Abstract
Large T antigen is the replicative helicase of simian virus 40. Its specific binding to the origin of replication and oligomerization into a double hexamer distorts and unwinds dsDNA. In viral replication, T antigen acts as a functional homolog of the eukaryotic minichromosome maintenance factor MCM. T antigen is also an oncoprotein involved in transformation through interaction with p53 and pRb. We obtained the three-dimensional structure of the full-length T antigen double hexamer assembled at its origin of replication by cryoelectron microscopy and single-particle reconstruction techniques. The double hexamer shows different degrees of bending along the DNA axis. The two hexamers are differentiated entities rotated relative to each other. Isolated strands of density, putatively assigned to ssDNA, protrude from the hexamer–hexamer junction mainly at two opposite sites. The structure of the T antigen at the origin of replication can be understood as a snapshot of the dynamic events leading to DNA unwinding. Based on these results a model for the initiation of simian virus 40 DNA replication is proposed.
Keywords: cryoelectron microscopy/DNA replication/helicases/large T antigen/SV40
Introduction
Large T antigen (Tag) of simian virus 40 (SV40) is a multifunctional protein involved in several processes that eventually lead to the replication of the virus inside eukaryotic cells. Among its functions Tag has the ability to interfere with key cellular regulatory pathways resulting in the deregulation of the cell cycle and tumorogenesis in part by blocking p53 and the retinoblastoma protein (pRB) function (Simmons, 2000).
SV40 has been extensively used as a model system for the study of replication in eukaryotes for two main reasons. First, the origin of replication for SV40 genome, unlike that of the eukaryotic cells, is very well characterized and is specifically recognized by Tag. Secondly, the host cell provides all the other components necessary to form the primosome for initiating DNA replication, which include single-stranded DNA binding proteins (Replication Protein A or RPA), topoisomerase I and DNA polymerase α-primase (for a review see Simmons, 2000). Tag can be envisaged as a functional homolog of eukaryotic MCM protein (minichromosome maintenance factor), since both unwind duplex DNA and assemble as double hexamers (Fletcher et al., 2003). However, there are differences between Tag and MCM, such as those regarding origin recruitment (Tye, 1999), which involve a series of different proteins that assemble into different complexes, making Tag a simpler and more accessible model system for the study of eukaryotic DNA replication.
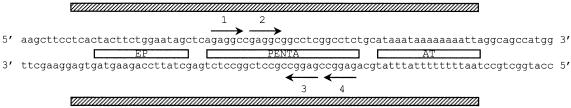
In the first step of replication, Tag performs key functions: it specifically binds to the viral origin of replication (ori) and then unwinds the DNA as a helicase (Stahl et al., 1986). There are several known steps during the process. First, Tag recognizes several 5′-GAGGC-3′ sequences located at the PENTA region of the ori (Figure 1); it is not clear whether it then assembles as a double hexamer in a cooperative process triggered by adenosine nucleotides (Deb and Tegtmeyer, 1987) or binds as a preformed ring (Uhlmann-Schiffler et al., 2002). As a result, one of the hexamers is bound to the early palindrome (EP) site of the ori (Deb and Tegtmeyer, 1987), stimulating the binding of the second hexamer on the adjacent AT-rich side. This ATP-dependent assembly of Tag at the SV40 ori induces structural distortions in the PENTA flanking regions. In two separate but concurrent reactions the EP melts and the AT-rich region untwists with no need for ATP hydrolysis (Borowiec and Hurwitz, 1988). Then, in a process coupled to ATP hydrolysis (Goetz et al., 1988), Tag further unwinds bidirectionally the DNA duplex with a 3′→5′ polarity. Thus Tag combines in a single protein several functions that in Escherichia coli are carried out by three different proteins in replication, namely DnaA, DnaB and DnaC (Kornberg and Baker, 1992).
Fig. 1. DNA probe used in this study that includes the minimal SV40 origin of replication (ori) marked with striped bars. The three main regions of the ori, outlined as open bars, are PENTA, with the four 5′-GAGGC-3′ sequences shown as arrows, the AT-rich region (AT) and the early palindrome region (EP).
A wide range of techniques has been used for the structural characterization of several aspects of Tag. These include scanning transmission microscopy (Mastrangelo et al., 1989), transmission electron microscopy (Wessel et al., 1992; San Martin et al., 1997; Valle et al., 2000; VanLoock et al., 2002) and atomic force microscopy (Mastrangelo et al., 1994). Atomic coordinates for several Tag domains are available: the DNA origin binding domain (Luo et al., 1996), the J domain (Kim et al., 2001) and the structure of a single hexamer of the Tag helicase domain (Li et al., 2003). The last of these, a DNA- free structure, has revealed the structural basis for hexamerization and has also given important clues to a possible mechanism for ori disruption and DNA unwinding.
