Abstract
The small multidrug resistance family of transporters is widespread in bacteria and is responsible for bacterial resistance to toxic aromatic cations by proton-linked efflux. We have determined the three-dimensional (3D) structure of the Escherichia coli multidrug transporter EmrE by electron cryomicroscopy of 2D crystals, including data to 7.0 Å resolution. The structure of EmrE consists of a bundle of eight transmembrane α-helices with one substrate molecule bound near the centre. The substrate binding chamber is formed from six helices and is accessible both from the aqueous phase and laterally from the lipid bilayer. The most remarkable feature of the structure of EmrE is that it is an asymmetric homodimer. The possible arrangement of the two polypeptides in the EmrE dimer is discussed based on the 3D density map.
Keywords: asymmetry/electron crystallography/membrane protein/multidrug/structure
Introduction
Multidrug resistance in bacteria is a significant problem throughout the world, with the appearance of pathogens that are resistant to common antibiotics, and even normally harmless bacteria can now lead to life-threatening primary or secondary infections (Wise et al., 1998; Cizman, 2003). Bacteria have evolved complex mechanisms for the extrusion of toxic compounds, many of which we now use routinely as antibiotics and antiseptics (Zgurskaya and Nikaido, 2002). The efflux of toxic molecules is driven either by ATP hydrolysis, as in the ABC transporter superfamily, or by coupling efflux to the inward movement of protons down their concentration gradient. This antiporter mechanism has evolved in many different transporter families (Paulsen et al., 1996), including the Major Facilitator Superfamily (MFS), the Resistance-Nodulation-cell Division (RND) family and the Small Multidrug Resistance (SMR) family. An atomic resolution structure of an RND family multidrug transporter has been described previously (Murakami et al., 2002; Elkins and Nikaido, 2003), but only the structures of multidrug transporter homologues in the ABC family (Chang and Roth, 2001; Locher et al., 2002; Chang, 2003) and transporters in the MFS family (Abramson et al., 2003; Hirai et al., 2002; Huang et al., 2003) have been obtained so far. This paper describes the three-dimensional (3D) structure of EmrE, a prototypic member of the SMR family.
EmrE has been subject to intensive investigation over the last decade (Schuldiner et al., 2001). EmrE is a multidrug transporter that catalyses the electrogenic efflux of one substrate molecule in exchange for two or more protons through a hydrophobic pathway in the protein (Lebendiker and Schuldiner, 1996; Mordoch et al., 1999; Yerushalmi and Schuldiner, 2000). EmrE is classified as a multidrug transporter because its substrates include a wide variety of cationic aromatic hydrocarbons of varying size, structure and charge (Yerushalmi et al., 1995). The amino acid sequence of EmrE is predicted to form four transmembrane α-helices, which is strongly supported by evidence from FTIR (Arkin et al., 1996) and heteronuclear NMR (Schwaiger et al., 1998). Four transmembrane α-helices are thought unlikely alone to be capable of forming a structure that could catalyse proton-linked multidrug efflux, so the oligomeric state of EmrE has received much attention. Negative-dominance studies, cross-linking, ligand binding and hetero-oligomer formation all confirm that EmrE functions as an oligomer (Yerushalmi et al., 1996; Muth and Schuldiner, 2000; Rotem et al., 2001; Soskine et al., 2002; Tate et al., 2003). Projection structures of EmrE determined by electron cryomicroscopy (cryo-EM) of 2D crystals showed that the repetitive unit in the crystal was composed of eight α-helices arranged in an asymmetric manner, indicating that the minimal functional unit for substrate binding is a dimer (Tate et al., 2001, 2003). Available data do not preclude the existence of higher functional oligomers in vivo. Here we describe the architecture of dimeric EmrE in the membrane, based on cryo-EM of 2D crystals, including the location of the TPP+ binding site and the translocation pathway within the dimer.
Results and discussion
Description of the structure
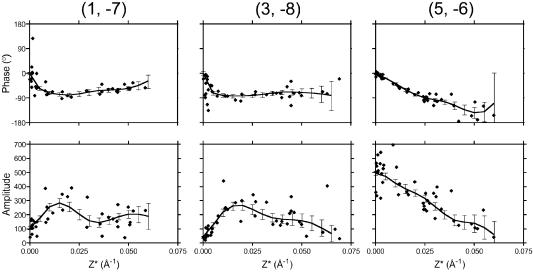
Tubular crystals of EmrE were obtained by dialysis of the purified transporter to remove detergent in the presence of the substrate TPP+. We chose to solve the structure of EmrE with TPP+ bound, because these crystals are more highly ordered than the native crystals, and we knew from previous work (Tate et al., 2003) that the structure of the TPP+-bound form is virtually identical in projection to the native structure. Samples for cryo-EM were prepared by drying briefly in 1% glucose followed by rapid freezing in liquid nitrogen. Images of the flattened tubes showed two easily distinguishable crystalline lattices from the top and bottom surfaces of the vesicle each with p2 symmetry and unit cell dimensions a = 72.1 Å, b = 86.8 Å, γ = 107.3°. Forty-seven images from samples tilted nominally up to 40° were processed and merged (Table I) using the Medical Research Council suite of programs (Crowther et al., 1996). The 2D lattice lines (Figure 1) were fitted and used to calculate a 3D density map of EmrE.
