Abstract
The protein FinO represses F-plasmid conjugative transfer by facilitating interactions between the mRNA of the major F-plasmid transcriptional activator, TraJ, and an antisense RNA, FinP. FinO is known to bind stem–loop structures in both FinP and traJ RNAs; however, the mechanism by which FinO facilitates sense–antisense pairing is poorly understood. Here we show that FinO acts as an RNA chaperone to promote strand exchange and duplexing between minimal RNA targets derived from FinP. This strongly suggests that FinO may function to destabilize internal secondary structures within FinP and traJ RNAs that would otherwise act as a kinetic trap to sense–antisense pairing. The energy for FinO-catalyzed base-pair destabilization does not arise from ATP hydrolysis but appears to be supplied directly from FinO RNA binding free energy. An analysis of the activities of mutants that are specifically deficient in strand exchange but not RNA-binding activity demonstrates that strand exchange is essential to the ability of FinO to mediate sense–antisense RNA recognition, and that this function also plays a role in repression of conjugation in vivo.
Keywords: antisense RNA/bacterial conjugation/RNA chaperone/strand exchange
Introduction
The F family of plasmids confer antibiotic resistance and virulence to a wide variety of enterobacteria. The transfer of F-like plasmids between bacterial species has been linked to the rapid acquisition of antibiotic resistance in strains of Escherichia coli that caused wide-spread outbreaks of antibiotic-resistant dysentery in post-World War II Japan (Watanabe and Fukasawa, 1961). These resistance or R factors were found to inhibit F plasmid transfer, a process termed fertility inhibition (fi+ or fin), found among most members of the IncF plasmid complex (Watanabe and Fukasawa, 1961). Many fin+ plasmids in the Enterobacteriaceae are related to F and R factors and have been associated with the acquisition of virulence operons and pathogenicity such as the Salmonella type I strains (Boyd and Hartl, 1997, 1998).
The RNA binding protein FinO, along with the 79-nt antisense RNA FinP, make up a two-component inhibition system for F-plasmid-mediated bacterial conjugation (Finnegan and Willetts, 1972). FinP is complementary to the 5′ untranslated region of traJ mRNA (Figure 1A) and is believed to block ribosomal entry when associated with this mRNA (Mullineaux and Willetts, 1985; van Biesen and Frost, 1994; Koraimann et al., 1996). TraJ is a transcriptional activator that is required for expression of the majority of conjugative protein components (Willetts, 1977; Cuozzo and Silverman, 1986). In the absence of FinO, FinP is rapidly degraded by RNases within bacterial cells, allowing TraJ to be readily synthesized (Lee et al., 1992). FinO binds FinP and traJ mRNA, stabilizing FinP against degradation (Lee et al., 1992; Jerome et al., 1999), and promoting extended duplex formation between the complementary RNA molecules (van Biesen and Frost, 1994). This, in turn, blocks TraJ translation and inhibits bacterial conjugation.
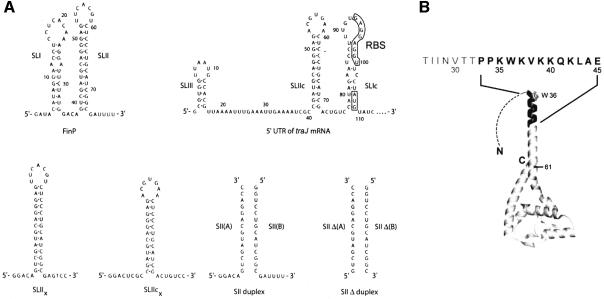
Fig. 1. The structures of FinO and its RNA targets. (A) The sequence and secondary structure of RNA molecules used in this study. The 5′ untranslated region of traJ mRNA (which duplexes with FinP) is shown with the start codon (AUG) and ribosomal binding site (RBS) boxed. The A and B strands of SII and SIIΔ RNA duplexes are aligned to show the regions of base-pair complementarity. (B) Ribbons representation of FinO. The structure of FinO(26–186) (Ghetu et al., 2000) is shown, with a dotted line representing the unstructured N-terminal 25 residues. The Trp36 side-chain is displayed and the N-terminal residues 33–46 are highlighted in black. The amino acid sequence of the N-terminal region of FinO that is critical for strand exchange activity with the RNA duplex is shown above the structure. Finally, position 61 is shown on the structure in reference to the C-terminal truncation fragment FinO(1–61).
Recent biochemical and crystallographic studies have begun to reveal how FinO interacts with its target RNAs. FinO recognizes RNA duplex stems that have 5′ and 3′ single-stranded tails at one end (Jerome and Frost, 1999). As this recognition is dependent on the structure but not the sequence of the RNA, FinO can bind several such tailed stem–loop structures in both FinP and traJ RNAs (Jerome and Frost, 1999). FinO adopts an elongated structure with a solvent exposed N-terminal helix extended from a C-terminal domain (Figure 1B) (Ghetu et al., 2000). Biochemical studies have demonstrated that FinO binds stem–loop structures as a monomer and that the positively charged N-terminal helix, as well as the globular body of the protein, directly contact RNA (Ghetu et al., 1999, 2002).
Pairing of FinP and traJ RNAs is believed to initiate with the formation of ‘kissing complexes’ between complementary loops in the two RNAs (Finlay et al., 1986; Koraimann et al., 1991; Gubbins et al., 2003), and it has been suggested that FinO stabilizes kissing complexes to facilitate sense–antisense RNA interactions (Ghetu et al., 2000). The 25 N-terminal amino acids of FinO enhance the rate of FinO mediated FinP-traJ duplexing 10-fold, but do not play a significant role in the binding of individual RNA targets (Ghetu et al., 1999, 2000). Based on these observations, it has been suggested that, upon binding stem–loop structures, the N-terminal region of FinO is positioned near the RNA loop to stabilize loop–loop pairing. A similar function has been demonstrated for Rom, which binds and stabilizes sense–antisense RNA kissing complexes to ultimately inhibit replication of the ColE1 plasmid (Eguchi and Tomizawa, 1991; Predki et al., 1995). Interestingly, both FinP and traJ mRNA contain stable stem–loop structures that would be expected to present a kinetic barrier to duplex formation (Figure 1A). It has been suggested previously that FinO might act to destabilize intramolecular stem–loop structures to allow the formation of sense–antisense interactions (Ghetu et al., 1999).
