Abstract
A method of direct lipid analysis by MALDI mass spectrometry in intact membranes, without prior extraction/separation steps, is described. The purple membrane isolated from the extremely halophilic archaeon Halobacterium salinarum was selected as model membrane. Lyophilized purple membrane were grinded with 9-aminoacridine (9-AA) as dry matrix, and the powder mixture was crushed in a mechanical die press to form a thin pellet. Small pieces of the pellet were then attached to the MALDI target and directly analyzed. In parallel, individual archaebacterial phospholipids and glycolipids, together with the total lipid extract of the purple membrane, were analyzed by MALDI-TOF/MS using 9-AA as the matrix in solution. Results show that 9-AA represents a suitable matrix for the conventional MALDI-TOF/MS analysis of lipid extracts from archaeal microorganisms, as well as for fast and reliable direct dry lipid analysis of lyophilized archaebacterial membranes. This method might be of general application, offering the advantage of quickly gaining information about lipid components without disrupting or altering the membrane matrix.
Keywords: glycerolipids (diphytanylglycerol ether analogs), glycerophospholipids (diphytanylglycerolphosphate ether analogs), Halobacterium salinarum, purple membrane, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
Today shotgun ESI-MS lipidomics represents one of the most powerful tools of analysis of the total lipid extract of biomembranes, allowing the detection of both zwitterionic and acidic phospholipids (1).
Recently, a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) approach, based on the use of 9-aminoacridine (9-AA) as matrix, has been developed for the rapid analysis of glycerophospholipids, allowing a rapid, sensitive, and highly informative approach for the direct analysis of cell lipid extracts (2). Notably the use of the novel matrix 9-aminoacridine in MALDI-TOF/MS enables the fast and reliable detection of anionic phospholipids and also cardiolipin species (2).
In our study we extended the use of 9-aminoacridine as a matrix for the MALDI-TOF/MS to glycerolipids and glycerophospholipids of extremely halophilic microorganisms belonging to the Archaea kingdom. The extremely halophilic archaeons are well adapted to life in NaCl- saturated aquatic ecosystems, where they are exposed to extremely low water activity and desiccation stress.
Glycerolipids and glycerophospholipids are defined as glycerol and glycerol-phosphate derivatives, respectively (3). Whereas in bacteria and eukaryotic cells, glycerolipids and glycerophospholipids are constituted by straight-chain fatty acids linked to glycerol by ester bonds, in the archaeal microorganisms, the hydrophobic chains are isopranoid chain alcohols linked to glycerol by ether bonds (4).
The stereochemistry of archaebacterial membrane lipids is also different from that of other membrane lipids. In archaebacterial lipids, the glycerol group is alkylated at the sn-2 and sn-3 positions, at variance from bacterial and eukaryotic lipids, whose chains are linked at the sn-1 and sn-2 positions (4). The ether bond and the absence of unsaturations in the hydrophobic tail of the phospholipids (and other polar lipids present in archaeal membranes) are considered adaptive traits of microorganisms able to thrive in harsh or extreme environments, such as saturated salt, anoxic, and high-temperature waters (5).
Besides phospholipids and cardiolipin analogs (6), a number of glycosyldiradylglycerols (i.e., glycolipids) have been described (4, 7, 8).
As many of the lipid components of archaeal cell membranes are not commercially available, we have isolated and purified individual archaeal lipids for MALDI-TOF/MS analysis. The purple membrane (PM) of the extreme halophile Hbt. salinarum was selected as a model membrane for our studies.
The PM domain has an important role in the bioenergetics of the microorganism, as it contains the photoactivated proton pump bacteriorhodopsin. Bacteriorhodopsin is able to convert green sunlight in a proton gradient across the cell membrane (more acid outside); the protons flowing back through ATP synthase allow ATP synthesis inside the cell. The purple membrane is constituted of a 2D crystalline lattice, formed by bacteriorhodopsin as only protein and a small number of lipid molecules (about 10 per each protein molecule). In the past we determined the PM lipid-protein stoichiometries by means of nuclear magnetic resonance (NMR) analysis (9).
In the present study we illustrate a new method of direct MALDI-TOF/MS analysis of lyophilized PM finely grinded with 9-AA as dry matrix. The simplified method of sample preparation for MALDI lipid analysis is also suitable for other archaebacterial membranes in lyophilized form. In parallel, to validate results obtained with the new method, individual lipid components and the total lipid extract of archaebacterial membranes were analyzed for the first time by MALDI-TOF/MS.