Our group has focused on the structural characterization of Tag double hexamers assembled on the SV40 ori, a complex formed in the first step of viral replication, using electron microscopy and image processing techniques. Our previous 2D structural studies (Valle et al., 2000) showed important features of this nucleoprotein complex, such as its overall shape and size, and provided the first mapping of its functional domains. Here we present the first 3D structure of the full-length Tag double hexamer binding the SV40 ori in the presence of ADP, determined using cryoelectron microscopy (cryo-EM) and single-particle reconstruction techniques. The 3D map shows an extremely flexible double-hexamer structure, with a prominent curvature along the DNA axis. The two Tag hexamers within the complex are not structurally identical, suggesting a DNA-induced conformational change. The fitting of the atomic coordinates of the Tag helicase domain (Li et al., 2003) into the 3D cryo-EM density map ascertained that the C-terminal domains are rotated relative to each other in the complex. Most interestingly, strands of density, attributable to single stranded DNA (ssDNA), exit mainly from two opposite sites at the interface of the hexamers and possibly make contact with the distal C-terminal domains. Taking into account our results and the biochemical data available, an integrated model for the initiation of viral DNA replication is proposed.
Results and discussion
Average structure of the Tag double hexamer on the SV40 origin of replication
The functional complexes formed by Tag double hexamers assembled on the SV40 ori in the presence of ADP were imaged as frozen-hydrated specimens in a cryoelectron microscope. A representative field is shown in Figure 2A. An average density map of the Tag–ori complex at 27.5 Å resolution (see Supplementary figure S1A available at The EMBO Journal Online), which includes 13 860 particle images, was calculated (Figure 2B–E) as described in Materials and methods.
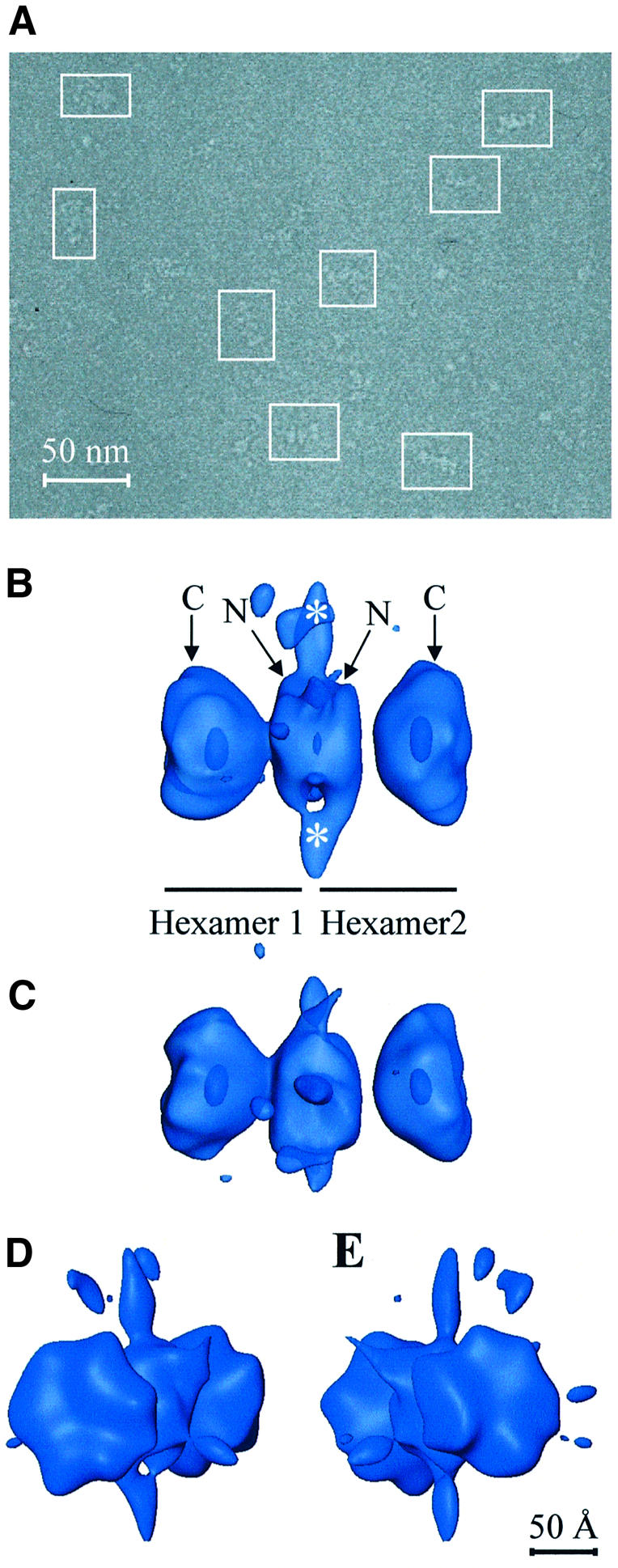
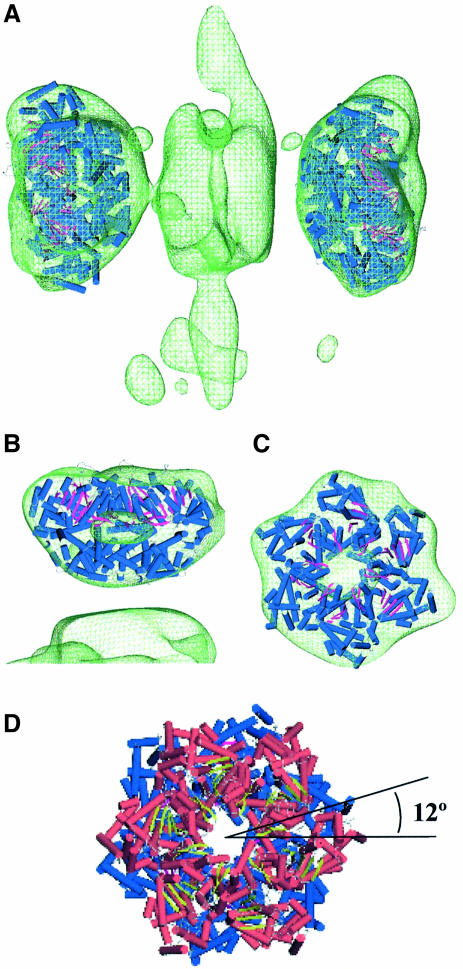
Fig. 2. Tag double hexamer on the SV40 origin of replication. (A) General field of one of the untilted micrographs used in this study, with some of the selected particles outlined. (B and C) Side views of the Tag–ori complex in a semitransparent display. Annotations indicate each of the two hexamers and their corresponding C-terminal (C) and N-terminal (N) domains. Masses protruding from the hexamer–hexamer junction are marked with white asterisks. Hexamers 1 and 2 have been arbitrarily assigned. (D and E) Oblique views showing a close-up of the two C-terminal domains. In all cases 125% of the expected protein mass is shown.