RNA chaperones are a class of RNA binding proteins that, unlike helicases, have the ability to remodel structured RNAs in an ATP-independent fashion. Chaperone functions include the resolution of kinetically trapped, or misfolded secondary or tertiary RNA structures, as well as RNA annealing, helix destabilization and strand exchange activities (Herschlag, 1995; Weeks, 1997; Cristofari and Darlix, 2002). In this study, we show that FinO acts as an RNA chaperone to promote strand exchange between minimal RNA targets from FinP. This activity strongly suggests that FinO destabilizes intramolecular base pairing in its bound substrate. The strand exchange does not require ATP hydrolysis or preferential binding and stabilization of single-stranded RNA. Instead, we suggest that FinO may use its own RNA binding free energy to destabilize a limited number of intramolecular base pairs in the stem–loop. We show that FinO mediates intermolecular pairing between FinP- and traJ-derived RNAs, and we suggest that destabilization of stem–loop base pairing facilitates duplexing between the sense and antisense target RNAs. Finally, we demonstrate that these activities are involved in FinO-mediated repression of conjugative plasmid transfer in vivo.
Results
FinO promotes RNA strand exchange
Several studies have revealed that FinO can facilitate FinP–traJ RNA interactions in vitro (van Biesen and Frost, 1994; Sandercock and Frost, 1998; Ghetu et al., 2000). However, both FinP and traJ RNAs contain large intramolecular duplex regions that are predicted to be very stable (Figure 1A). For example, the free energy of unfolding at 37°C for the stem–loop I (SLI) and stem–loop II (SLII) structures in FinP are predicted to be –10 and –28 kcal/mol, respectively (Mathews et al., 1999). We therefore wondered whether FinO might overcome kinetic barriers to sense–antisense RNA interactions through the specific destabilization of intramolecular secondary structures within the target RNAs. To test this idea, we used an RNA strand exchange assay based on previous methods used to characterize the RNA unwinding properties of ATP-dependent helicases (Wagner et al., 1998; Wang et al., 1998) (Figure 2A). In these assays, we used an RNA duplex (SII) that mimics the structure of FinP SLII (Figure 1A) and is a high affinity binding substrate for FinO (Table I; for preparation of protein and RNA substrates see Supplementary data, available at The EMBO Journal Online). One strand of the duplex, SII(A), was labeled with 32P, and strand dissociation in the presence or absence FinO was monitored by gel electrophoresis. In this assay, SDS is added to the sample loading buffer to specifically denature protein and release the RNA species prior to electrophoresis. To prevent the 32P-labeled strand from re-associating with the complementary strand after release from the duplex, a large molar excess of an unlabeled version of SII(A) was added at the initiation of the reaction. Control experiments demonstrated that the single-stranded SII(A), unlike SII, does not form a complex with FinO that is detectable by electrophoretic mobility shift assay (EMSA) (data not shown).
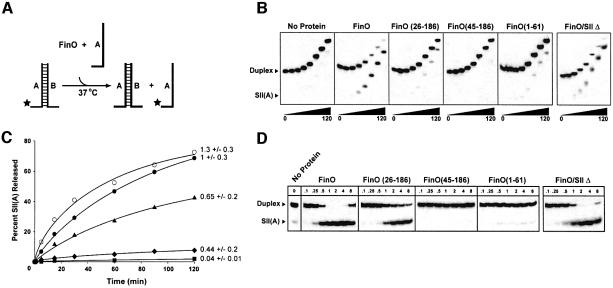
Fig. 2. FinO can catalyze strand exchange in duplex RNA substrates. (A) Schematic diagram of the RNA strand exchange assay. The SII RNA duplex is 5′-labeled with 32P on the A strand (star) and is incubated with FinO and a molar excess of the unlabeled SII(A) strand at 37°C. Release of 32P-labeled SII(A) strand is monitored by native gel electrophoresis over a 2 h time-course. (B) Comparison of strand exchange efficiencies between FinO and various N- and C-terminal truncated fragments. SII RNA was incubated with either FinO, FinO(26–186), FinO(45–186) or FinO(1–61) (each at a final concentration of 1 µM), or no protein. SIIΔ RNA was incubated with FinO at a final concentration of 1 µM. Aliquots were taken at 0, 1, 5, 15, 30, 60, 90 and 120 min after the start of the reaction and loaded directly onto a continuously running gel. (C) The percentage of 32P-labeled SII(A) strand released from the duplex was plotted as a function of time and the apparent first order rate constant, k1, was determined from this plot for FinO (filled circles), FinO(26–186) (filled triangles), no protein (filled squares) and FinO(1–61) (filled diamonds) (see Materials and methods). The percentage of 32P-labeled SIIΔ(A) strand released from the SIIΔ duplex was similarly plotted as a function of time for FinO (open circles). The relative rates of strand exchange as a fraction of FinO, and their standard deviations (derived from at least three independent rate determinations) are shown. (D) Strand exchange of SII RNA after a 120-min reaction was measured as a function of protein concentration for FinO and the indicated mutants. Also shown is strand exchange of SIIΔ RNA after a 120-min incubation with FinO. The protein concentrations (in µM) are indicated at the top of each lane.
Table I. Effect of FinO mutations on rates of strand-exchange, duplexing, and conjugative inhibition.
| Protein | Relative affinitiesa,b (Ka) RNA substrate SLII | Relative rate of strand-exchangea,b (k1) SII | SIIΔ | Relative rate of duplexinga,b (k2) SLII/SLIIc | FinP–traJ mRNA | Relative mating efficiencyc |
|---|---|---|---|---|---|---|
| None | – | 0.04 (±0.01) | – | <0.01 | 0.02 (±0.008) | 1 |
| FinO3 | 1 (±0.1) | 1 (±0.3) | 1.3 (±0.3) | 1 (±0.4) | 1 (±0.4) | 0.05 (±0.03) |
| 26–186 | 4 (±0.3) | 0.65(±0.2) | – | 0.11 (±0.07) | 0.12 (±0.08) | 0.87 (±0.20) |
| 45–186 | 20 (±2.4) | <0.04 | – | <0.01 | 0.02 (±0.001) | 1.0 (±0.17) |
| 1–61 | 0.004 (±0.0006) | 0.44 (±0.2) | – | 0.01(±0.0008) | – | – |
| W36A | 5 (±0.9) | – | – | – | 0.48 (±0.2) | 0.36 (±0.1) |
| K37A/V38A | – | – | – | – | 0.6 (±0.3) | 0.06 (±0.01) |
| K39A/K40A | 1.37 (±0.2) | – | – | – | 0.4 (± 0.2) | 0.02 (0.003) |
aAll rates are as a percentage of FinO.
bFor FinO; Ka = 5 (±1) × 107/M; k1 = 1.1 (±0.3) × 10–2/s; k2 (SIIx and SIIcx) = 1.4 (±0.2) × 105/M/s; k2 (FinP–traJ mRNA) = 2.5 (±1) × 107/M/s.
cEfficiencies are as a percentage of mating in the absence of protein.