Table I. Electron crystallographic data.
| Plane group | p2 |
|---|---|
| Cell dimensions | |
| a (Å) | 72.1 |
| b (Å) | 86.8 |
| c (Å) | 200.0 (arbitrary) |
| α = β (°) | 90.0 |
| γ (°) | 107.3 |
| Number of imagesa | 47 |
| Resolution limit for merging (Å) | 7.0 |
| Effective resolution of 3D data setb (Å) | |
| In-plane | 7.5 |
| Perpendicular to the membrane | 16.0 |
| Total number of observations | 6782 (IQ <7) |
| Number of structural factors | 2198 |
| Completeness (%) | |
| 0–40° | 93 |
| 0–90° | 60 |
| Overall weighted R-factorc (%) | 22.6 |
| Overall weighted phase residualc (°) | 16.3 |
aThirteen at 0°, eight at 20°, 20 at 30° and six at 40° (nominal tilts).
bAs calculated from a point-spread function of the experimental data.
cFrom program LATLINEK.
Fig. 1. Lattice line data. Plots of amplitudes (lower panels) and phases (upper panels) along the z* axis for three selected reflections. The fitted lattice lines were produced by weighted least squares fitting and the resulting errors are shown.
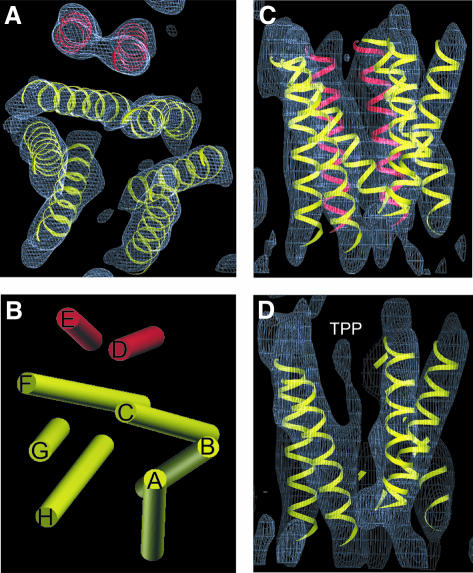
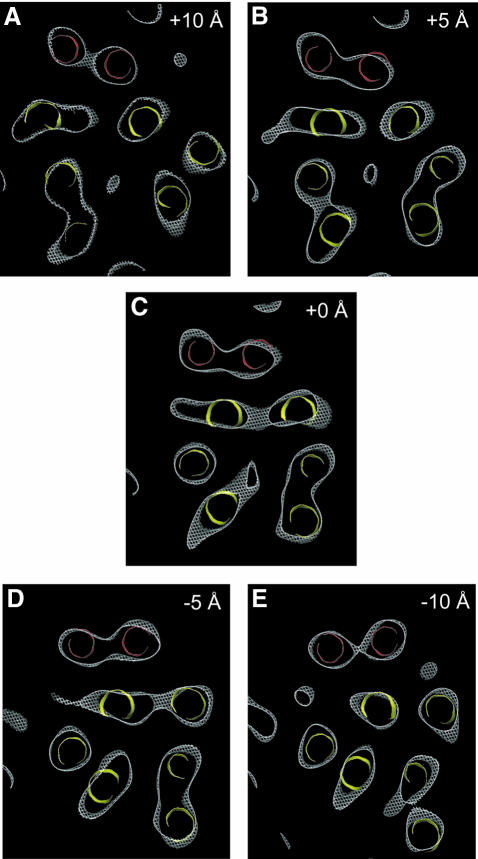
The structure of the EmrE dimer (Figure 2) is composed of eight transmembrane α-helices. Six α-helices, coloured yellow (Figure 2B, helices A–C and F–H), with tilt angles of 12–38° relative to the membrane normal, are packed to form the wall of a chamber. The other two α-helices D and E (coloured red), which are more nearly perpendicular to the membrane with tilt angles of 11 and 12°, respectively, are separated from the chamber by two of the highly tilted helices C (24° tilt) and F (38° tilt) in the wall. Slices of the map at different heights (Figure 3A–E) show that this chamber extends from one surface of the membrane to just past the membrane centre, where it appears to be closed by the convergence of helices F and H. Density connects these two helices in our map (Figure 5B), strongly suggesting that they are joined by a loop at the bottom of the structure. The chamber constitutes the binding site in EmrE for substrate, because the density associated with TPP+, identified previously by comparing the native and the TPP+-bound forms (Tate et al., 2003), is found near its centre (Figure 3A–C). The density for TPP+ appears as a triangular mass merging to the main density for helix H (Figure 2D). The limited vertical resolution of our structure and the fact that TPP+ has a tetrahedral structure suggests that we are observing the density for only the part of TPP+ that is immobilized by direct interactions with amino acid residue side-chains in neighbouring helices. The substrate binding chamber has two openings, one facing the aqueous