The distinctive characteristics of this initial reconstructed volume confirmed our previous 2D analysis performed on negatively stained specimens (Valle et al., 2000). Clearly, the Tag–ori complex is a bilobed structure made up of two hexamers arranged in a head-to-head orientation. Our previous assignment of the N-terminal and C-terminal domains, based on monoclonal antibody labeling (Valle et al., 2000), can be directly transferred to the reconstructed volume (Figure 2B). Thus each lobule, a single hexamer, consists of two different parts: a larger and distal one with a clear inner cavity, which corresponds to the C-terminal domain, and a smaller one in the region where both hexamers interact with each other, which corresponds to the N-terminal domain. The C-terminal domain interacts with p53 and DNA polymerase α-primase and has the ATPase and helicase domains, while the N-terminal domain contains the RPA binding domain and the DNA origin binding domain that recognizes and binds to the pentanucleotides of the SV40 ori (Simmons, 2000).
The structures of the C-terminal domains in the double hexamer are not completely identical, despite the fact that both hexamers are made up of six copies of the same polypeptide. This could indicate that either the structure of this region might be dependent on the DNA sequence with which it is interacting or that it possesses intrinsic structural flexibility, as was observed in X-ray studies of this domain (Li et al., 2003).
The transition from the N-terminal to the C-terminal domains in each hexamer goes through a low-density region (Figure 2B and C). According to the recently reported structure of the C-terminal region of Tag (Li et al., 2003), this connection corresponds to amino acids 255–266 in the sequence of the protein, a putative flexible linker between the origin binding and helicase domains of Tag. In our map, this connection is clearly seen in one of the hexamers (e.g. hexamer 1 in Figure 2B) while it is lost in the other (e.g. hexamer 2 in Figure 2B). It seems that through this flexible linker the N-terminal and C-terminal domains of the Tag hexamer achieve nearly complete structural independence, perhaps reflecting the assumed functional separation between ori binding and melting/untwisting of the flanking regions based on the observation that mutation of the Tag ori binding domain does not disrupt origin melting (Chen et al., 1997) and that the C-terminal domain has helicase activity (Li et al., 2003).
Strikingly, at the interface between the hexamers, in the connecting region between the N-terminal domains, clear strands of density protrude away (labeled with asterisks in Figure 2B). These are seen mainly at two opposite points (data not shown).
During the image processing procedure no symmetry was imposed. The analysis of the rotational power spectra of sections taken along the longitudinal axis of the C-terminal domain of each hexamer (data not shown) revealed a preferential 2-fold symmetry, with some small areas of 3-fold symmetry. No pure 6-fold symmetry was found in the complex, underlining the unsuitability of imposing 6-fold symmetry during the reconstruction process (VanLoock et al., 2002). The existence of a preferential 2-fold symmetry and a minor 3-fold symmetry in the C-terminal domains of the complex would indicate that at least at this level the Tag polypeptide would arrange as a trimer of dimers. Interestingly, Li and coworkers (Li et al., 2003) noted in their X-ray crystallography studies of the DNA-free helicase domain that three dimers come together around the crystallographic 3-fold axis to form a hexamer.
Fitting of the helicase domain in the cryo-EM map
The high-resolution structures of several Tag domains are known, including the solution structure of the DNA origin binding domain as a monomer (Luo et al., 1996), the J domain crystallized as a monomer in complex with a portion of the Rb protein (Kim et al., 2001) and the X-ray structure of the hexamer formed by six C-terminal (helicase) domains (Li et al., 2003). During the fitting, we found that it is only possible to fit the hexameric helicase domain into our structure with high confidence because the N-terminal regions, where the DNA origin binding and J domains are localized, are not completely defined in our 3D map, and there are no restrictions in shape that could shed light to the way in which these domains are arranged in the complex.
The atomic coordinates of the hexamer formed by the Tag helicase domain (Li et al., 2003), residues 266–627, were fitted into the C-terminal domains of the double-hexamer electron density map (Figure 3A). The overall agreement in shape between each C-terminal domain and the fitted atomic coordinates is high, as revealed by the cross-correlation values of 0.83 and 0.89 in hexamer 1 and hexamer 2, respectively. Moreover, the measure of resolution using the C-terminal domain of hexamer 2 and the fitted atomic coordinates in this hexamer also reflect this concordance (see Supplementary figure S1B for more details). Figure 3B and C presents a close-up of the fitting performed on hexamer 2, which has a higher cross-correlation value for the fitting. The central cavities observed in the map correspond well to the cavity described in the X-ray structure and localize in the same region (Figure 3B and C). The N-terminal end of the X-ray structure points to the mass at the center of the double hexamer where Tag N-terminal domains are located. Owing to the differences in resolution it is not possible to observe the six lateral exit holes described in the X-ray structure and therefore a comparison between these two structures at this level is not feasible.