The results of these experiments show that FinO is capable of catalyzing strand exchange of the SII RNA substrate in a time-dependent manner (Figure 2B and C). The reactions were carried out at 37°C, well below the Tm of the SII RNA substrate (>50°C; data not shown), suggesting that FinO destabilizes base pairs within SII as a first step in the strand exchange reaction. Consistent with this suggestion, factors that stabilize double-stranded RNA, such as reductions in temperature or the addition of Mg2+, reduced FinO-catalyzed strand exchange rates (data not shown).
While it seems clear that FinO-catalyzed base-pair destabilization within SII must occur to allow strand exchange, it is also possible that the rate of binding of the single-stranded SII(A) RNA to its complementary strand is also involved in determining the overall rate of strand exchange. Consistent with this idea, strand exchange efficiency is moderately enhanced with increasing concentrations of SII(A), suggesting that it might be involved in the rate limiting step of the reaction (data not shown). It is unlikely that SII(A) is directly recognized by FinO, because we have not been able to observe a stable FinO–SII(A) complex in gel mobility shift experiments (data not shown).
Strand exchange activity is also highly dependent on FinO concentration (Figure 2D). The dramatic increase in strand exchange activity between 0.25 and 1 µM FinO concentrations suggests that multiple FinO molecules might cooperate in strand exchange. However, as the concentration of FinO is further increased beyond 2 µM, strand exchange activity gradually decreases. The reduction of strand exchange activity at the higher concentrations may be due to additional, non-specific interactions between FinO and RNA. Indeed, gel EMSAs indicate that FinO can form large, non-specific aggregates on RNA at similar concentrations (data not shown).
Since it has been shown previously that the single-stranded spacer and tail regions are important for specific binding of FinO to SLII (Jerome and Frost, 1999), we wanted to test whether the same portions of the SII duplex were involved in strand exchange. A duplex was designed that lacked the 5′ and 3′ single-stranded tail regions (SIIΔ; Figure 1A). It had been shown previously that removing the spacer and tail from the SLII hairpin reduced specific binding of GST–FinO by 25-fold (Jerome and Frost, 1999). In this study, EMSAs performed under different conditions [4°C in this study, compared with room temperature in Jerome and Frost (1999)], showed that FinO did not bind stably to the SIIΔ duplex, producing smeary shifts and aggregates at protein concentrations >0.7 µM (data not shown). In spite of the significantly lower binding affinity, FinO was nevertheless able to catalyze strand exchange between SIIΔ and a complementary single strand with a rate roughly equivalent to the rate observed with SII (Figure 2B and C). This indicates that recognition of the double-stranded RNA substrate by FinO is not rate limiting in strand exchange. In addition, this also suggests that the exchange reaction is not initiated by interactions between one of the single-stranded tails and its complementary region on the unlabeled SII(A).
RNA strand exchange activity is associated with the N-terminal region of FinO
We have shown previously that the N-terminal 25 residues of FinO are dispensable for specific interactions with a single RNA target but nevertheless play an important role in its ability to promote duplex formation between FinP and traJ mRNA (Ghetu et al., 2000). To test the role of N-terminal regions of FinO in RNA strand exchange, we compared the exchange activities of full-length FinO, the N-terminal deletion mutants FinO(26–186) and FinO(45–186), and the C-terminal deletion mutant FinO(1–61) (Figure 2B and C). A schematic diagram illustrating the positions of the deletions within the FinO(26–186) structure is shown in Figure 1B. Data from strand exchange time-courses were used to calculate apparent first order strand exchange rate constants for each of the proteins (Figure 2C; Table I; Supplementary data). FinO(26–186) is able to catalyze strand exchange of SII at a modestly decreased level compared with the rate observed for full-length FinO at 1 µM protein concentration (Figure 2B and C). However, at higher protein concentrations (>2 µM), there appears to be little difference in the strand exchange activities of the two proteins (Figure 2D), indicating that the N-terminal 25 residues of FinO plays a relatively minor role in strand exchange and duplex destabilization. However, unlike wild-type FinO, FinO(26–186) strand exchange is not inhibited at higher protein concentrations (Figure 2D). This indicates that the first 25 amino acids may be important in forming non-specific FinO–RNA aggregates at higher protein concentrations. In stark contrast to FinO(26–186), FinO(45–186) appears to be completely deficient in RNA strand exchange activity at all concentrations tested, and may even stabilize the duplex form of SII. These results show that the N-terminus of FinO is essential for RNA strand exchange activity, and suggest that the region between residues 26 and 44 is particularly critical.
Consistent with the importance of the N-terminal region for strand exchange, FinO(1–61) showed a small but reproducible strand exchange activity (Figure 2B and C), in spite of the fact that the fragment binds SII with very low affinity (Table I).
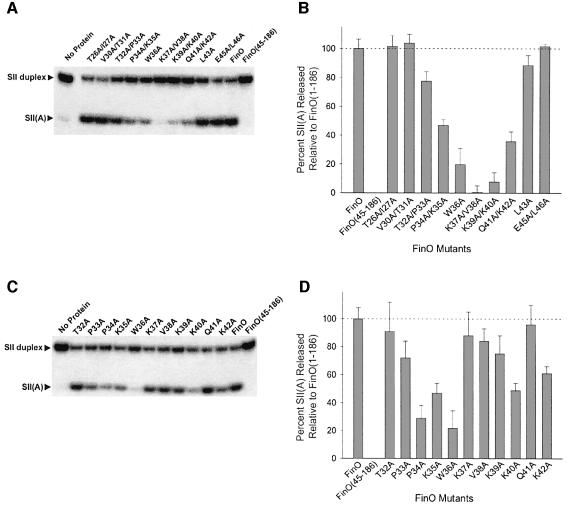
While residues 1–32 are apparently unstructured in the free protein, residues 33–44 constitute the end of a solvent exposed helix that directly contacts RNA (Figure 1B) (Ghetu et al., 2002). To test the role of this helix in more detail, we constructed a series of FinO mutant proteins with double amino acid to alanine substitutions throughout the region from residue 26 to 44, and measured the abilities of these mutants to promote strand exchange after a 2 h incubation at 37°C. The results of these experiments (Figure 3A and B) show that the extreme N-terminus of the helix is critical for RNA strand exchange. Strand exchange was most dramatically reduced for the K37A/V38A and K39A/K40A mutants. The mutants T32A/P33A, P34A/K35A and Q41A/K42A also had decreased strand exchange activity compared with wild-type FinO. To identify the individual residues most critical for strand exchange, we constructed a second set of single amino acid to alanine substitution mutants and tested their strand exchange activities (Figure 3C and D). The single site substitution mutants that showed the most significant decrease in RNA strand exchange activity were P34A, K35A, W36A and K40A, with W36A showing the most dramatic effect. A couple of single point mutants, P37A and V38A, did not have as severe an effect on strand exchange individually compared with when they were in a double mutation.