medium and the other laterally facing the lipid bilayer, which would allow hydrophobic substrates in one leaflet of the Escherichia coli inner membrane to diffuse into the EmrE binding site. In addition, it is possible that access to the lipid bilayer between α-helices A and H is required for larger substrates too big to fit into the binding region defined by TPP+ in the structure, thus allowing EmrE to transport a larger range of substrates. It is noticeable that there appears to be no access to the substrate binding site from the other leaflet of the membrane in this conformation of the EmrE dimer (Figure 3D and E).
Fig. 2. Top and side view of the 3D density map of EmrE. (A) Top view perpendicular to the membrane plane of the density contoured at 1.2 σ. (B) Schematic view perpendicular to the membrane plane of the architecture of EmrE with all helices (A–H) approximated as straight cylinders. (C) Side view along the membrane plane of the density contoured at 1.2 σ. (D) Side view of a slice along the membrane plane of the density contoured at 0.8 σ, to emphasize the location of the substrate TPP+. The eight idealized helices were placed manually into the map and were not subjected to refinement. The helices are grouped into two sets: those coloured yellow form the substrate binding pocket and those coloured red are separated from the binding pocket by helices C and F. The maps were analysed and the idealized helices generated in the environment of O (Jones et al., 1991).
Fig. 3. Horizontal slices through the density map: 3Å-deep slices through the density map (contoured at 0.8 σ) perpendicular to the membrane plane separated by 5 Å. The panels show sections above (A and B) and below (D and E) the centre (C). The six yellow helices form the wall of the substrate binding pocket, whereas the two red helices are separated from the binding pocket. The density at the centre of the yellow helices in sections A–C is believed to represent the substrate TPP+.
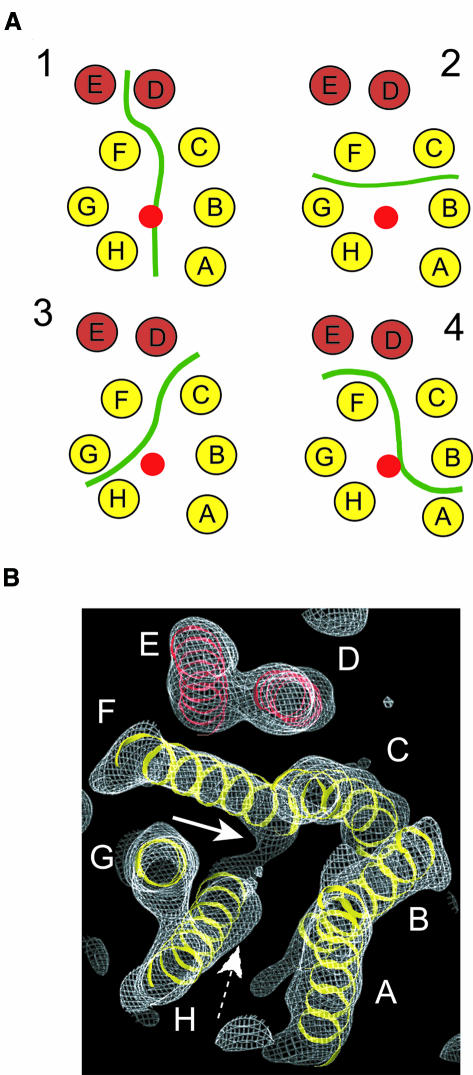
Fig. 5. Possible arrangements of monomers in the dimer. (A) Four possible monomer boundaries in the EmrE dimer are shown. The transmembrane helices are depicted as circles distributed as in the central section of the structure shown in Figure 3C. A red circle depicts the location of the substrate TPP+. A continuous green line illustrates the hypothetical interface between monomers. (B) View of the density map contoured at 1.0 σ to show the connection of density between helix H and helix F (white arrow). The density for TPP+ is also shown for reference (dashed arrow). The view is along helix G and from the same side as in Figure 2.