Fig. 3. Fitting of the C-terminal helicase domain into the average volume. (A) The crystal structure of Tag hexameric helicase domain (shown in terms of its secondary structure elements, cylinder for α-helix and arrow for β-strand) was fitted into the cryo-EM map of the double hexamer (green wire-mesh). (B and C) Side view and top view of the close-up for the fitting on hexamer 2, where an outer section of the EM map has been removed for clearer visualization of the inner cavity. (D) The atomic coordinates of Tag helicase domain fitted into hexamer 1 (in red) and hexamer 2 (in blue) without the cryo-EM map to depict the relative rotation of around 12° among the C-terminal domains. In all cases the Tag–ori complex is shown in translucent wire-mesh representations accounting for 125% of the expected mass.
Relative rotation of the C-terminal domains
The fitted atomic coordinates of Tag helicase domains clearly showed a relative rotation (Figure 3D). The measurement of such rotation gave an angle of 12° (data not shown). In the cryo-EM map, when slices corresponding to the cavity regions from both hexamers were compared, a relative rotation of around 10°–15° between the two C-terminal domains was also found (data not shown). Since the resolution of the cryo-EM map does not allow a straightforward interpretation of this rotation, the following two scenarios are possible: (i) each hexamer binds the ori in an interlocked way, i.e. the N-terminal domains are not in register, or (ii) the N-terminal domains are in register and so the rotation comes from the C-terminal domain and/or the linker region. In the first case, the phase difference between the hexamers is probably due to the recognition of the pair of pentanucleotides in the center of the ori. The initial binding of Tag to ori is preferentially supported by the interaction with pentanucleotide pairs 1–3 or 2–4 (Joo et al., 1998). The pentanucleotides in each pair are situated in the same face of the double-stranded DNA (dsDNA), but with a phase shift of ∼72° (considering 10 bp per turn or 36° per nucleotide). Taking into account the fact that the six monomers of Tag are indistinguishable, a rotation of 72° would be equivalent to an apparent relative rotation of 12° between the hexamers. In the second case, the N-terminal domains would bind in register, and so the rotation of the C-terminal domains might either be the result of opposed torsions in the flexible linkers or a genuine relative rotation of the distal parts of the C-terminal domains with respect to the proximal parts.
Flexibility of the Tag–ori complex
The cryo-EM map described in Figure 2B–E reached only a moderate resolution despite the high number of CTF-corrected images used. Furthermore, the Tag–ori complex was known to display an intrinsic variability in its degree of bending and its total length along the main particle axis (Valle et al., 2000). These facts led us to consider possible sources of structural heterogeneity within the sample, such as different structural conformations or intrinsic flexibility of the nucleoprotein complex. The way to ascertain this issue is to calculate volumes from different types of particles (bent or straight) using the random conical tilt (RCT) scheme (Radermacher et al., 1987). This technique allows de novo computation of a volume without any prior knowledge of its structure. If these two types of particles, bent or straight, give two different RCT reconstructions, then it conclusively indicates that there is intrinsic structural heterogeneity in the sample. If, on the other hand, the two RCT volumes look the same, then the bent and straight particles are merely different views of the same structure.
Thus two RCT reconstructions were calculated using pairs of images recorded at 0° and 50° tilt. The appearance of the particles in the 0° images (bent or straight) was used as the basis for the classification, while the particle projections in the 50° images were used to calculate the reconstructions (see Supplementary figure S2 for details). Set 1 comprised those untilted images that showed straight particles, whereas Set 2 comprised those that showed bent particles. The reconstruction calculated from Set 1 is straight when visualized from any point of view, and all projection images generated from this reconstruction show a straight structure. In contrast, the volume calculated from Set 2 shows a bent structure when viewed from adequate angles, and is also bent in some of its projections. This is a conclusive proof for the presence of structural heterogeneity. Therefore the total population of particles, which was used to generate the average volume, clearly consists of various subpopulations in different states of bending.
To analyze these variations in three dimensions, a classification approach was followed based on the KerDenSOM algorithm (Pascual-Montano et al., 2001) with a subgroup, the side views, of the total particle population used to calculate the reconstruction shown in Figure 2 (see Materials and methods). A total of some 4200 side views (i.e. particles lying flat on the grid) were initially considered and fed to the KerDenSOM algorithm. The corresponding output map is shown in Figure 4A, where different degrees of bending along the longitudinal axis of the double hexamer can be observed. No mask (rectangular or any kind) was applied to the particles in the calculation of this map. The algorithm looks for heterogeneities within the particle’s perimeter, including the whole of the particle plus as little as possible of the background so as to avoid a classification based on spurious features that might be present in the background of the entire image (see Materials and methods). In order to define homogeneous ‘classes’ by grouping images corresponding to similar code vectors from the output map of the KenDerSOM algorithm, the cross-correlation coefficient among neighboring code vectors was computed, grouping only those images corresponding to code vectors which were strongly correlated (see Materials and methods). Four different classes were obtained in this way (containing varying numbers of particles, with 352 in the least populated and 961 in the highest populated). The average images of the selected classes are presented in Figure 4B. Separate reconstructions were calculated for each of the four groups using the previously assigned alignment parameters with regard to the average volume (Figure 2B). These preliminary volumes were then used in a 3D multireference alignment performed on a subset of 7615 particles that had values of the angle θ between 70° and 110° (particles that lay flat on the grid with a rocking of ±20°). This resulted in four newly defined groups, with populations ranging from 2580 to 1410 particles. New separate reconstructions were calculated and then used as reference for a complete angular search followed by an angular refinement procedure (see Materials and methods). The reconstructions had improved resolution and good angular coverage (see Supplementary figures S1A and S3).