Fig. 3. Sequences near the N-terminus of FinO α1 are critical for RNA strand exchange Samples were incubated for 120 min with the indicated double (A and B) or single (C and D) alanine point mutants and strand exchange was detected by the release of 32P-labeled SII(A) strand from the SII RNA duplex. The percentage of 32P-labeled SII(A) strand released from duplex is presented in graphical form for the double (B) and single (D) alanine point mutants (standard deviations represent results from at least three independent experiments). As controls, strand exchange of SII RNA in the presence of FinO, FinO(45–186) or no protein was performed in parallel.
Residues 1–44 of FinO function to promote FinP–traJ RNA duplex formation
To test our proposal that FinO-catalyzed RNA destabilization of intramolecular stem–loop structure reduces kinetic barriers to sense–antisense duplex formation, we probed the ability of strand exchange deficient FinO mutants to facilitate sense–antisense RNA recognition. Previously, studies employing an in vitro RNA duplexing assay to measure the rate of binding of 32P-labeled FinP to its complementary sequence within traJ mRNA showed that wild-type FinO enhances the rate of FinP–traJ RNA duplexing 50-fold compared with the no protein control (Table I) (van Biesen and Frost, 1994; Sandercock and Frost, 1998; Ghetu et al., 2000). FinO(26–186) was able to facilitate RNA duplexing, albeit at a 10-fold reduced rate compared with wild type FinO (Ghetu et al., 2000). In this study, we tested the ability of FinO(45–186) to mediate sense–antisense interactions between FinP and traJ mRNA. Reactions performed in the presence of FinO(45–186) were indistinguishable from the no protein control (Table I).
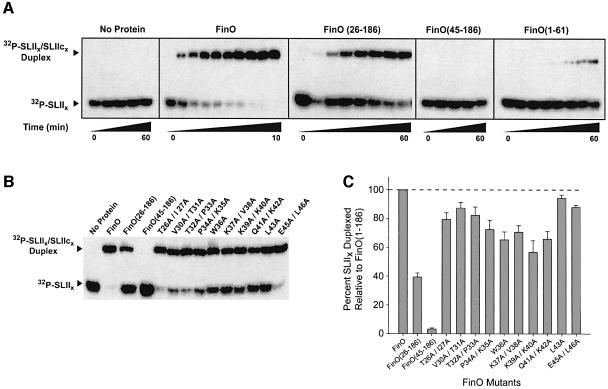
We also analyzed the ability of FinO to promote duplex formation between two complementary stem–loops (SLIIx and SLIIcx) derived from SLII of FinP and SLIIc of traJ mRNA (Figure 1A). It was shown previously that the SLII hairpins are the primary FinO binding structures of FinP and traJ mRNA (Jerome and Frost, 1999). We found that SLIIx and its complementary sequence, SLIIcx, do not associate over a 1 h reaction. FinO, however, dramatically facilitates their pairing such that the reaction is complete within 10 min, with a second-order rate constant (k2) of 1.4 × 105/M/s (Figure 4A; Table I; Supplementary data). FinO can therefore promote duplex formation between minimal stem–loop targets and does not require full-length FinP and traJ mRNA. Consistent with the results obtained with FinP and traJ RNA, the rate of SLIIx–SLIIcx duplex formation in the presence of FinO(26–186) was an order of magnitude lower than with full-length FinO and there was no duplex formation in the presence of FinO(45–186). Thus, the N-terminal 44 residues of FinO, which are required for its strand exchange activity, are essential to promote the association of FinP and traJ mRNA, as well as minimal stem–loop targets. We wondered whether the C-terminal mutant, FinO(1–61), had duplexing activity since it was capable of facilitating strand exchange. As shown in Figure 4A, FinO(1–61) is able to duplex the complementary hairpins, albeit at a reduced level compared with wild-type FinO. This confirms the importance of the N-terminal region for duplexing and indicates that the weak protein–RNA interactions provided by FinO(1–61) are sufficient for both strand exchange and duplexing activities. FinO(1–61) is able to bring together complementary RNAs and contains the N-terminal region necessary for strand exchange. More efficient strand exchange and duplexing would require the rest of the protein.
Fig. 4. FinO RNA strand exchange mutants are deficient in facilitating sense–antisense RNA interactions. (A) FinO, FinO(26–186) and FinO(45–186), each at a final concentration of 1 µM, and a no protein control were tested for their ability to facilitate sense–antisense pairing between SLIIx and SLIIcx RNAs. Aliquots were taken at 0, 0.25, 0.5, 0.75, 1, 1.5, 2, 3, 4 and 10 min for FinO; 1, 2, 4, 8, 15, 30 and 60 min for FinO(26–186) and FinO(1–61); and 0, 10, 20, 30 and 60 min for FinO(45–186) and the no protein control. (B) Two-hour duplexing reactions were performed in the presence of FinO and the various FinO mutants indicated. (C) The amount of SLIIx duplexed at 2 h, expressed as a percentage of the amount of duplex generated for wild-type FinO, is displayed in graphical form. Error bars represent standard deviations calculated from at least three independent experiments.
To further assess the correlation between strand exchange and RNA duplexing, we tested the ability of the strand exchange deficient FinO double-point mutants to promote duplex formation between SLIIx and a SLIIcx under the same conditions used in the strand exchange assays. We found that there is a significant decrease in the duplexing activity of several double-point mutants in comparison with wild-type FinO (Figure 4B and C). Alanine substitutions between residues 34 and 42 caused the greatest reduction in duplexing activity, demonstrating that the same residues that are required for efficient RNA strand exchange are also important for RNA duplexing. However, the magnitude of the effect of the mutations on duplexing is much less than the effect on strand exchange. This suggests that while the strand exchange and duplexing mechanisms share common features involving the region of FinO between residues 34 and 42, additional residues in the N-terminal region of FinO play roles that are specific to the duplexing reaction.