EXPERIMENTAL PROCEDURES
Materials
DNase I was obtained from Sigma Aldrich. 9-Aminoacridine hemilhydrate was purchased from Acros Organics (Morris Plains, NJ). Archaebacterial lipid standards were isolated and purified from archaeons cultured in our laboratory as described in the following sections. The following archaebacterial lipids (i.e., diphytanylglycerol ether analogs) were used in the present study: PG, phosphatidylglycerol; PGP-Me, phosphatidylglycerophosphate methyl ester; PGS, phosphatidylglycerosulfate; BPG, bisphosphatidylglycerol; S-TGD-1, (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]; and S-TGD-1-PA, (3′-sulfo)Galpβ1- 6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]6-[phospho- sn-2,3-di-O-phytanylglycerol], all obtained from Hbt. salinarum. S-DGD-5-PA, (2'-sulfo)Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol], was isolated from a Halorubrum sp. (MdS1 strain). The commercial glycerophospholipids (used as standards)—1,1',2,2'-tetratetradecanoyl cardiolipin; 1,1'2,2'-tetra-(9Z-octadecenoyl) cardiolipin; 1,2-ditetradecanoyl-sn-glycero-3-phosphate; 1,2-ditetradecanoyl-sn-glycero-3-phospho-(1'-rac-glycerol); and 1,2-ditetradecanoyl-sn-glycero-3-phospho-L-serine; 1,2-di-(9Z-hexadecenoyl)- sn-glycero-3-phosphoethanolamine—were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). All organic solvents used in extraction and MS analysis were commercially distilled and of the highest available purity and were purchased from Sigma Aldrich, J. T. Baker, or Carlo Erba. TLC plates (TLC Silica gel 60 A, glass plates), obtained from Merck, were washed twice with chloroform/methanol (1:1, v/v) and activated at 120°C before use.
Microorganism growth conditions
The engineered high-producing BR strain of Hbt. salinarum used in this study was kindly provided by Richard Needleman (10). The Hbt. salinarum cells were grown in light at 37°C in liquid growth medium containing neutralized peptone (L34, Oxoid), prepared as previously described (11). The MdS1 strain, a representative of Halorubrum sp., was isolated from the salterns Margherita di Savoia and grown in our laboratory as previously described (12).
Isolation and lyophilization of archaebacterial membranes
Purple membranes were isolated from the high-producing BR strains of Hbt. salinarum as previously described (11) and when indicated were used in lyophilized form. The red membranes of the extremely halophilic microorganism MdS1 were isolated as previously described (13) and lyophilized before lipid analysis.
Lipid extraction of the purple membrane
Total lipids of the PM were extracted using the Bligh and Dyer method (14), as modified for extreme halophiles (15). The extracts were carefully dried under N2 before weighing, and then dissolved in chloroform (10 mg/ml).
Thin-layer chromatography
Total lipid extracts were analyzed by TLC (Merck 10 × 20 cm × 0.25 mm thick layer) with Solvent A (chloroform/methanol/90% acetic acid, 65:4:35, v/v). Lipid detection was carried out by spraying with 5% sulfuric acid in water, followed by charring at 120°C for 45 min.
Isolation and purification of individual lipids from the total extract
The lipid components of PM were separated by preparative TLC (Merck 20 × 20 cm × 0.5 mm thick layer) in Solvent A. Lipids were visualized by staining with iodine vapor and were eluted and recovered from the scraped silica as previously described (15). Isolated and purified phospholipids were dissolved in chloroform at the concentration of 1 mg/ml; glycolipids and cardiolipin analogs in the mass range 1000–2000 amu were resuspended at the final concentration of 2 mg/ml.
Preparation of lipid samples in solution for MALDI-TOF/MS
Samples were prepared as previously described (2). Briefly, the total PM lipid extract (10 mg/ml) and individual lipid components (1–2 mg/ml) were diluted from 20 to 200 µl with isopropanol/acetonitrile (60/40, v/v). Next, 10 µl of diluted sample was mixed with 10 µl of 9-aminoacridine (10 mg/ml; dissolved in isopropanol/acetonitrile (60/40, v/v)). Then 0.25 µl of the mixture was spotted on the MALDI target (Micro Scout Plate, MSP 96 ground steel target).
Sample preparation of lyophilized archaebacterial membranes for MALDI-TOF/MS
The procedure for sample preparation is analogous to the preparation of KBr samples for infrared analysis (16, 17). Lyophilized archaebacterial membranes (PM and red membrane of MdS1) and dry 9-AA matrix were mixed in 2:1 ratio (w/w). Lyophilized membranes were grinded with 9-AA in an agate mortar, and the powder mixture was crushed in a mechanical die press to form a thin pellet disc (less than 1 mm thickness). Small pieces of the disc were then fixed to the MALDI target with double-sided adhesive tape and directly analyzed.
MALDI-TOF/MS
MALDI-TOF mass spectra were acquired on a Bruker Microflex mass spectrometer (Bruker Daltonics, Bremen, Germany). The system utilizes a pulsed nitrogen laser, emitting at 337 nm; the extraction voltage was 20 kV. 600 single laser shots were averaged for each mass spectrum. In the analysis of lipids in solution, the spectrum was obtained by moving the laser within the spot. The laser fluence was kept at about 80% of maximum value to have a good signal-to-noise ratio. At variance in the analysis of lipids in intact membranes, it was not necessary to move the laser during the acquisition of the spectrum, thanks to the thickness of the pellet (see below). It was necessary to keep the laser fluence at 100% for good desorption and ionization of the sample in the pellet; at lower laser intensity, it was not possible to have good signals. All spectra were acquired in negative ion mode using the delayed pulsed extraction. Spectral mass resolutions and signal-to-noise ratios were determined by the software for the instrument, “Flex Analysis 3.0” (Bruker Daltonics). When indicated, spectra were acquired with a Bruker Autoflex mass spectrometer (Bruker Daltonics) in reflector mode; the laser fluence was kept about 10% above threshold to optimize the signal-to-noise ratio.