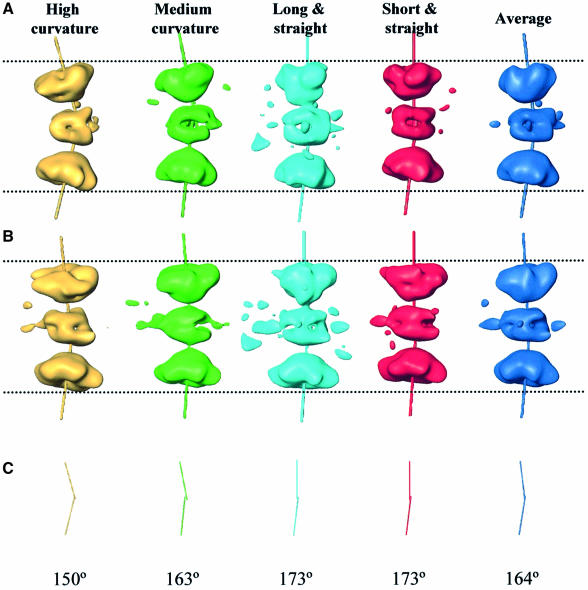
Fig. 4. 2D classification of Tag–ori side views using a KerDenSOM algorithm. (A) Self-organizing map of Tag–ori side views. The four groups used to reconstruct the four different volumes are outlined in different colors. (B) 2D averages of the images coming for the groups outlined in (A). Note that in the case of the most bent and medium bent groups two views are shown. The color code is the same as in Figure 5.
These four reconstructions show structures with different lengths as well as different degrees of bending along their main longitudinal axis (Figure 5A and B). The C-terminal domain of each hexamer within the volume had its own rotational symmetry axis. The angle between these two axes within a double hexamer varies, ranging from 150° to 173° (Figure 5C).
Fig. 5. Flexibility and structural heterogeneity of the Tag–ori complex. (A) The four volumes reconstructed after 3D multireference alignment and angular refinement. The rotational symmetry axis for each C-terminal domain is also shown. (B) A 90° view when rotating along the main longitudinal axis. (C) Angles formed by the two rotational symmetry axes within each complex. The volumes are displayed showing 100% of the expected protein mass.
The four reconstructions represent the Tag double hexamer assembled at the SV40 ori. They portray the macromolecular assembly responsible for the initiation of DNA replication as an extremely flexible and structurally heterogeneous complex. In fact, the ori DNA undergoes extensive remodeling (untwisting of the AT region and melting of the EP region) upon Tag binding prior to the initiation of replication (Borowiec and Hurwitz, 1988). Therefore it is not surprising that the protein complex would be able to accommodate and sustain changes that occur in ori DNA through its great flexibility and capacity to bend.
The four structures calculated in this study have been obtained such that the specimens that contribute to them are in approximately the same structural conformation. However, at the level of resolution of this study, it is impossible to rule out some degree of heterogeneity within each class. Indeed, the most likely situation is that the bending of Tag occurs along a path with more or less continuous curvature, so that the volumes shown (Figure 5A) would then correspond to four ‘snapshots’ taken along a rather continuous variable trend.
ssDNA around the Tag–ori complex
Preliminary studies carried out in our laboratory indicated the possibility of the presence of ssDNA outside the Tag–ori complex. In an attempt to calculate a 3D reconstruction of a complex that was free of any loose ssDNA, the Tag–ori samples were treated, prior to the vitrification step, with Mung Bean nuclease, an enzyme that specifically degrades ssDNA. However, this treatment resulted in the complete disassembly of the dodecameric complex, as no double hexamers were observed in the electron microscope in the nuclease-treated samples. This observation strongly suggests that ssDNA is important in the maintenance of the structural integrity of the nucleoprotein complex, and also that this ssDNA is positioned somewhere outside the double hexamer since it was accessible to the nucleolytic enzyme. In our five reconstructions (i.e. the one from the total pool and the individual ones, for each of the four classes defined), where the complex has not been treated with nucleases, several low-density masses were observed projecting away from the junction between the two hexamers in the Tag–ori complex (Figures 2B and C and 5A). These masses are not artifacts due to a poor orientation sampling in Fourier space because the angular coverage is extremely good in the average reconstruction and more than adequate in the class reconstructions (see Supplementary data for more details). We interpret these density protrusions as exiting ssDNA at the interface between the two hexamers. The rest of the nucleic acid would not be visible further away from the hexamer owing to its natural flexibility. It is worth noting that when the volume is contoured at a lower density value, protrusions seem to appear from the distal part of the C-terminal domain, although, at this resolution, we refrain from considering it as a proven fact, but rather treat it as a possibility to be further investigated and confirmed.