FinO may utilize its RNA-binding energy to destabilize RNA duplexes
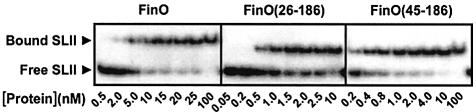
DNA and RNA helicases hydrolyze ATP to provide the free energy needed to destabilize base pairing (von Hippel and Delagoutte, 2001). FinO bears no overall structural similarity to known ATP-dependent helicases and its strand exchange activity is not dependent on nucleoside triphosphates (data not shown). We wondered whether FinO might instead use its free energy of RNA binding to destabilize RNA base pairs. To test this idea, we compared the RNA binding affinities of FinO and the strand exchange deficient mutants using an EMSA (Figure 5). The results show that the strand exchange deficient mutants all bind SLII RNA with high affinity. Furthermore, we observed an inverse correlation between the RNA binding and strand exchange activities of the set of mutants such that the mutant with the lowest degree of strand exchange, FinO(45–186), displayed a 20-fold enhanced affinity for SLII over wild-type FinO. FinO(26–186) and FinO(W36A), which display an intermediate strand exchange activity, bind SLII ∼4- to 5-fold tighter than wild-type FinO (Table I). We suggest that the overall RNA binding energy is the sum of favorable energy terms derived from protein–RNA contacts, and unfavorable terms derived from the destabilization of base pairing. Removal of the N-terminal regions that are responsible for base-pair destabilization reduces the unfavorable energy terms, while leaving most of the favorable interactions intact, resulting in a more favorable overall free energy of RNA binding. It must be noted, however, that the conditions of the binding assay differ significantly from those of the strand exchange and duplexing assays. The binding assays are carried out at 4°C, compared with 37°C for the strand exchange/duplexing assays. Previous results indicate that FinO–RNA binding affinity decreases with increasing temperature [compare the results of Jerome and Frost (1999) with results presented here in Table I], but increases in temperature do not appear to affect relative binding affinities. In addition, control gel shift assays at 4°C show that FinO binding affinities are not affected by the presence of excess cold SII(A) strand, which is present in the strand exchange assay (data not shown).
Fig. 5. RNA strand exchange deficient FinO mutants bind RNA more tightly than wild-type FinO. Representative gel EMSAs for FinO, FinO(26–186) or FinO(45–186) binding to SLII RNA. Samples containing 50 pM SLII were incubated with protein at the concentrations indicated. These and similar experiments were used to determine the relative FinO–RNA association constants shown in Table I.
The RNA strand exchange and duplexing activities of FinO are involved in the repression of bacterial conjugation
To determine the physiological relevance of the RNA strand exchange and duplexing activities of FinO, we assayed the ability of N-terminal mutants of FinO (expressed as GST fusion proteins) to repress the transfer of F plasmids from donor to recipient E.coli cells. The donor cells contained an F-derived plasmid, pOX38-Km (Chandler and Galas, 1983), bearing a kanamycin resistance gene, as well as a FinO expression plasmid. pOX38-Km does not express FinO and is dependent on FinO supplied in trans for efficient inhibition of transfer. Donor and spectinomycin-resistant recipient cells were mixed, and transconjugant recipient cells were selected for resistance to both kanamycin and spectinomycin. Mating efficiencies were calculated as the ratio of transconjugants to donor cells.
As expected, mating was severely inhibited by FinO, with only 5% mating efficiency compared with no protein (Table I). FinO(26–186) was a much less effective repressor of conjugation and displayed 87% of the mating efficiency compared with no protein, while FinO(45–186) did not inhibit mating at all. These results reveal that the N-terminal region of FinO that contains the RNA strand exchange activity plays an essential role in the ability of FinO to block bacterial conjugation.
The alanine substitution mutants used in the strand exchange assays were also tested in the mating assay. Of all the mutants tested, only W36A showed a significant loss of repression. The W36A mutant also showed the most dramatic loss of strand exchange activity of all the single amino acid substitutions tested, consistent with an important role for RNA base-pair destabilization in the repression of conjugation. However, the K37A/V38A, K39A/K40A and Q41A/K42A mutants all exhibited defects in strand exchange and duplexing comparable to W36A, yet none of the double mutants showed a significant loss of repressor activity in vivo. This observation may indicate that Trp36 is also important for some other function, in addition to RNA strand exchange and sense–antisense RNA pairing, which ultimately contributes to the repression of plasmid transfer.
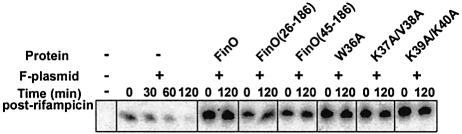
FinO has also been shown to stabilize FinP against endonucleolytic degradation by RNase E, a function that contributes to the repression of conjugation (Frost et al., 1989; Lee et al., 1992; Jerome et al., 1999). To test the effects of the N-terminal FinO mutations on FinP stability, we introduced the mutants into a FinP-expressing E.coli strain and measured the stability of FinP transcripts isolated from these strains at various times after rifampicin-induced blockage of transcription initiation (Figure 6). Consistent with previous results, FinP is degraded in cells that do not express FinO (t1/2 ∼50 min); however, in cells that express wild-type FinO, the half-life of FinP is extended to well beyond 2 h. As a result of the stabilization of FinP, its steady state level (measured at the 0 time point; Figure 6) is significantly higher in cells expressing FinO than in its absence. Similar levels of FinP stabilization are observed for all the N-terminal FinO mutants. We therefore conclude that the deficiencies of conjugation repression observed in these mutants is a direct consequence of their inability to facilitate RNA strand exchange, and therefore facilitate FinP–traJ RNA interactions, and cannot be explained by changes in FinP stabilization.
Fig. 6. In vivo stabilization of FinP by FinO and FinO derivatives. Stabilization of FinP in cells expressing FinP and the indicated FinO proteins was examined at the given times after the addition of rifampicin by northern blot analysis. As controls, FinP stability was examined in the absence of FinO, and hybridization was performed on RNA extracted from cells not harboring the F-plasmid.
Discussion
In this study we have demonstrated that FinO acts as an RNA chaperone, catalyzing strand exchange as well as promoting the duplexing of two complementary stem–loops. Our results strongly suggest that FinO destabilizes otherwise stable stem structures in FinP and traJ mRNA, and that this destabilization is required for the ability of FinO to promote duplex formation between FinP and traJ mRNA. The base-pair destabilization activity, together with the previously demonstrated ability of FinO to bind and protect FinP against endonucleolytic degradation is responsible for the 20-fold enhancement of FinP-mediated conjugation repression in vivo.
The strand exchange activity does not require ATP and appears to involve the recognition and destabilization of the SII duplex substrate followed by exchange with the single-stranded SII(A) RNA. It is not clear whether the single-stranded RNA is directly recognized by FinO, since EMSAs reveal that FinO does not form a stable complex with SII(A) RNA at concentrations relevant to strand exchange. However, it does appear that SII(A) RNA may be involved in the rate limiting step in this process since strand exchange rates are moderately enhanced with increasing concentrations of single strand.
While the precise molecular mechanism that underlies FinO-mediated RNA duplex destabilization is not yet clear, the data presented here, together with previous biochemical and structural data on the FinOP system, indicate critical features of this process. Our deletion study reveals that FinO residues 1–44 are absolutely required for RNA strand exchange, but not for high-affinity interactions with RNA. In contrast, further N-terminal or C-terminal deletions significantly decrease the affinity and specificity of FinO for its cognate RNA targets (Ghetu et al., 1999), indicating that the N-terminal region plays a particularly critical and direct role in RNA strand exchange. The N-terminal FinO(1–61) fragment is still able to catalyze strand exchange and duplexing, even though it binds rather poorly to RNA. However, more efficient strand exchange/duplexing requires the presence of the remaining C-terminal portion of the protein.
