipidomics analysis that would include both minor and modified species [i.e. oxidized phospholipids (32)] and individual molecular species resolution could enhance the initial analysis reported herein.
An ER plasmalogen that is a source of arachidonic acid
Ether-linked phospholipids are highly abundant in the ER (Fig. 2D), and among these lipids, a relatively long-chain and unsaturated PE (P-38:4/O-36:5) is one of the most abundant components (Fig. 3). We speculate that a large fraction of this lipid contains arachidonic acid at the sn2 position; specifically, this lipid is likely 1-stearoyl, 2-archidonyl plasmalogen PE(P-36:4). Consistent with this hypothesis, the lipid decreases by a factor of two upon activation of macrophages (Fig. 9), a situation in which large quantities of arachidonic acid are released into the medium or are converted into prostaglandins (33). Thus, our data support prior suggestions regarding the role of plasmalogens as a source of arachidonic acid in macrophages (34, 35), and do not support the opposing view (36).
ER-specific increase in the levels of phosphatidic acid species (Fig. 6) may indicate site-specific activation of a phospholipase D (or phospholipase C in combination with diacylglycerol kinase), a hypothesis we are currently testing further.
CONCLUSION
On this first mapping of the subcellular macrophage lipidome, we focused on the major lipid species and the major organelles. However, local effects of the lipids that modulate protein functions may be unresolved at the level of organelles and manifest themselves at the level of particular membranes (e.g., outer vs. inner membranes of mitochondria), parts of membranes (basal vs. apical parts of plasmalemma), or even small clusters of lipids (lipid rafts). This report represents the first step on a long road to the complete subcellular lipid mapping of the cell.
Supplementary Material
Footnotes
Abbreviations:
- CL
- cardiolipin
- CoQ
- coenzyme Q
- ER
- endoplasmic reticulum
- IPA
- isopropanol
- KLA
- Kdo2-lipid A [(3-deoxy-D-manno-octulosonic acid)2-lipid A]
- LPS
- lipopolysaccharide
- MRM
- multiple reaction monitoring
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- ROS
- reactive oxygen species
- TLR-4
- Toll-like receptor 4
This study was supported by the LIPID MAPS Large Scale Collaborative Grant (GM-069338) from the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of 12 figures and one table.
REFERENCES
- 1.Wymann M. P., Schneiter R. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176. [DOI] [PubMed] [Google Scholar]
- 2.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls D. G. 2006. The physiological regulation of uncoupling proteins. Biochim. Biophys. Acta. 1757: 459–466. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins C. M., Cedars A., Gross R. W. 2009. Eicosanoid signalling pathways in the heart. Cardiovasc. Res. 82: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen O. S., Koeppe R. E., II 2007. Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36: 107–130. [DOI] [PubMed] [Google Scholar]
- 7.Dowhan W., Mileykovskaya E., Bogdanov M. 2004. Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim. Biophys. Acta. 1666: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666: 62–87. [DOI] [PubMed] [Google Scholar]
- 9.Andreyev A. Y., Shen Z., Guan Z., Ryan A., Fahy E., Subramaniam S., Raetz C. R., Briggs S., Dennis E. A. 2010. Application of proteomic marker ensembles to subcellular organelle identification. Mol. Cell. Proteomics. 9: 388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnovsky M. L., Lazdins J. K. 1978. Biochemical criteria for activated macrophages. J. Immunol. 121: 809–813. [PubMed] [Google Scholar]
- 11.North R. J. 1978. The concept of the activated macrophage. J. Immunol. 121: 806–809. [PMC free article] [PubMed] [Google Scholar]
- 12.Cohn Z. A. 1978. Activation of mononuclear phagocytes: fact, fancy, and future. J. Immunol. 121: 813–816. [PubMed] [Google Scholar]
- 13.Mosser D. M., Edwards J. P. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8: 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmelzer K., Fahy E., Subramaniam S., Dennis E. A. 2007. The lipid maps initiative in lipidomics. Methods Enzymol. 432: 171–183. [DOI] [PubMed] [Google Scholar]