Loops of ssDNA have also been observed in Tag helicase assays using a plasmid containing SV40 ori DNA as a substrate, monitored by electron microscopy (Wessel et al., 1992; Seinsoth et al., 2003). In these studies the two loops of ssDNA appear to emerge from, and return to, the helicase and they were described as ‘rabbit-ear-containing structures’. Although helicase activity is not possible in our samples, since they were prepared in the presence of ADP, these observations lend further support to our interpretation of the protruding masses as ssDNA.
An integrated model for the initiation of DNA replication
It is very well established that, during the initiation of SV40 DNA replication, the ori region undergoes extensive Tag-dependent structural remodeling, a process that is not yet fully understood and for which data are sometimes contradictory. This process renders the DNA accessible to the rest of the proteins of the replication machinery.
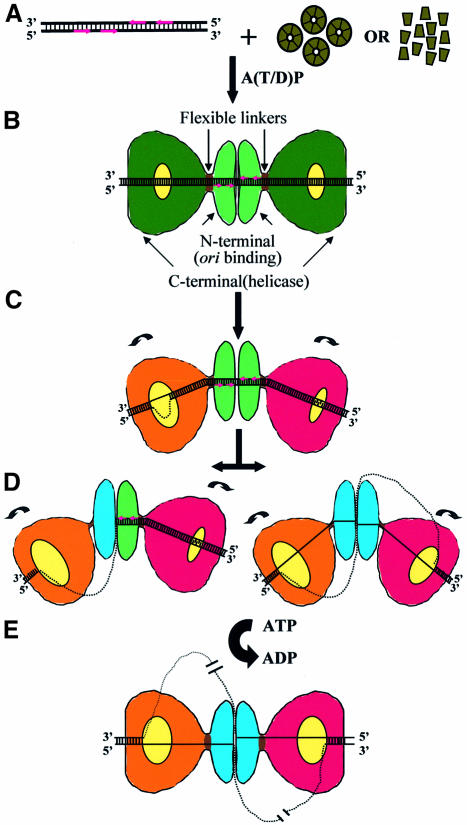
Our results on the 3D structural analysis of the Tag–ori complex and the known biochemical and other structural data are integrated in a model for the initiation of SV40 DNA replication (Figure 6). Tag initiates SV40 DNA replication by specific recognition and binding to the four pentanucleotides in the presence of adenosine nucleotides, either as a preformed hexamer (Uhlmann-Schiffler et al., 2002) (a ring-opening mechanism would then be needed) or as a succession of monomers (Simmons, 2000) (Figure 6A). The result is a double hexamer assembled on the origin of replication arranged in a head-to-head orientation (Figure 6B). In this way the N-terminal (ori binding) domains of each hexamer would cover the appropriate pairs of pentanucleotides, while the C-terminal (helicase) domains would cover the flanking EP and AT-rich regions. Upon double-hexamer formation, the EP region would melt and the AT region would undergo local untwisting (Borowiec and Hurwitz, 1988), events thought to precede strand separation in the pentanucleotide region (Figure 6C). The initial alteration of the flanking regions could be mediated by the intrinsic dynamic properties observed in the C-terminal domains of Tag (Li et al., 2003; this work) and facilitated by the capacity of the complex to adopt varying degrees of bending along the DNA axis. At this point the two hexamers would have already adopted different conformations to perform the task of disrupting the DNA (indicated by the coloring scheme in Figure 6). It is also possible that the inner chambers within each C-terminal domain might change their shape and size to accommodate the emerging modifications in the DNA structure. The putative capability of the C-terminal domains to rotate might also help to destabilize the DNA flanking regions. At this stage, the pentanucleotides would remain as dsDNA. The next step or steps would lead to the structure shown in this work, in which masses attributable to ssDNA are observed exiting the interhexamer junction mainly at two opposite sites, in the absence of ATP hydrolysis. This would imply (Figure 6D) that at least part of the core ori must have changed from dsDNA to ssDNA, since the mass assigned to ssDNA emerges from the N-terminal domains interface where the pentanucleotides (the specific binding sites) are located. Probably, the angle between the two hexamers changes as the ori remodeling proceeds.
Fig. 6. Model of the initiation of SV40 DNA replication. See text for details.
It is not known how the ori achieves unwinding. Perhaps the remodeling of the flanking regions induces a rearrangement of the N-terminal domains that results in a change in the binding mode from dsDNA to ssDNA. As a result, the N-terminal domain (which might have changed its conformation, as indicated by the different coloring in Figure 6D) encircles a piece of ssDNA, while the other DNA strand is displaced from the hexamer. This consideration would imply that at least one of the adjacent flanking regions, likely the EP region, must have previously progressed in its remodeling to the form of fully ssDNA. The corresponding C-terminal domain would then also encircle one piece of ssDNA through sugar–phosphate contacts (SenGupta and Borowiec, 1994), probably via the positively charged residues present in the C-terminal chamber (Li et al., 2003). As a result, the other DNA strand is displaced from the hexameric channel at the C-terminal level through an as yet unknown mechanism. The six side-holes observed in the wall of the chamber within the helicase domain (Li et al., 2003) might provide exiting points for the ssDNA from the C-terminal domain. The extensive structural flexibility of the Tag–ori complex as well as the possible ability of the C-terminal domains to rotate in the absence of energy would, again, be vital for the promotion and maintenance of this sequence of movements.