The N-terminal ∼32 residues of FinO appear to be disordered either in the free protein, or in complex with RNA; however, the adjacent region (residues 33–61) becomes more structured upon binding RNA (Ghetu et al., 1999) and direct interactions between this region and RNA have recently been demonstrated by site-specific protein–RNA crosslinking (Ghetu et al., 2002). The structure of the free protein, crystallized at low temperature, revealed that residues 34–67 form a solvent exposed helix. Alanine point mutations within the lysine-rich N-terminal end of this helix revealed that, in addition to the lysines, several hydrophobic residues, most notably, Trp36, as well as Pro34 and Val38, also mediate RNA strand exchange. We propose that the N-terminal tip of this helix is docked to duplex RNA via electrostatic interactions between the lysine residues and the phosphodiester backbone of the RNA. This docking may position the critical hydrophobic residues in a groove of the RNA duplex, where they could potentially intercalate between adjacent bases and thereby disrupt base pairing. The stacking of aromatic amino acids with unpaired bases is a common feature of many protein–RNA interactions, and appears to play a role in the ATP-dependent disruption of base-pairing by helicases (Kim et al., 1998; Velankar et al., 1999; Marians, 2000).
While FinO-catalyzed RNA destabilization is critical to the ability of FinO to facilitate sense–antisense RNA interactions, we believe that FinO must also bring the complementary RNA molecules into close proximity. Otherwise, the unwound RNAs would simply ‘snap back’ to their intramolecular stem–loop conformations before contact could be made with the complementary RNA partner. This may explain why the strand exchange reaction, which occurs over a 2-h time period, is much slower than the duplexing reaction, which occurs within a few minutes under identical conditions. Sense–antisense RNA interactions that regulate a variety of aspects of plasmid replication and transfer are thought to proceed via loop–loop or kissing complex intermediates enroute to the final, paired state (Franch et al., 1999). For example, the ColE1 protein Rom directly interacts with and stabilizes a kissing complex to facilitate RNA–RNA pairing (Eguchi and Tomizawa, 1990; Predki et al., 1995), and while Rom and FinO are not structurally related, it is tempting to speculate that FinO may carry out a similar function. Indeed, many finP alleles contain a 5′-YUNR-3′ (Y, pyrimidine; N, any base; R, any purine) sequence in the loop, which has been shown to be a major recognition element in a number of antisense RNAs (Franch et al., 1999). Tethering FinP and traJ RNA together in this way would facilitate pairing of the two RNAs, once destabilization of the internal secondary structures had taken place.
While we have demonstrated that FinO can facilitate RNA–RNA interactions between full-length FinP and traJ mRNA, as well as between minimal SLII–SLIIc substrates, the interactions between the larger RNAs occur almost 100-fold faster than with the minimal RNAs. The larger RNAs contain SLI and SLIc, which are less stable than SLII and SLIIc due to their shorter length and the presence of base-pair mismatches in the stem region near the loops. Thus, the less stable SLI and SLIc, as well as the additional single-stranded regions present in the larger RNAs, may play a critical role in sense–antisense association. Interestingly, the destabilization of stem structures by bulges proximal to loops has been shown to play a key role in the control of plasmid replication through the duplexing of CopA and CopT RNAs (Kolb et al., 2001). In this system, the antisense CopT and its target CopA contain several base-pair mismatches in a pair of stem–loop structures that form an intermolecular kissing complex. These bulges are necessary for propagation of the intermolecular base pairing from the initial kissing complex to a more fully paired and therefore more stable structure. In analogy to CopA/CopT, it appears that the bulges in SLI and SLIc are important in facilitating duplexing between these two stem–loops, since mutations altering the purine:purine mismatch in SLI and SLIc to more stable A:U base pairs decreased FinO-mediated duplex formation relative to wild-type SLI and SLIc (Gubbins et al., 2003). Therefore, it is probable that SLI and SLIc contribute to the enhanced rates of stable association between full-length FinP and traJ mRNA.
ATP-dependent DNA helicases often unwind large tracts of duplex DNA with significant energy input from ATP hydrolysis. In contrast, most RNA helicases only need to unwind short segments of duplex, and therefore the processivity afforded by continuous ATP hydrolysis may not be absolutely required for many RNA remodeling processes. This is the case for a class of proteins called RNA chaperones, which have the ability to remodel RNA structure without energy input from ATP. One of the most critical functions inherent in these proteins is the ability to lower the energy barrier needed to resolve kinetically trapped, misfolded secondary or tertiary RNA structures in vivo (Herschlag, 1995; Weeks, 1997; Cristofari and Darlix, 2002; Lorsch, 2002). Recently, a number of RNA chaperones have been found to have helix destabilizing activities and the ability to promote strand annealing. Examples include the N-terminal domain of the hepatitis delta antigen (NdAg) (Huang et al., 2003) and the E.coli host factor I (Hfq) (Moller et al., 2002; Zhang et al., 2002). Perhaps the best characterized candidate for a functional homolog for FinO is the nucleocapsid protein (NCp7) of HIV-1. This nucleic acid chaperone mediates a conformational change in the viral RNA genome to the mature, more stable dimeric state. It also facilitates binding of the primer tRNA to its binding site to initiate reverse transcription, and is subsequently involved in two strand-transfer reactions leading to the final DNA copy of the genome (Rein et al., 1998). Like FinO, NCp7 can bind kissing stem–loop structures and facilitate their transition into an extended duplex; however, in contrast to FinO, it can stably bind single-stranded nucleic acids leading to annealing or strand exchange events (Takahashi et al., 2001; Urbaneja et al., 2002). Recently, it has been shown that NCp7 has helix destabilizing properties in vitro that are crucial for annealing of the minus-strand DNA to the 3′ terminus of the RNA genome, and preventing self-priming reactions (Bernacchi et al., 2002; Hong et al., 2003). These melting events are limited to a few base pairs, enough to initiate annealing to the complementary strand. It is probable that FinO might act in a similar fashion to NCp7, destabilizing a small portion of FinP and traJ mRNA, allowing the two complementary stem–loops to duplex. Like NCp7, the mechanism of FinO-mediated RNA–RNA interactions is not fully understood. Further structural studies using NMR and crystallographic strategies are underway to investigate possible helix destabilization activities and sense–antisense interactions.
Materials and methods
Preparation of proteins and RNA substrates
See Supplementary data.