A mix containing: 1,1',2,2'-tetratetradecanoyl cardiolipin, 1,1',2,2'-tetra-(9Z-octadecenoyl) cardiolipin, 1,2-ditetradecanoyl-sn-glycero-3-phosphate, 1,2-ditetradecanoyl-sn-glycero-3-phospho- (1'-rac-glycerol), 1,2-ditetradecanoyl-sn-glycero-3-phospho-L- serine, 1,2-di-(9Z-hexadecenoyl)-sn-glycero-3-phosphoethanolamine and the archaebacterial glycerolipid (2'-sulfo)Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol] (S-DGD-5-PA) was always spotted next to the sample as external standard. An external calibration was performed before each measurement.
RESULTS
Application of MALDI-TOF/MS to the analysis of archaebacterial lipids in solution
Phospholipids (diphytanylglycerolphosphate ether analogs).
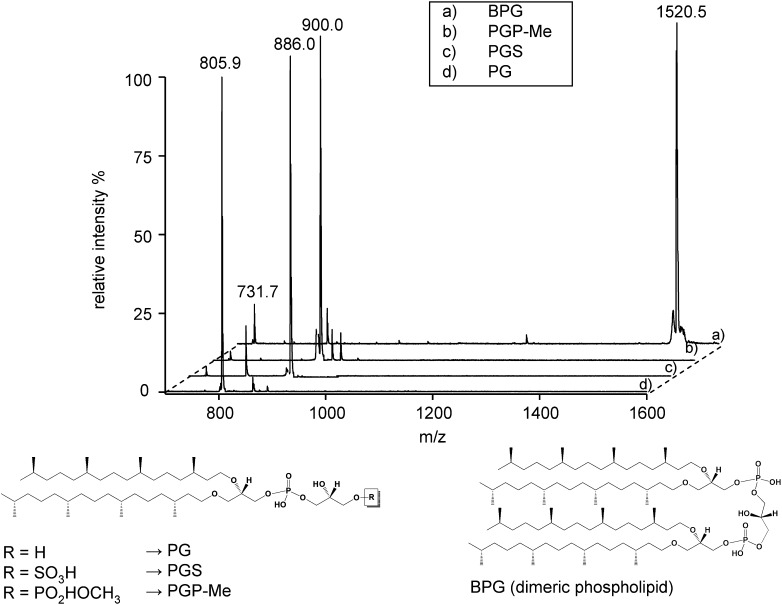
The MALDI-TOF mass spectra of some archaebacterial phospholipids present in the membranes of the extremely halophilic archaeal microorganisms are reported in Fig. 1. In particular the following lipids were analyzed: PG, PGP-Me, PGS, and BPG. The chemical structures of analyzed phospholipids are illustrated at the bottom of the Fig. 1. All these archaebacterial phospholipid standards have been isolated and purified from the lipid extract of Hbt. salinarum. PGP-Me is the major phospholipid in all extreme halophiles and extreme haloalkalophiles as well (4). BPG is the archaeal analog of eukaryotic or bacterial cardiolipin, an interesting kind of dimeric phospholipid having four phytanyl chains in the hydrophobic tail.
Fig. 1.
MALDI-TOF MS analysis of phospholipids standards (diphytanylglycerolphosphate ether analogs) isolated and purified from extremely halophilic archaeal microorganisms. Shown are the peaks of the molecular ions [M-H]−: BPG at m/z 1520.5 (A); PGP-Me at m/z 900.0 (B); PGS at m/z 886.0 (C); and PG at m/z 805.9 (D). In addition to the peaks of the molecular ions, some fragments that were produced during the desorption/ionization process are shown: the fragment at m/z 731.7 corresponding to PA is present in all spectra, while the fragment at m/z 805.9 in the PGS spectrum (C) corresponds to PG. Under the spectra, chemical structures of the archaebacterial phospholipids are reported.
The basic unit of all archaebacterial phospholipids is represented by the archaetidyl group or phosphatidic acid, which is generally detected only in traces in the cellular extracts as biosynthetic intermediates or is produced by molecular fragmentation in mass spectra. Table 1 reports formulas, exact masses, and MALDI-TOF/MS signals of archaeal phospholipids.
TABLE 1.
Detected MALDI-TOF/MS ion peaks of archaeal lipids of purple membrane
| Monoisotopic Mass [M-H] |
|||
|---|---|---|---|
| Lipid Species | Molecular Formula | Theoretical | Experimental |
| BPG | C89H182O13P2 | 1520.3 | 1520.5 |
| PG | C46H95O8P | 805.7 | 805.9 |
| PGS | C46H95O11PS | 885.6 | 886.0 |
| PGP-Me | C47H98O11P2 | 899.7 | 900.0 |
| S-TGD-1 | C61H118O21S | 1217.8 | 1218.1 |
| S-TGD-1-PA (GlyC) | C104H205O26PS | 1932.4 | 1932.8 |
BPG, bisphosphatidylglycerol; GlyC, glycocardiolipin; PG, phosphatidylglycerol; PGP-Me, phosphatidylglycerophosphate methyl ester; PGS, phosphatidylglycerosulfate; PM, purple membrane; S-TGD-1, (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]; S-TGD-1-PA, (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]-6-[phospho-sn-2,3-di-O-phytanylglycerol].