- 15.Raetz C. R., Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rietschel E. T., Kirikae T., Schade F. U., Mamat U., Schmidt G., Loppnow H., Ulmer A. J., Zahringer U., Seydel U., Di Padova F., et al. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8: 217–225. [DOI] [PubMed] [Google Scholar]
- 17.Raetz C. R., Garrett T. A., Reynolds C. M., Shaw W. A., Moore J. D., Smith D. C., Jr., Ribeiro A. A., Murphy R. C., Ulevitch R. J., Fearns C., et al. 2006. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 47: 1097–1111. [DOI] [PubMed] [Google Scholar]
- 18.Aderem A., Ulevitch R. J. 2000. Toll-like receptors in the induction of the innate immune response. Nature. 406: 782–787. [DOI] [PubMed] [Google Scholar]
- 19.Visintin A., Halmen K. A., Latz E., Monks B. G., Golenbock D. T. 2005. Pharmacological inhibition of endotoxin responses is achieved by targeting the TLR4 coreceptor, MD-2. J. Immunol. 175: 6465–6472. [DOI] [PubMed] [Google Scholar]
- 20.Akashi S., Saitoh S., Wakabayashi Y., Kikuchi T., Takamura N., Nagai Y., Kusumoto Y., Fukase K., Kusumoto S., Adachi Y., et al. 2003. Lipopolysaccharide interaction with cell surface Toll-like receptor 4-MD-2: higher affinity than that with MD-2 or CD14. J. Exp. Med. 198: 1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff M., Gay N. J. 2004. MD-2: the Toll ‘gatekeeper’ in endotoxin signalling. Trends Biochem. Sci. 29: 294–300. [DOI] [PubMed] [Google Scholar]
- 22.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., Brown H. A. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 23.Milne S., Ivanova P., Forrester J., Alex Brown H. 2006. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 39: 92–103. [DOI] [PubMed] [Google Scholar]
- 24.Garrett T. A., Guan Z., Raetz C. R. 2007. Analysis of ubiquinones, dolichols, and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 432: 117–143. [DOI] [PubMed] [Google Scholar]
- 25.Sullards M. C., Allegood J. C., Kelly S., Wang E., Haynes C. A., Park H., Chen Y., Merrill A. H., Jr 2007. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol. 432: 83–115. [DOI] [PubMed] [Google Scholar]
- 26.McDonald J. G., Thompson B. M., McCrum E. C., Russell D. W. 2007. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 432: 145–170. [DOI] [PubMed] [Google Scholar]
- 27.Ikonen E., Jansen M. 2008. Cellular sterol trafficking and metabolism: spotlight on structure. Curr. Opin. Cell Biol. 20: 371–377. [DOI] [PubMed] [Google Scholar]
- 28.MacKichan M. L., DeFranco A. L. 1999. Role of ceramide in lipopolysaccharide (LPS)-induced signaling. LPS increases ceramide rather than acting as a structural homolog. J. Biol. Chem. 274: 1767–1775. [DOI] [PubMed] [Google Scholar]
- 29.Morel F., Doussiere J., Vignais P. V. 1991. The superoxide-generating oxidase of phagocytic cells. Physiological, molecular and pathological aspects. Eur. J. Biochem. 201: 523–546. [DOI] [PubMed] [Google Scholar]
- 30.Andreyev A. Y., Kushnareva Y. E., Starkov A. A. 2005. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc.). 70: 200–214. [DOI] [PubMed] [Google Scholar]
- 31.Kushnareva Y., Murphy A. N., Andreyev A. 2002. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 368: 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyurin V. A., Tyurina Y. Y., Jung M. Y., Tungekar M. A., Wasserloos K. J., Bayir H., Greenberger J. S., Kochanek P. M., Shvedova A. A., Pitt B., et al. 2009. Mass-spectrometric analysis of hydroperoxy- and hydroxy-derivatives of cardiolipin and phosphatidylserine in cells and tissues induced by pro-apoptotic and pro-inflammatory stimuli. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2863–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., Dennis E. A. 2007. TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem. 282: 22834–22847. [DOI] [PubMed] [Google Scholar]
- 34.Ford D. A., Gross R. W. 1989. Plasmenylethanolamine is the major storage depot for arachidonic acid in rabbit vascular smooth muscle and is rapidly hydrolyzed after angiotensin II stimulation. Proc. Natl. Acad. Sci. USA. 86: 3479–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chilton F. H., Connell T. R. 1988. 1-ether-linked phosphoglycerides. Major endogenous sources of arachidonate in the human neutrophil. J. Biol. Chem. 263: 5260–5265. [PubMed] [Google Scholar]
- 36.Gaposchkin D. P., Farber H. W., Zoeller R. A. 2008. On the importance of plasmalogen status in stimulated arachidonic acid release in the macrophage cell line RAW 264.7. Biochim. Biophys. Acta. 1781: 213–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.