Strand exchange assay
Strand exchange assays were set up on ice in 12 µl reaction volumes consisting of 6 µl reaction buffer (50 mM Tris pH 8.1, 100 µg/ml BSA, 10% glycerol, 0.1% β-mercaptoethanol and 20 mM NaCl), 1 µl of 20 U/µl RNAguard (Amersham Biosciences) (in 50 mM KCl, 20 mM HEPES–KOH pH 7.6 and 5 mM dithiothreitol), 1.5 µl protein (in 50 mM MES pH 6.5, 0.1% β-mercaptoethanol and 150 mM NaCl; 8 µM to give a final concentration of 1 µM, unless otherwise indicated), 1 µl of the labeled SII duplex (5 nM to give a final duplex concentration of ∼400 pM), 2 µl of the unlabeled SII(A) strand (1.25 µM to give a final concentration of ∼200 nM) and 0.5 µl of distilled water. Strand exchange assays were initiated by placing reaction tubes at 37°C. For time-course experiments, aliquots were taken from a scaled up reaction mixture at various time points and were stopped by addition of an equal volume of stop solution (5% glycerol, 0.4% SDS and 20 mM EDTA). Samples were subjected to 15% non-denaturing PAGE at room temperature to separate the free and duplexed 32P-labeled strands. For rate determination assays, samples were loaded onto a continuously running gel. To visualize bands corresponding to duplexed and single-stranded 32P-labeled RNA, a Molecular Dynamics storage phosphor screen was exposed to the gels for ∼12 h and scanned using a Storm 840 PhosphorImager (Molecular Dynamics). Bands were quantified with ImageQuant software (Molecular Dynamics). See Supplementary data for determination of apparent first order strand exchange rates.
Duplexing assays
Duplexing between SLIIx and SLIIcx was performed in 50 µl reaction mixtures containing 25 µl of 2× reaction buffer (50 mM Tris pH 8.1, 100 µg/ml BSA, 10% glycerol, 0.1% β-mercaptoethanol and 40 mM NaCl), 5 µl of protein at 10 µM, in 20 mM MES pH 6.5, 0.1% β-mercaptoethanol and 60 mM NaCl, 5 µl of SLIIcx at 1 µM, 5 µl of 32P-labeled SLIIx at 50 nM and 10 µl of ddH2O. To initiate duplexing, labeled SLIIx, preincubated at 37°C, was added to reaction mixtures, also preincubated at 37°C and lacking SLIIx. Aliquots (5 µl) were taken at various time points and added to 5 µl of cold stop buffer (5% glycerol, 0.4% SDS and 20 mM EDTA). Samples were subjected to 10% non-denaturing PAGE for 2 h prior to visualization and quantification of the labeled SLIIx using ImageQuant software, as described above. FinP–traJ RNA duplexing assays with FinO(45–186) were performed as described previously (Ghetu et al., 2000). See Supplementary data for determination of second order duplexing rates.
EMSAs
To determine the association constants of FinO and FinO-derived proteins for SLII or SII, EMSAs were performed with increasing concentrations of protein at 4°C as described previously (Ghetu et al., 1999), with the following modification. Binding reactions contained 5 µl of the reaction buffer used in the strand exchange assays (with an additional 10% glycerol and 1 mM EDTA), 4 µl of protein in 50 mM MES (pH 6.5), 0.1% β-mercaptoethanol, 450 mM NaCl and 100 µg/ml BSA and 1 µl of labeled RNA at a concentration of 500 pM.
Mating assays
Mating assays were performed essentially as described previously (Sandercock and Frost, 1998) using E.coli MC4100 cells bearing the F derivative plasmid pOX38-Km (Chandler and Galas, 1983) and various pGEX-FinO plasmids. The presence of both plasmids was confirmed by agarose gel electrophoresis of plasmid DNA isolated from the E.coli strains, and GST–FinO protein expression levels were assayed by western blot analysis using anti-GST antibodies (Sigma) and anti-FinO antiserum. All GST–FinO proteins used in these studies were expressed at similar levels, within ∼20% of wild type. The ratio of transconjugants to donors was calculated, allowing mating efficiency to be compared with the control of conjugal transfer of pOX38-Km alone.
Northern blot analysis to determine FinP half-life
The half-life of FinP RNA isolated from the GST–FinO-expressing E.coli MC4100 strains used for the mating assays was assessed by northern blot analysis as described previously (Sandercock and Frost, 1998; Jerome et al., 1999). Equivalent amounts (35 µg) of total RNA were loaded in each lane of the gel used for the northern analysis. After measurement of FinP band intensities, the blots were stripped and re-probed for the control RNA (tRNASer), and the FinP intensities were normalized based on the amounts of tRNASer detected in same lanes.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank members of the Glover laboratory and Andrew MacMillan for helpful discussions and critical reading of the manuscript. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J.N.M.G. and L.S.F., and Alberta Heritage Foundation for Medical Research (AHFMR) to J.N.M.G. D.C.A. was supported by an AHFMR studentship, and R.A.E. was supported by an AHFMR postdoctoral fellowship. M.J.G. was supported by studentships from CIHR and AHFMR.