In some spectra, fragmentation ions are present together with the molecular ion; fragments produced in the desorption/ionization process are generally similar to those produced in the course of ESI-MS analysis. In the BPG spectrum (Fig. 1A) the fragmentation ion at m/z 731.7 represents the archaetidyl group of archaebacterial phospholipids, i.e., phosphatidic acid. In the case of PGS (Fig. 1C), MALDI-TOF/MS analysis reveals the presence of a main fragment ion at m/z 805.9, produced after the neutral loss of SO3 group and corresponding to PG.
Glycolipids (diphytanylglycerol ether analogs).
In the genus Halobacterium, the major glycolipid is the sulfated triglycosyl archaeol (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol] (S-TGD-1) (7); in addition the sulfated tetraglycosyl archaeol (3′-sulfo)Galpβ1-6Manpα3- 1Galfα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol] (S-TeGD) has also been found (18). S-TGD-1 is the main glycolipid of the purple membrane, while S-TeGD is present in minor amount very likely as a contaminant.
Furthermore a glycosylated cardiolipin is present in the purple membrane of Hbt. salinarum (15). Glycosyl derivatives of cardiolipin have also been described in bacteria (8, 19).
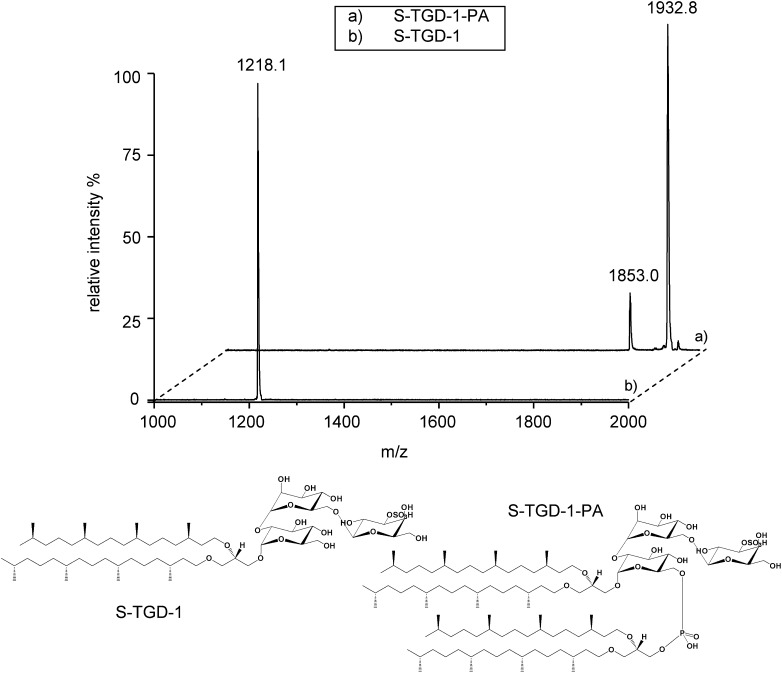
Fig. 2 shows the MALDI-TOF mass spectra of isolated and purified S-TGD-1 and S-TGD-1-PA and, at the bottom, their chemical structures.
Fig. 2.
MALDI-TOF/MS analysis of glycolipids standards (diphytanylglycerol ether analogs) isolated and purified from extremely halophilic archaeal microorganisms. The peaks of the molecular ions [M-H]− of S-TGD-1 (B) and S-TGD-1-PA (A) are at m/z 1218.1 and m/z 1932.8, respectively. Present in the spectrum of S-TGD-1-PA (A) is a peak at m/z 1853.0, corresponding to the fragment [M-HSO3]− produced during the desorption/ionization process. Under the spectra, the previously determined (7, 15) chemical structures of the archaebacterial glycolipids are shown.
The glycosylated cardiolipin of PM has the structure of a complex phosphosulfoglycolipid, (3′-sulfo)Galpβ1- 6-Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]6- [phospho-sn-2,3-di-O-phytanylglycerol] (S-TGD-1-PA) that, having two diphytanylglycerol moieties, can be considered a dimeric phospholipid. The structure of S-TGD-1-PA was first determined in our laboratory by combining TLC, NMR, ESI-MS, and MS/MS analysis of the isolated and purified glycolipid and its hydrolysis products (15). It has also been shown that S-TGD-1 is the precursor of S-TGD-1-PA (20).
The lipid extract of the purple membrane of Hbt. salinarum.