The most remarkable feature of the EmrE homodimer is that the eight transmembrane α-helices are arranged in an asymmetric manner, with no 2-fold axis relating the two monomers, confirming our previous observations from the projection map of native EmrE to 7.0 Å resolution (Tate et al., 2001). It should be noted that the 3D structure of EmrE determined from the 2D crystals represents functional protein. The detergent-solubilized EmrE used for crystallization binds 3H-TPP+ with high affinity, with a stoichiometry of one TPP+ molecule per EmrE dimer (Tate et al., 2003). The native EmrE crystals that do not contain bound TPP+ can also bind 3H-TPP+ with high affinity (Kd = 3.2 ± 0.3 nM) (C.G.Tate, unpublished data), inducing the crystals to fragment (Tate et al., 2003). A comparison between projection maps of native EmrE and TPP+-bound EmrE suggest only a minor conformational change between the two structures (Tate et al., 2003). These data all suggest that the asymmetric structure of dimeric EmrE is not a consequence of crystallization and is therefore directly relevant to the mechanism of multidrug transport.
Arrangement of EmrE dimers in the crystals
Comparison of 2D EmrE crystals of three different planar space groups (Tate et al., 2003) shows that EmrE crystallizes in the membrane, with the dimers arranged in two distinct ways to form two different tetramers (Figure 4). In projection, one crystallographic tetramer (dimers 1 and 3) is related by a 2-fold axis in the plane of the membrane, with the two dimers closely interacting, but in opposite orientations across the membrane. The other tetramer (dimers 1 and 2) is related by a 2-fold axis perpendicular to the membrane and is formed by the interaction of helices E and D from two adjacent dimers, which create a four-helix bundle. As the two dimers would have the same orientation in the membrane, it is likely that this tetramer could well exist in vivo in the inner bacterial membrane.
Fig. 4. Packing of EmrE dimers in the 2D crystals. The two different tetrameric arrangements of EmrE dimers as observed in the p2 crystals are shown with all helices approximated as straight cylinders. Each dimer is numbered and dimer 1 is identical to the one shown in Figure 2B. The 2-fold axes perpendicular and parallel to the membrane plane are shown in white.
The oligomeric state of EmrE in vivo is unproven. The detergent-solubilized EmrE used for 2D crystallization is a dimer as determined by analytical ultracentrifugation experiments (P.J.G.Butler and C.G.Tate, unpublished data) and is fully competent to bind TPP+ with high affinity at a stoichiometry of one TPP+ per dimer (Tate et al., 2003). However, there is some evidence for an oligomeric order higher than a dimer in the membrane in vivo (Yerushalmi et al., 1996). Given that the minimal functional unit for TPP+ binding is a dimer, then a tetramer is the most likely oligomeric state of EmrE in vivo, but there is no biophysical data on membrane-reconstituted EmrE to support this. It remains to be seen whether there is a functional role for a tetramer in vivo, or whether the dimer is fully capable of catalysing drug efflux.
Structural models for EmrE
To understand how the two EmrE monomers associate to form an asymmetric structure, we need to interpret the medium-resolution 3D structure in terms of which density represents each α-helix in the protein sequence. However, at the current resolution of our map we cannot directly assign the amino acid sequence to each transmembrane helix. Is there sufficient information from the structural data and biochemical data to define a likely model unambiguously? From the structure data we present here, there is only one connection between helices H and F that is sufficiently convincing to be used as a constraint in model building (Figure 5). Another constraint is that Glu14 in helix 1 of one EmrE monomer should be close to Glu14 of the other monomer because they are proposed to bind simultaneously to the substrate and the helices have been shown to be parallel in spin-labelling studies (Koteiche et al., 2003). The only other data that could be used is cross-linking data, which suggests that helix 4 in one monomer is close and parallel to helix 4 in another monomer, and that helix 1 from one monomer can cross-link with helix 4 from another monomer (Soskine et al., 2002). Unfortunately the published cross-linking data cannot be used to discriminate between likely models of EmrE; under the conditions used for the cross-linking experiments (unpurified detergent-solubilized EmrE), it is not clear whether EmrE is a tetramer, which is the proposed state of EmrE in the membrane, or a dimer, which is the oligomeric state of EmrE after purification in detergent. We will therefore not discuss detailed helix assignments, but we will consider how the monomers could pack together to form the asymmetric dimer.