Our integrated model proposes that the C-terminal (helicase) domains on each end of the Tag double hexamer and the N-terminal (origin binding) domains are able to distort at least half of the SV40 ori (and possibly all of it) to produce fully ssDNA during the initiation of replication. The ssDNA loop(s) generated would be released through the sides of the C-terminal domains and the interface between the N-terminal domains, explaining the so-called ‘rabbit-ear-containing structures’ observed in the electron microscope by Wessel and coworkers (Wessel et al., 1992) as intermediates during DNA helicase reactions. The bubble of ssDNA outside the Tag–ori complex is then available for the access of other components of the replication machinery and the complex is ready for the next stage: the beginning of the helicase activity and the subsequent opening of the DNA in an ATP-fueled process (Figure 6E).
During DNA unwinding, the two hexamers could become separate entities that migrate in the 3′→5′ direction. Or, as suggested in Figure 6E, the complex would remain as a double hexamer with the ssDNA threaded through it, implying that the helicase cannot move along the DNA. Instead, the DNA would be translocated through the complex, which is consistent with the observation that replisomes are bound to the nuclear scaffold at replication centers (Cook, 1999; Falaschi, 2000). Also consistent with this notion, it has been reported that the double hexamers form a stable ternary complex with nucleolin and topoisomerase I (Seinsoth et al., 2003) and that the ternary complex has a ‘helicase-swivelase’ activity thought to be essential for DNA replication.
The reconstructed volume of the double hexamer of Tag on the SV40 ori presented here is the first 3D structure described for this macromolecular complex. Other structural studies of the Tag on DNA have been reported (VanLoock et al., 2002), but using a synthetic DNA probe that has no sequence or structure analogy to SV40 ori and thus of unknown physiological relevance. The results presented here show a highly flexible Tag double hexamer that is able to promote and accommodate all the structural rearrangements that the ori must undergo prior to replication. Our structure provides new evidence for displaced ssDNA exiting the complex from the inte-hexamer junction formed by the N-terminal domains at a very early time during the initiation of SV40 DNA replication. We suggest that these structures represent a gallery of 3D snapshot of the earliest stages in SV40 DNA replication.
Materials and methods
Formation of Tag–DNA complexes
Tag was purified as described (Simanis and Lane, 1985). The DNA probe containing the complete SV40 ori (Figure 1) was obtained by PCR amplification with the primers Oligo-NcoI (5′-CCA-TGG-CTG-ACT-AAT-TTT-TTT-TAT-TTA-TGC-AG-3′) and Oligo-HindIII (5′-AAG-CTT-TCT-CAC-TAC-TTC-TGG-AAT-AGC-3′), using pOR1 as a template (DeLucia et al., 1986). A solution of 922 nM Tag and 39 nM DNA was incubated in the presence of 3 mM ADP, in 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM KCl and 2 mM MgCl2 buffer for 1 h at 37°C, and then dialyzed against the incubation buffer.
Cryoelectron microscopy
The sample was vitrified on a carbon-coated copper grid that was previously glow-discharged. Micrographs were taken under low-dose conditions on a Tecnai F20 field emission gun at 200 kV, at a magnification of 64 200×. Images taken at 50°, 10° and 0° tilt, with defocus ranging from 1.2 to 4.5 µm, were used for the final reconstruction. Micrographs were digitized on a SCAI flatbed scanner (Zeiss, Germany) with a sampling window corresponding to 3.27 Å per pixel on the object scale. The image processing was performed using Xmipp (Marabini et al., 1996) and SPIDER (Frank et al., 1996) software packages.
Initial reconstruction using the random conical tilt scheme
A preliminary reconstruction was obtained using the random conical tilt (RCT) scheme (Radermacher et al., 1987). Pairs of images were collected at 50° and 0°. The 0° images were analyzed with a novel neural-network-based self-organizing map algorithm termed KerDenSOM. This algorithm is able to produce a sampling of the probability density function of the image particle population in a quantitative and defined manner (Pascual-Montano et al., 2001). In this way, a homogeneous population of around 500 particles was selected. This group was then translationally aligned by cross-correlation using a low-pass filtered disk with a diameter close to the longitude of the particle, and rotationally aligned by cross-correlation with a low-pass filtered average image obtained with a reference-free alignment algorithm (Marco et al., 1996). This procedure yielded the in-plane rotation angle. The translational alignment of the 50° tilted images was done by cross-correlation between the untilted partner and the tilted-specimen image stretched by a factor of 1/cos θt (where θt is the tilt angle) in the direction perpendicular to the tilt axis. The tilted-specimen projections, thus aligned, were used to calculate a 3D volume using ART with blobs (Marabini et al., 1998) with an imposed 6-fold symmetry to improve the signal-to-noise ratio.