References
- Bernacchi S., Stoylov,S., Piemont,E., Ficheux,D., Roques,B.P., Darlix,J.L. and Mely,Y. (2002) HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol., 317, 385–399. [DOI] [PubMed] [Google Scholar]
- Boyd E.F. and Hartl,D.L. (1997) Recent horizontal transmission of plasmids between natural populations of Escherichia coli and Salmonella enterica. J. Bacteriol., 179, 1622–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E.F. and Hartl,D.L. (1998) Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics, 149, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Galas,D.J. (1983) IS1-mediated tandem duplication of plasmid pBR322. Dependence on recA and on DNA polymerase I. J. Mol. Biol., 165, 183–190. [DOI] [PubMed] [Google Scholar]
- Cristofari G. and Darlix,J.L. (2002) The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol., 72, 223–268. [DOI] [PubMed] [Google Scholar]
- Cuozzo M. and Silverman,P.M. (1986) Characterization of the F plasmid TraJ protein synthesized in F¢ and Hfr strains of Escherichia coli K-12. J. Biol. Chem., 261, 5175–5179. [PubMed] [Google Scholar]
- Eguchi Y. and Tomizawa,J. (1990) Complex formed by complementary RNA stem–loops and its stabilization by a protein: function of CoIE1 Rom protein. Cell, 60, 199–209. [DOI] [PubMed] [Google Scholar]
- Eguchi Y. and Tomizawa,J. (1991) Complexes formed by complementary RNA stem–loops. Their formations, structures and interaction with ColE1 Rom protein. J. Mol. Biol., 220, 831–842. [DOI] [PubMed] [Google Scholar]
- Finlay B.B., Frost,L.S., Paranchych,W. and Willetts,N.S. (1986) Nucleotide sequences of five IncF plasmid finP alleles. J. Bacteriol., 167, 754–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. and Willetts N. (1972) The nature of the transfer inhibitor of several F-like plasmids. Mol. Gen. Genet., 119, 57–66. [DOI] [PubMed] [Google Scholar]
- Franch T., Petersen,M., Wagner,E.G., Jacobsen,J.P. and Gerdes,K. (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol., 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Frost L., Lee,S., Yanchar,N. and Paranchych,W. (1989) finP and fisO mutations in FinP anti-sense RNA suggest a model for FinOP action in the repression of bacterial conjugation by the Flac plasmid JCFL0. Mol. Gen. Genet., 218, 152–160. [DOI] [PubMed] [Google Scholar]
- Ghetu A.F., Gubbins,M.J., Oikawa,K., Kay,C.M., Frost,L.S. and Glover,J.N. (1999) The FinO repressor of bacterial conjugation contains two RNA binding regions. Biochemistry, 38, 14036–14044. [DOI] [PubMed] [Google Scholar]
- Ghetu A.F., Gubbins,M.J., Frost,L.S. and Glover,J.N. (2000) Crystal structure of the bacterial conjugation repressor finO. Nat. Struct. Biol., 7, 565–569. [DOI] [PubMed] [Google Scholar]
- Ghetu A.F., Arthur,D.C., Kerppola,T.K. and Glover,J.N. (2002) Probing FinO–FinP RNA interactions by site-directed protein–RNA crosslinking and gelFRET. RNA, 8, 816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbins M.J., Arthur,D.C., Ghetu,A.F., Glover,J.N. and Frost,L.S. (2003) Characterizing the structural features of RNA/RNA interactions of the F-plasmid. FinOP fertility inhibition system. J. Biol. Chem., 278, 27663–27671. [DOI] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- Hong M.K., Harbron,E.J., O’Connor,D.B., Guo,J., Barbara,P.F., Levin,J.G. and Musier-Forsyth,K. (2003) Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer. J. Mol. Biol., 325, 1–10. [DOI] [PubMed] [Google Scholar]
- Huang Z.S., Su,W.H., Wang,J.L. and Wu,H.N. (2003) Selective strand annealing and selective strand exchange promoted by the N-terminal domain of hepatitis delta antigen. J. Biol. Chem., 278, 5685–5693. [DOI] [PubMed] [Google Scholar]
- Jerome L.J. and Frost,L.S. (1999) In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J. Biol. Chem., 274, 10356–10362. [DOI] [PubMed] [Google Scholar]
- Jerome L.J., van Biesen,T. and Frost,L.S. (1999) Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol., 285, 1457–1473. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]
- Kolb F.A., Westhof,E., Ehresmann,C., Ehresmann,B., Wagner,E.G. and Romby,P. (2001) Bulged residues promote the progression of a loop–loop interaction to a stable and inhibitory antisense–target RNA complex. Nucleic Acids Res., 29, 3145–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann G., Koraimann,C., Koronakis,V., Schlager,S. and Hogenauer,G. (1991) Repression and derepression of conjugation of plasmid R1 by wild-type and mutated finP antisense RNA. Mol. Microbiol., 5, 77–87. [DOI] [PubMed] [Google Scholar]
- Koraimann G., Teferle,K., Markolin,G., Woger,W. and Hogenauer,G. (1996) The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol. Microbiol., 21, 811–821. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Frost,L.S. and Paranchych,W. (1992) FinOP repression of the F plasmid involves extension of the half-life of FinP antisense RNA by FinO. Mol. Gen. Genet., 235, 131–139. [DOI] [PubMed] [Google Scholar]
- Lorsch J.R. (2002) RNA chaperones exist and DEAD box proteins get a life. Cell, 109, 797–800. [DOI] [PubMed] [Google Scholar]
- Marians K.J. (2000) Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure Fold. Des., 8, R227–R235. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- Moller T., Franch,T., Hojrup,P., Keene,D.R., Bachinger,H.P., Brennan,R.G. and Valentin-Hansen,P. (2002) Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell, 9, 23–30. [DOI] [PubMed] [Google Scholar]
- Mullineaux P. and Willetts,N. (1985) Promoters in the transfer region of plasmid F. Basic Life Sci., 30, 605–614. [DOI] [PubMed] [Google Scholar]
- Predki P.F., Nayak,L.M., Gottlieb,M.B. and Regan,L. (1995) Dissecting RNA–protein interactions: RNA–RNA recognition by Rop. Cell, 80, 41–50. [DOI] [PubMed] [Google Scholar]
- Rein A., Henderson,L.E. and Levin,J.G. (1998) Nucleic-acid–chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci., 23, 297–301. [DOI] [PubMed] [Google Scholar]
- Sandercock J.R. and Frost,L.S. (1998) Analysis of the major domains of the F fertility inhibition protein, FinO. Mol. Gen. Genet., 259, 622–629. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Baba,S., Koyanagi,Y., Yamamoto,N., Takaku,H. and Kawai,G. (2001) Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem., 276, 31274–31278. [DOI] [PubMed] [Google Scholar]
- Urbaneja M.A., Wu,M., Casas-Finet,J.R. and Karpel,R.L. (2002) HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity and annealing activity. J. Mol. Biol., 318, 749–764. [DOI] [PubMed] [Google Scholar]
- van Biesen T. and Frost,L.S. (1994) The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA and promotes duplex formation. Mol. Microbiol., 14, 427–436. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H. and Delagoutte,E. (2001) A general model for nucleic acid helicases and their ‘coupling’ within macromolecular machines. Cell, 104, 177–190. [DOI] [PubMed] [Google Scholar]
- Wagner J.D., Jankowsky,E., Company,M., Pyle,A.M. and Abelson,J.N. (1998) The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J., 17, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wagner,J.D. and Guthrie,C. (1998) The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol., 8, 441–451. [DOI] [PubMed] [Google Scholar]
- Watanabe T. and Fukasawa,T. (1961) Episome-mediated transfer of drug resistance in Enterobacteriaceae. I. Transfer of resistance factors by conjugation. J. Bacteriol, 81, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K.M. (1997) Protein-facilitated RNA folding. Curr. Opin. Struct. Biol., 7, 336–342. [DOI] [PubMed] [Google Scholar]
- Willetts N. (1977) The transcriptional control of fertility in F-like plasmids. J. Mol. Biol., 112, 141–148. [DOI] [PubMed] [Google Scholar]
- Zhang A., Wassarman,K.M., Ortega,J., Steven,A.C. and Storz,G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell, 9, 11–22. [DOI] [PubMed] [Google Scholar]