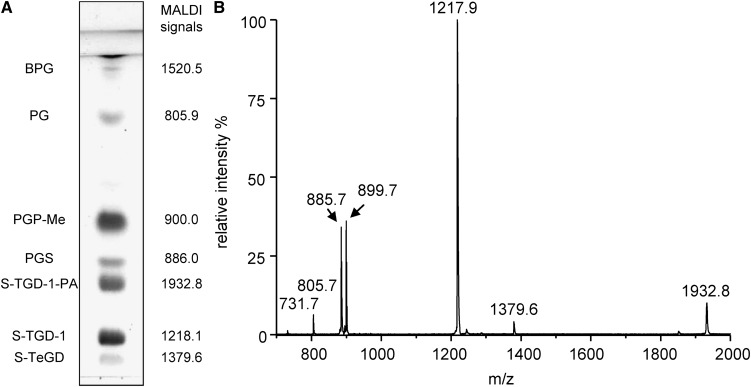
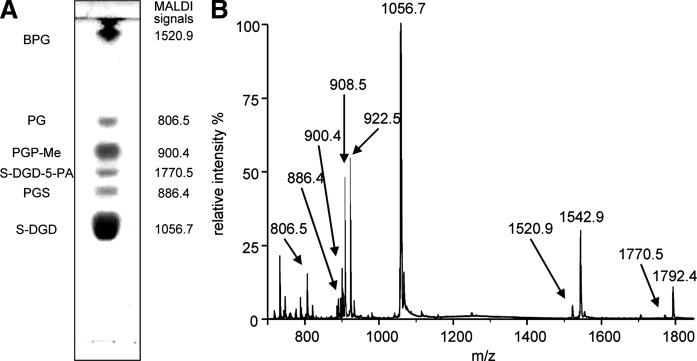
The lipid extract of the purple membrane was analyzed in parallel by TLC and MALDI-TOF/MS. The names of individual lipid components in the total lipid extract of PM are reported on the side of TLC plate in Fig. 3A, together with the monoisotopic molecular masses obtained by MALDI-TOF/MS analysis. In Fig. 3B, the MALDI-TOF/MS profile of the total lipid extract of purple membrane is reported. The signals in the mass spectrum have been assigned to the various lipids in the extract on the basis of MALDI-TOF/MS analysis of individual archaeal lipid standards previously analyzed.
Fig. 3.
A: TLC lipid profile of the total lipid extract of purple membranes of Halobacterium salinarum. Lipid abbreviations and molecular ion masses are reported. Lipid assignments are based on previous studies (15, 20). B: MALDI-TOF mass spectrum of the total lipid extract of purple membranes of Hbt. salinarum acquired in the negative ion mode using 9-AA as the matrix.
Besides the signals of molecular ions of the main lipid species present in the total lipid extracts, signals arising from molecular fragmentation are present. The signal at m/z 1218.1 is attributable to the main glycolipid of purple membrane S-TGD-1, while the peak at m/z 1379.6 corresponds to the minor glycolipid S-TeGD containing four sugar units in the polar head. The glycosylated cardiolipin named S-TGD-1-PA is represented by the signal at 1932.8 ([M-H]−). The signal of the phospholipid PGP-Me is at m/z 900.0 ([M-H]−), of PGS at m/z 886.0, of PG at m/z 805.9; finally the small peak at m/z 731.7 corresponds to PA, a lipid fragment. At variance from ESI-MS lipid profile, in MALDI-TOF mass spectrum the peak of BPG can be barely detected by enlarging the y axis, although it gives a good signal when analyzed individually (see mass spectrum in Fig. 1).
It has been shown that BPG is only a minor lipid component of PM, likely a contaminant, while S-TGD-1-PA is present in stoichiometric ratio 1:1 with bacteriorhodopsin (9).
MALDI-TOF/MS lipid analysis of equimolar mixtures of archaebacterial phospholipids, including BPG, showed that the BPG signal is always very small compared with those of other phospholipids (data not shown).
Lipid analysis of intact lyophilized archaebacterial membranes dry mixed with 9-aminoacridine
Recently MALDI-TOF/MS analysis of insoluble organic compounds of high molecular weight has also been performed by dry mixing the analyte and matrix, and then crushing it in a mechanical die press to form a thin disc or pellet that can be directly scanned by the MALDI laser beam (16, 17). We tested the possibility of applying this approach to the direct lipid analysis of isolated membranes in lyophilized form, thus avoiding the steps of membrane disruption, lipid solubilization, and phase partition.
It is well known that the purple membrane of Hbt. salinarum has an extraordinary thermal and chemical stability; we have verified that is possible to keep PM in lyophilized form at room temperature for months without changing their lipid composition (unpublished observations).
Lyophilized purple membranes were finely mixed with the powder of 9-AA. The dry mixture was kept under high pressure in a die press to form a thin disc, similar to those obtained by mixing substrates and KBr for IR analysis. Fig. 4 shows the fragment of PM/9-AA pellet attached to the MALDI target.
Fig. 4.
Photograph of fragments of PM/9-AA pellet attached to the MALDI-TOF target. Dimensions of the target are approximately 53 × 41 mm. MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; PM, purple membranes; 9-AA, 9-aminoacridine.