A first consideration is how the eight helices in the 3D map are allocated to the two monomers. Since those assignments involving helices interpenetrating between monomers are unlikely, we can envisage four possible boundaries between the monomers (Figure 5). In the arrangement in model 1, densities A-B-C-D comprise one monomer, whilst densities E-F-G-H comprise the other monomer. The other possibilities divide the densities as follows: C-D-E-F with A-B-G-H (model 2); A-B-C-H with D-E-F-G (model 3); and B-C-D-E with A-F-G-H (model 4). Models 2 and 3 seem less likely because our map shows that helices F and H are connected by a loop, indicating that these two helices are in the same monomer. Although model 4 fulfils the F to H connection, in our map helix A is seen to have no contacts except with helix B (Table II), suggesting that this configuration is unlikely. Even though we cannot exclude the other possibilities with any certainty, model 1 is the most likely based on the above criteria. Furthermore, there is significant similarity in packing between helices A-B-C-D and E-F-G-H (Table II). The inter-helical packing angles between pairs A/B, B/C and C/D compared to H/G, G/F and F/E are quite similar, and the sequence of signs of the crossover angles are the same.
Table II. Packing angle and axis-to-axis distance between helices.
| Helices | Packing angle (°) | Axis-to-axis distance (Å) | Level of closest approach | |
|---|---|---|---|---|
| A | B | +19 | 9 | Centre |
| B | C | –43 | 9 | Centre |
| C | D | +16 | 9 | Top (+Z) |
| H | G | +18 | 9 | Centre |
| G | F | –32 | 9 | Centre |
| F | E | +31 | 11 | Centre |
| D | E | +15 | 9 | Extended contact |
| C | F | –13 | 9 | Bottom (–Z) |
| B | G | –34 | 17 | Bottom (–Z) |
| A | H | –48 | 14 | Centre |
| B | H | –53 | 9 | Bottom (–Z) |
| A | G | –32 | 19 | Top (+Z) |
| F | H | –32 | 9 | Bottom (–Z) |
In considering the alternatives in helix identity in the monomers A-B-C-D and E-F-G-H, we will apply the constraint that those faces of helices involved in helix–helix contacts in a given interaction should not face lipid in another. At least two possible arrangements satisfy the above criteria for most of the interactions, and we will refer to them as the ‘upside-down’ and the ‘rotated’ model; they are discussed below. If the above constraint is weakened, then a plausible ‘side-by-side’ model can be constructed in which density A in one polypeptide corresponds to density H in the other, and density D corresponds to density E. The highly hydrophobic nature of EmrE means that the surfaces of the α-helices could either face the lipid exterior or the hydrophobic binding chamber, but it is unexpected that the surfaces might all be used so asymmetrically. This possibility therefore cannot be ruled out given the novelty of the EmrE asymmetric homodimer.
In the ‘upside-down’ model, the two monomers are related by a rotation of ∼180° about an axis roughly in the membrane plane. In this arrangement the helix represented by density A in one monomer corresponds to H in the other, but is inverted in the membrane. Such a model proposes a novel type of packing arrangement within a membrane protein homodimer. Support for this model comes from the striking conservation of packing between the densities F, G and H compared with A, B and C after rotation about an axis roughly in the membrane plane. After this transformation, F-G-H can be superimposed onto A-B-C, but a different geometrical transformation is required to superimpose D on to E. Evidence that membrane proteins can exist in vivo in opposite orientations has been described previously both in vivo (ductin; Dunlop et al., 1995) and in designed membrane proteins (Nilsson and von Heijne, 1990; Gafvelin and von Heijne, 1994). Neither is there any distinctive distribution of positive charge in EmrE to suggest an orientation more likely to face the cytoplasm according to the ‘positive-inside’ rule (von Heijne, 1986). It is also intriguing that in some atomic resolution structures of channels and transporters, there are structural motifs that appear to have evolved by a gene duplication event followed by an inversion of topology of the membrane protein domain, resulting in an antiparallel architecture (Fu et al., 2000; Dutzler et al., 2002). The ‘upside-down’ model is thus a tantalizing possibility and, if this were the case, the tetramer could comprise dimers related by an in-plane 2-fold symmetry axis (dimers 1 and 3; Figure 4).