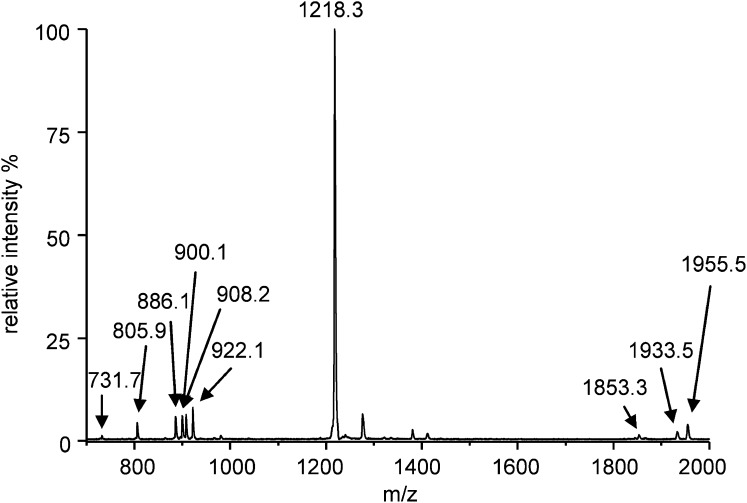
A representative MALDI-TOF mass spectrum obtained by analyzing the PM/9-AA pellet is reported in Fig. 5. The MALDI-TOF/MS lipid profile obtained in these experimental conditions corresponds quite well to the one reported in Fig. 3 that was obtained by analyzing the PM lipid extract. The glycosylated cardiolipin S-TGD-1-PA is represented by the signal at m/z 1955.5, 1933.5, and 1853.3, corresponding to the sodium adduct of the molecular ion, the molecular ion, and the desulfated glycolipid, respectively. The signals at m/z 1380.4 and 1218.3 correspond well to the glycolipid S-TeGD and S-TGD-1, respectively. PGS and PGP-Me, both phospholipids having two acidic groups in the polar head, are represented by a mixture of molecular ions and sodium adducts, whereas PG is represented only by its molecular ion. The mass deviation in the high range of lipid masses might be due to differences in the sample height in the dry mixture of the pellet.
Fig. 5.
MALDI-TOF mass spectrum of pellets obtained with lyophilized PM and 9-AA (2:1, w/w) as described in “Experimental Procedures.” The m/z signals corresponding to the different PM lipid components are PG, [M-H]− at 805.9; PGS, [M-H]− at 886.1; PGS sodium adduct, [M-H]− + Na+ at 908.2; PGP-Me, [M-H]− at 900.1; PGP-Me sodium adduct, [M-H]− + Na+ at 922.1; S-TGD-1, [M-H]− at 1218.3; S-TGD-1-PA, [M-H]− at 1933.5; S-TGD-1-PA, [M-HSO3]− at 1853.3; and S-TGD-1-PA sodium adduct, [M-H]− + Na+ at 1955.5.
The possibility of extending the novel method of MALDI-TOF/MS lipid analysis to other archaebacterial membranes in lyophilized form was also verified. Fig. 6 illustrates the MALDI-TOF/MS analysis of lyophilized red membranes isolated from the strain MdS1, another extremely halophilic microorganism of the Archaea world. MdS1 red membranes are highly enriched in BPG (12, 13).
Fig. 6.
A: TLC lipid profile of the total lipid extract of red membranes of Halorubrum sp. MdS1 strain. Lipid abbreviations and molecular ion masses are reported. Lipid assignments are based on previous studies (12, 13). B: MALDI-TOF mass spectrum of pellets obtained with lyophilized red membranes of Halorubrum sp. MdS1 strain and 9-AA (2:1, w/w) as described in “Experimental Procedures.” Most lipid components are mainly represented by their sodium adducts. The spectrum was acquired on a Bruker Autoflex mass spectrometer (Bruker Daltonics).
The lyophilized MdS1 red membranes and dry 9-AA were mixed in 2:1 ratio, obtaining a thin red pellet. A fragment of the pellet was used for MALDI-TOF/MS analysis. The resulting MALDI-TOF/MS lipid profile was perfectly superimposable onto the lipid profile obtained by previous TLC and ESI-MS analysis (12, 13). Fig. 6A reports the TLC lipid profile of red membranes and lipid assignments based on data in previous literature and mass spectrometric analysis. Experimental lipid masses obtained by MALDI-TOF/MS analysis correspond well to calculated lipid masses and to those previously obtained by ESI-MS analysis (12, 13). It can be seen that in the MALDI spectra of membrane/matrix pellets most of archaebacterial lipids are represented by their sodium adducts. The excellent quality of the MALDI-TOF/MS spectrum of lyophilized MdS1 membranes in Fig. 6 is likely due to the optimal homogeneity of the red membranes/9-AA pellet.
It is noteworthy that the BPG signal at m/z 1542.9 is comparable to that of other membrane lipid components; therefore, the novel method of lipid analysis offers the advantage of overcoming the problem of difficult ionization of BPG when it is mixed with other phospholipids in solution.
DISCUSSION
Recent applications of MALDI-TOF/MS to the analysis of cellular lipids have shown the potential of a technique long considered of practical utility only for protein analysis (2, 21).
It has been shown that MALDI mass spectrometry allows the analysis of glycerolipids, glycerophospholipids, triglycerides, cholesterol, and cholesterol derivatives, while reports describing the application of MALDI to the analysis of archaebacterial lipids are not yet available.
In this study, we illustrate for the first time the application of MALDI-TOF/MS to the analysis of archaebacterial lipids in solution, and we show that it is possible to directly analyze lipids of membranes isolated from archaeal microorganisms without prior extraction/separation steps.
We have analyzed archaeal membrane lipid standards and the total lipid extract of the archaeal membranes isolated from halophilic microorganisms, obtaining results comparable with those obtained by ESI-MS analysis.