In the ‘rotation’ model, a rotation of ∼180° about an axis approximately perpendicular to the membrane plane relates the region A-B-C-D to the region E-F-G-H, such that A is related to E, B is related to F, C to G and D to H. The interactions between helices within each of these monomers are similar but not identical. Furthermore, the interactions between the monomers will be asymmetric in that whilst E is close to D, and F is close to C throughout their lengths, the related pairs A–H and B–G will be separated by the binding pocket for TPP+. Therefore, the surfaces of helices involved in these two types of interactions would need to be capable of two different types of contact. We have one constraint from the fact that helix 1 from each monomer should be located at the interface between monomers to donate residue Glu14 to the TPP+ binding chamber, which means that helix 1 is placed at either B and F or at C and G.
It should be emphasized that in none of the models presented above do the monomers have identical structures, nor do they make entirely symmetrical interactions with the adjacent monomer. It seems reasonable to assume that the structure of free monomeric EmrE is not the same as that of either monomer in the dimer, and that the observed asymmetry arises with dimer formation. Further work will be required in order to identify which helix corresponds to which density, and to understand the nature of the helix–helix interactions that underlie the asymmetry of the EmrE homodimer.
Mechanism of transport
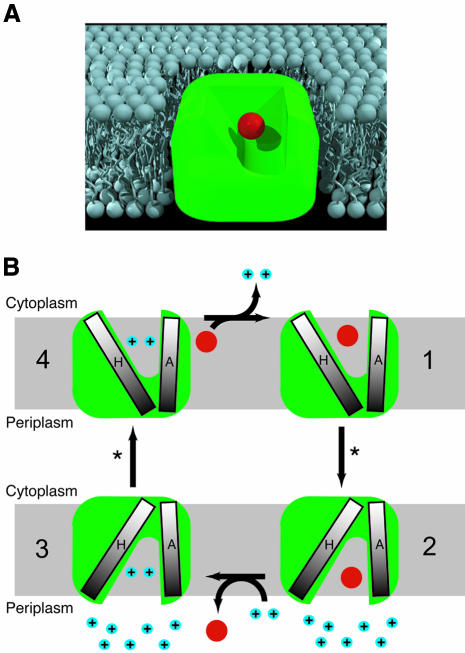
The axes of the six helices (A–C and F–H) forming the substrate binding chamber are all within 9–13 Å of the centre of the density attributed to TPP+, and are probably all involved in substrate recognition and translocation. We have crystallized EmrE in a single conformation, but it is not possible to define whether the binding chamber opens towards the periplasm or cytoplasm, because the resolution is insufficient to directly assign densities to amino acid sequence. Thus the structure of EmrE presented here could represent either the outward-facing conformation awaiting the displacement of TPP+ by protons, or it could represent the cytoplasmic-facing structure awaiting a conformational change to re-orient the binding chamber to open towards the periplasm. The proposed mechanism of transport involves the binding of TPP+ by the two glutamate residues (Glu14) located in helix 1 of adjacent monomers in the asymmetric dimer (Figure 6). The substrate enters EmrE either from the inner leaflet of the cytoplasmic membrane or directly from the cytoplasm; direct binding of protons to both Glu14 residues induces the release of substrate at the periplasmic surface. The chamber where TPP+ binds is sealed at the top by helix H; the creation of an opening on the opposite side of the membrane from the chamber entrance would require the movement of this helix, and the proximity of helices G and F could mean they move as a group.
Fig. 6. Transport mechanism in EmrE. (A) Cartoon of EmrE based upon the 3D structural analysis, with TPP+ (depicted by a red sphere) bound in the centre of a cavity open to the cytoplasm and one leaflet of the lipid bilayer. (B) Schematic indicating the minimal structural changes during electrogenic TPP+ efflux suggested by the 3D structure. EmrE binds TPP+ from either the inner leaflet of the membrane or from the cytoplasm (1) and a conformational change (*) involving the change in tilt of helix H relative to helix A occurs to open the binding chamber to the periplasm (2). Proton binding displaces TPP+ into the periplasm (3), and a second proton-driven conformation change (*) re-orients the binding site to face the cytoplasm (4). TPP+ is represented as a red ball and protons as blue balls, labelled ‘+’.
What could be the relevance of EmrE asymmetry for transport? One possibility is that the two Glu14 residues essential for transport have slightly different environments in an asymmetric structure, and this non-equivalence is important in the mechanism. Another possibility is that one conformation of a monomer during the transport cycle is unstable, and therefore requires the other monomer to stabilize it; a similar rationale has been proposed for the asymmetric heterodimer formed in the HIV-1 reverse transcriptase (Wang et al., 1994). Higher resolution structures of different conformational states will be required to elucidate the importance of the asymmetry for proton-linked drug efflux by EmrE.