Most of the phospholipids and glycolipids of extreme halophilic archaeons are anionic, so that their negatively charged groups would impart a high negative charge density to the halophile membranes. No nitrogenous base-containing phospholipids, such as phosphatidylserine or phosphatidylethanolamine, are present in extreme halophiles. This may be characteristic of the halophilic Archaea, in contrast to the methanogenic and thermophilic Archaea. Information on lipid biosynthesis in Archaea is available thanks to labeling studies with whole cells (5, 22).
In a previous study, we analyzed the lipids of the purple membrane by combining TLC and NMR analysis. The results obtained allowed the estimation of stoichiometric ratio of individual lipid component and the sole protein of the purple membrane bacteriorhodopsin (9).
In this study, we considered the purple membrane as a model membrane to investigate the potential of MALDI-TOF/MS in the analysis of lipids of archaeal microorganisms. Results indicate that 9-AA in solution is a suitable matrix for the desorption/ionization of archaeal phospholipids and glycolipids. The method, originally developed by Sun G. et al. (2), allows good ionization and easy detection of the dimeric archaeal phospholipids or cardiolipins. The sensitivity of the method is particularly high for the archaeal sulfated glycolipids, which in analogy with sulfatides of eukaryotic cells (23), exhibit an extremely high tendency to ionize in the presence of 9-AA.
The MALDI-TOF/MS profile of the lipid extract of the purple membranes contains the same main signals present in the ESI mass spectrum (15). All the lipids in the extract can be easily revealed by this technique of analysis, with the exception of the minor PM lipid component BPG, whose signal is very low.
Interestingly we also show that it is possible to perform a direct lipid analysis of the intact lyophilized archaebacterial membranes by MALDI-TOF/MS dry mixed with the matrix. We recommend care in the preparation of the membrane/matrix pellets, because the incomplete homogeneity of the samples might cause some inaccuracy in the measurements.
The study of lipid composition of cell membranes normally requires two separate steps: 1) extraction of lipids by harsh organic solvents and 2) combined chromatographic and mass spectrometry analysis. The novel approach to analyze lipids of intact membranes is particularly useful if doubts about the presence of artifacts introduced by the lipid extraction procedure are raised (e.g., highly polar lipids as sulfated polyglycolipids may preferentially partition into the methanol-water phase and therefore be absent in the chloroform phase). In a previous study of 31P NMR analysis of solubilized purple membrane, the phosphorus signal of the glycosylated cardiolipin S-TGD-1-PA could be not detected, raising doubts about the presence of the archaeal cardiolipins in the PM (24). In the present study, our data confirm that archaeal cardiolipins are true components of the membrane matrix of the purple membrane.
The direct lipid analysis of lyophilized membranes appears to be of general applicability for archaebacterial lipids, whose structures are chemically stable even in extreme environmental conditions. During the dehydration process, the polar heads of phospholipids lose the associated water, whereas sodium ions likely remain bound to membrane. This might explain why most of highly polar lipids of archaebacterial membranes appear as sodium adducts in the MALDI spectra of lyophilized membranes.
The method of MALDI-TOF/MS lipid analysis of lyophilized membranes is particularly suited to the analysis of the complex archaebacterial phospholipid BPG, allowing an easier detection of it. The sodium adduct of BPG represents one of the main signals in the MALDI-TOF/MS lipid profile in Fig. 6, although it appears difficult to obtain the ionization of BPG in solution in the presence of other phospholipids and glycolipids.
The novel method we describe might be of general applicability. It offers the advantage of quickly gaining information about membrane lipid components without disrupting or altering the membrane matrix. Experiments are in progress to evaluate the potential of this novel method in the study of lipid composition of bacterial and eukaryotic membranes.
Acknowledgments
The authors thank Prof. Francesco Naso for the use of MALDI-TOF/MS instrument Bruker Microflex in the Chemistry Department of the University of Bari, Prof. Loreto Gesualdo for the use of MALDI-TOF/MS instrument Bruker Autoflex in the Proteomic Facility of the University of Foggia, and Dr. Patrizia Lopalco for preparing the archaebacterial lipid standards and membranes.
Footnotes
Abbreviations:
- BPG
- bisphosphatidylglycerol (diphytanylglycerol ether analog)
- Hbt. salinarum
- Halobacterium salinarum
- MALDI-TOF/MS
- matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- NMR
- nuclear magnetic resonance
- PG
- phosphatidylglycerol (diphytanylglycerol ether analog)
- PGP-Me
- phosphatidylglycerophosphate methyl ester (diphytanylglycerol ether analog)
- PGS
- phosphatidylglycerosulfate (diphytanylglycerol ether analog)
- PM
- purple membrane
- S-DGD-5-PA
- (2'-sulfo)Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]
- S-TeGD
- (3′-sulfo)Galpβ1-6Manpα3-1Galfα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]
- S-TGD-1
- (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]
- S-TGD-1-PA
- (3′-sulfo)Galpβ1-6Manpα1-2Glcpα1-1-[sn-2,3-di-O-phytanylglycerol]-6-[phospho-sn-2,3-di-O-phytanylglycerol]
- 9-AA
- 9-aminoacridine
This work was supported by Ministero Italiano dell'Università e della Ricerca (Fondi Ateneo), Ministero Italiano della Difesa (Contract No. 1999 and Regione Puglia (Grant code 15, Sens&MicroLab).