Materials and methods
EmrE was overexpressed in E.coli and purified in dodecylmaltoside as described previously (Muth and Schuldiner, 2000; Tate et al., 2001). Two-dimensional crystals of EmrE were obtained by the addition of dimyristoylphosphatidylcholine and dialysis to remove detergent for 10–14 days at 25°C against buffer containing 20 µM TPP+. The resulting tubular p2 crystals have been described previously (Tate et al., 2003). The crystals were placed on molybdenum grids coated with 4-day-old carbon, blotted in 1% glucose, briefly dried, and rapidly frozen in liquid nitrogen. The grids were transferred to a liquid nitrogen-cooled Gatan 626 cold stage, and low-dose electron micrographs of specimens tilted from 0 to 40° were recorded on a Tecnai F30 electron microscope at an accelerating voltage of 300 keV. Images recorded on Kodak S0-163 film were collected either with flood beam or spot-scan illumination at a nominal magnification of ×57 400 and underfocus ranging from 2000 to 16 000 Å. Crystal quality was assessed by optical diffraction and the best images were digitised using a Zeiss SCAI scanner with a 7-µm step size. The MRC image-processing package (Crowther et al., 1996) was used to analyse 47 individual images by correcting for lattice distortions and the effects of the contrast transfer function. Image data were then merged in plane group p2, and the tilt geometries and phase origin were refined using information to a resolution of 7.0 Å through an iterative process to produce lattice lines and a 3D map. All images were then processed a second time using projections calculated from the refined 3D model corresponding to the unrefined 3D map as the reference for a more accurate correction of the lattice distortions as described previously (Kunji et al., 2000). The procedure did not alter any of the gross features in the 3D structure, but it significantly improved the vertical resolution and reduced the background noise. The final 3D density map was calculated to 7.0 Å resolution after two more cycles of geometry, phase origin and contrast function refinement. The 3D density map was analysed in the environment of O (Jones et al., 1991).
Acknowledgments
Acknowledgements
We are indebted to R.Henderson for support and help throughout this work, J.Berriman for advice on electron microscopy and J.Li for assistance with graphics procedures. A European Molecular Biology Organisation Long-Term Fellowship funded I.U.-B. S.S. is supported by grants from the National Institutes of Health (NS16708) and from the Israel Science Foundation (463/00).
References
- Abramson J., Smirnova,I., Kasho,V., Verner,G., Kaback,H.R. and Iwata,S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science, 301, 610–615. [DOI] [PubMed] [Google Scholar]
- Arkin I.T., Russ,W.P., Lebendiker,M. and Schuldiner,S. (1996) Determining the secondary structure and orientation of EmrE, a multi-drug transporter, indicates a transmembrane four-helix bundle. Biochemistry, 35, 7233–7238. [DOI] [PubMed] [Google Scholar]
- Chang G. (2003) Structure of MsbA from Vibrio cholera: a multidrug resistance ABC transporter homolog in a closed conformation. J. Mol. Biol., 330, 419–430. [DOI] [PubMed] [Google Scholar]
- Chang G. and Roth,C.B. (2001) Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science, 293, 1793–1800. [DOI] [PubMed] [Google Scholar]
- Cizman M. (2003) The use and resistance to antibiotics in the community. Int. J. Antimicrob. Agents, 21, 297–307. [DOI] [PubMed] [Google Scholar]
- Crowther R.A., Henderson,R. and Smith,J.M. (1996) MRC image processing programs. J. Struct. Biol., 116, 9–16. [DOI] [PubMed] [Google Scholar]
- Dunlop J., Jones,P.C. and Finbow,M.E. (1995) Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J., 14, 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzler R., Campbell,E.B., Cadene,M., Chait,B.T. and MacKinnon,R. (2002) X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature, 415, 287–294. [DOI] [PubMed] [Google Scholar]
- Elkins C.A. and Nikaido,H. (2003) 3D structure of AcrB: the archetypal multidrug efflux transporter of Escherichia coli likely captures substrates from periplasm. Drug Resist. Updat., 6, 9–13. [DOI] [PubMed] [Google Scholar]
- Fu D., Libson,A., Miercke,L.J., Weitzman,C., Nollert,P., Krucinski,J. and Stroud,R.M. (2000) Structure of a glycerol-conducting channel and the basis for its selectivity. Science, 290, 481–486. [DOI] [PubMed] [Google Scholar]
- Gafvelin G. and von Heijne,G. (1994) Topological ‘frustration’ in multispanning E. coli inner membrane proteins. Cell, 77, 401–412. [DOI] [PubMed] [Google Scholar]
- Hirai T., Heymann,J.A.W., Shi,D., Sarker,R., Maloney,P.C. and Subramaniam,S. (2002) Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol., 9, 597–600. [DOI] [PubMed] [Google Scholar]