REFERENCES
- 1.Han X., Gross R. W. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- 2.Sun G., Yang K., Zhao Z., Guan S., Han X., Gross R. W. 2008. Matrix assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80: 7576–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Jr. Merrill A. H., Murphy R. C., Raetz C. R. H., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 4.Kates M. 1993. The Biochemistry of Archaea (Archaebacteria). Kates M., Kushner D. J., Matheson A. T., Elsevier Science Publishers, Amsterdam. [Google Scholar]
- 5.Kamekura M., Kates M. 1988. Lipids of Halophilic Archaebacteria. Halophilic bacteria. Rodriguez Valera F., editor CRC press, Boca Raton, Florida, FL: Vol. II, pp. 25–54. [Google Scholar]
- 6.Corcelli A. 2009. The cardiolipin analogues of Archaea. Biochim. Biophys. Acta. 1788: 2101–2106. [DOI] [PubMed] [Google Scholar]
- 7.Kates M., Deroo P. W. 1973. Structure determination of the glycolipid sulfate from the extreme halophile Halobacterium cutirubrum. J. Lipid Res. 14: 438–445. [PubMed] [Google Scholar]
- 8.Sprott G. D., Larocque S., Cadotte N., Dicaire C. J., McGee M., Brisson J. I. 2003. Novel polar lipids of halophilic eubacterium Planococcus H8 and archaeon Haloferax volcanii. Biochim. Biophys. Acta. 1663: 179–188. [DOI] [PubMed] [Google Scholar]
- 9.Corcelli A., Lattanzio V. M. T., Papadia P., Fanizzi F. P. 2002. Lipid/protein stoichiometries in a crystalline biological membrane: NMR quantitative analysis of the lipid extract of the purple membrane. J. Lipid Res. 43: 132–140. [PubMed] [Google Scholar]
- 10.Ni B. F., Chang M., Duschl A., Lanyi J. K., Needlemann R. 1990. An efficient system for the synthesis of bacteriorhodopsin in Halobacterium halobium. Gene. 90: 169–172. [DOI] [PubMed] [Google Scholar]
- 11.Oesterhelt D., Stoeckenius W. 1974. Isolation of cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31: 667–678. [DOI] [PubMed] [Google Scholar]
- 12.Lopalco P., Lobasso S., Babudri F., Corcelli A. 2004. Osmotic shock stimulates de novo synthesis of two cardiolipins in an extreme halophilic archaeon. J. Lipid Res. 45: 194–201. [DOI] [PubMed] [Google Scholar]
- 13.Corcelli A., Lobasso S., Palese L. L., Sublimi Saponetti M., Papa S. 2007. Cardiolipin is associated with the terminal oxidase of an extremely halophilic archaeon. Biochem. Biophys. Res. Commun. 354: 795–801. [DOI] [PubMed] [Google Scholar]
- 14.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 15.Corcelli A., Colella M., Mascolo G., Fanizzi F. P., Kates M. 2000. A novel glycolipid and phospholipid in the purple membrane. Biochemistry. 39: 3318–3326. [DOI] [PubMed] [Google Scholar]
- 16.Skelton R., Dubois F., Zenobi R. 2000. A MALDI sample preparation method suitable for insoluble polymers. Anal. Chem. 72: 1707–1710. [DOI] [PubMed] [Google Scholar]
- 17.Babudri F., Cardone A., Chiavarone L., Ciccarella G., Farinola G. M., Naso F., Scamarcio G. 2001. Synthesis and characterization of poly(2,3,5,6-tetrafluoro-1,4-phenylenevinylene). Chem. Commun. (Camb.). 1940–1941. [DOI] [PubMed] [Google Scholar]
- 18.Smallbone B. W., Kates M. 1981. Structural identification of minor glycolipids in Halobacterium cutirubrum. Biochim. Biophys. Acta. 665: 551–558. [Google Scholar]
- 19.Fischer W. 1976. Newly discovered lipids from Streptococci. Lipids. Paoletti R., Porcellati G., Jacini G., Raven, New York: 255–266. [Google Scholar]
- 20.Lobasso S., Lopalco P., Lattanzio V. M., Corcelli A. 2003. Osmotic shock induces the presence of glycocardiolipin in the purple membrane of Halobacterium salinarum. J. Lipid Res. 44: 2120–2126. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs B., Schiller J. 2009. Application of MALDI-TOF mass spectrometry in lipidomics. Eur. J. Lipid Sci. Technol. 111: 83–98. [Google Scholar]
- 22.Koga Y., Morii H. 2005. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69: 2019–2034. [DOI] [PubMed] [Google Scholar]
- 23.Cheng H., Sun G., Yang K., Gross R. W., Han X. 2010. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J. Lipid Res. 51: 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renner C., Kessler B., Oesterhelt D. 2005. Lipid composition of integral purple membrane by 1H and 31P NMR. J. Lipid Res. 46: 1755–1764. [DOI] [PubMed] [Google Scholar]