- Huang Y., Lemieux,M.J.S.,J., Auer,M. and Wang,D.-N. (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science, 301, 616–620. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Koteiche H.A., Reeves,M.D. and McHaourab,H.S. (2003) Structure of the substrate binding pocket of the multidrug transporter EmrE: site-directed spin labeling of transmembrane segment 1. Biochemistry, 42, 6099–6105. [DOI] [PubMed] [Google Scholar]
- Kunji E.R., von Gronau,S., Oesterhelt,D. and Henderson,R. (2000) The three-dimensional structure of halorhodopsin to 5 Å by electron crystallography: a new unbending procedure for two-dimensional crystals by using a global reference structure. Proc. Natl Acad. Sci. USA, 97, 4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebendiker M. and Schuldiner,S. (1996) Identification of residues in the translocation pathway of EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem., 271, 21193–21199. [DOI] [PubMed] [Google Scholar]
- Locher K.P., Lee,A.T. and Rees,D.C. (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science, 296, 1091–1098. [DOI] [PubMed] [Google Scholar]
- Mordoch S.S., Granot,D., Lebendiker,M. and Schuldiner,S. (1999) Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J. Biol. Chem., 274, 19480–19486. [DOI] [PubMed] [Google Scholar]
- Murakami S., Nakashima,R., Yamashita,E. and Yamaguchi,A. (2002) Crystal structure of bacterial multidrug efflux transporter AcrB. Nature, 419, 587–593. [DOI] [PubMed] [Google Scholar]
- Muth T.R. and Schuldiner,S. (2000) A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J., 19, 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. and von Heijne,G. (1990) Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell, 62, 1135–1141. [DOI] [PubMed] [Google Scholar]
- Paulsen I.T., Skurray,R.A., Tam,R., Saier,M.H.,Jr, Turner,R.J., Weiner,J.H., Goldberg,E.B. and Grinius,L.L. (1996) The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol., 19, 1167–1175. [DOI] [PubMed] [Google Scholar]
- Rotem D., Sal-man,N. and Schuldiner,S. (2001) In vitro monomer swapping in EmrE, a multidrug transporter from Escherichia coli, reveals that the oligomer is the functional unit. J. Biol. Chem., 276, 48243–48249. [DOI] [PubMed] [Google Scholar]
- Schuldiner S., Granot,D., Mordoch,S.S., Ninio,S., Rotem,D., Soskin,M., Tate,C.G. and Yerushalmi,H. (2001) Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. News Physiol. Sci., 16, 130–134. [DOI] [PubMed] [Google Scholar]
- Schwaiger M., Lebendiker,M., Yerushalmi,H., Coles,M., Groger,A., Schwarz,C., Schuldiner,S. and Kessler,H. (1998) NMR investigation of the multidrug transporter EmrE, an integral membrane protein. Eur. J. Biochem., 254, 610–619. [DOI] [PubMed] [Google Scholar]
- Soskine M., Steiner-Mordoch,S. and Schuldiner,S. (2002) Crosslinking of membrane-embedded cysteines reveals contact points in the EmrE oligomer. Proc. Natl Acad. Sci. USA, 99, 12043–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C.G., Kunji,E.R., Lebendiker,M. and Schuldiner,S. (2001) The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 Å resolution. EMBO J., 20, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C.G., Ubarretxena-Belandia,I. and Baldwin,J.M. (2003) Conformational changes in the multidrug transporter EmrE associated with substrate binding. J. Mol. Biol., 332, 229–242. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1986) The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J., 5, 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Smerdon,S.J., Jager,J., Kohlstaedt,L.A., Rice,P.A., Friedman,J.M. and Steitz,T.A. (1994) Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl Acad. Sci. USA, 91, 7242–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R., Hart,T., Cars,O., Streulens,M., Helmuth,R., Huovinen,P. and Sprenger,M. (1998) Antimicrobial resistance. Is a major threat to public health. Br. Med. J., 317, 609–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi H. and Schuldiner,S. (2000) A model for coupling of H(+) and substrate fluxes based on ‘time-sharing’ of a common binding site. Biochemistry, 39, 14711–14719. [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Lebendiker,M. and Schuldiner,S. (1995) EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem., 270, 6856–6863. [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Lebendiker,M. and Schuldiner,S. (1996) Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem., 271, 31044–31048. [DOI] [PubMed] [Google Scholar]
- Zgurskaya H.I. and Nikaido,H. (2002) Mechanistic parallels in bacterial and human multidrug efflux transporters. Curr. Protein Pept. Sci., 3, 531–540. [DOI] [PubMed] [Google Scholar]