Three-dimensional reconstructions of Tag/ori complexes
A total of 14 140 particles from micrographs recorded at 50° (944), 10° (1839) and 0° (11 357) tilt were translationally aligned using Radon transforms (Radermacher, 1994), with the preliminary RCT reconstruction low-pass filtered to 40 Å used as a reference. An initial search for projection orientations was done using Radon transforms in increments of 12° for the three Eulerian angles over the complete angular range. At the end of each search, the volume was updated and the process was started again. Six of these rounds were performed. Since the images were taken at several defoci, a correction of the contrast transfer function (CTF) was performed (Radermacher et al., 2001). The astigmatism and defocus of each micrograph was calculated, and the CTF was corrected by flipping the sign of the Fourier components. A new reconstruction was calculated using the corrected images, and was then used as a starting reference for a final set of refinement steps. The angular increment and the search intervals in the three angles was gradually decreased until more than 95% of the images had stable values of angles and the increments reached 1°. The angular coverage is shown in Supplementary figure S3A. Some particles were discarded during the refinement procedure, so that 13 860 particles were finally used.
The final 3D reconstruction was calculated using ART with blobs (Marabini et al., 1998), using a λ value of 0.01 and a radius of 47 pixels, and then filtered to the estimated final resolution. No symmetry was applied. The resolution was estimated by randomly splitting the dataset in two groups, calculating separate volumes and comparing them in Fourier space. The resolution was based on a 0.5 cut-off for the Fourier shell correlation curve (Saxton and Baumeister, 1982), with a final value of 27.5 Å (see Supplementary figure S1A). The threshold values containing the different percentages of the predicted molecular mass were calculated assuming a value of 1.33 g/cm3 as the average density of a protein (Squire and Himmel, 1979).
Study of the structural heterogeneity
In order to sort out the total particle image population into more homogeneous structural classes the following procedure was used. First, all the side views used to calculate the Tag–ori reconstruction were selected (a side view was defined as a view coming from a projection direction such that the largest dimension of the particle, its ’axis’, made an angle between 80° and 100° with the electron beam). Secondly, these selected particles were subjected to a KerDenSOM variability analysis. Thirdly, four groups were defined in the output map on the basis of the degree of bending and the total length of the side views. The numbers of particles in each group were 961, 904, 688 and 352, respectively. Finally, separate reconstructions were calculated for each of the groups using the previously assigned alignment parameters. These four reconstructions were used as reference volumes for a 3D multireference alignment using a dataset of 7614 particles which θ angles were between 70° and 110°. In this procedure, each of these particles was confronted with the four volumes in a complete angular search, and then the decision as to which volume they best belonged to was made according to the size of the best cross-correlation coefficient. After regrouping the particles into new four subsets, an angular refinement procedure was done to allow each particle to find its best position in the new volume. The distribution of φ angles (the rotation angle around the main longitudinal axis of the particle) in each volume after all the cycles of angular alignment provides a good indication of the achieved angular coverage (see Supplementary figure S3B–E). The four volumes shown in Figure 5 were calculated using ART with blobs and λ = 0.062. Each volume had a different number of particles: 2579 for the volume with highest curvature, 2048 for the one with medium curvature, 1411 for the straight and long, and 1576 for the straight and short. The resolution (calculated as previously described) was 30.5 Å for the straight and longest volume and 29.5 Å for the others (Supplementary figure S1A).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We specially thank Dr Michael Radermacher and Dr Ellen Fanning for their comments on the manuscript. Helpful discussions with our colleagues Dr Yolanda Robledo, Dr Carmen San Martín and Dr Montserrat Bárcena are very much appreciated. This work was partially supported by grant CAM 07B/0032/2002 from Comunidad de Madrid (to J.M.C.), grant BIO2001–1237 from the Spanish CICYT (to J.M.C.) and grant R37 GM29169 from the NIH (to J.F.). M.G.G.-L. has a postdoctoral fellowship from the Consejería de Educación of the Comunidad de Madrid, Spain. L.E.D. and M.V. hold contracts from the ‘Ramón y Cajal’ program implemented by Ministerio de Ciencia y Tecnología, Spain.
References
- Borowiec J.A. and Hurwitz,J. (1988) Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J., 7, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Joo,W.S., Bullock,P.A. and Simmons,D.T. (1997) The N-terminal side of the origin-binding domain of simian virus 40 large T antigen is involved in A/T untwisting. J. Virol., 71, 8743–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- Deb S.P. and Tegtmeyer,P. (1987) ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J. Virol., 61, 3649–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A.L., Deb,S., Partin,K. and Tegtmeyer,P. (1986) Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J. Virol., 57, 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A. (2000) Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet., 16, 88–92. [DOI] [PubMed] [Google Scholar]
- Fletcher R.J., Bishop,B.E., Leon,R.P., Sclafani,R.A., Ogata,C.M. and Chen,X.S. (2003) The structure and function of MCM from archaeal M.thermoautotrophicum. Nat. Struct. Biol., 10, 160–167. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher,M., Penczek,P., Zhu,J., Li,Y., Ladjadj,M. and Leith,A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Goetz G.S., Dean,F.B., Hurwitz,J. and Matson,S.W. (1988) The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem., 263, 383–392. [PubMed] [Google Scholar]
- Joo W.S., Kim,H.Y., Purviance,J.D., Sreekumar,K.R. and Bullock,P.A. (1998) Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol., 18, 2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., Ahn,B.Y. and Cho,Y. (2001) Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J., 20, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A. (1992) DNA Replication. W.H.Freeman, New York. [Google Scholar]
- Li D. et al. (2003) Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature, 423, 512–518. [DOI] [PubMed] [Google Scholar]
- Luo X., Sanford,D.G., Bullock,P.A. and Bachovchin,W.W. (1996) Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat. Struct. Biol., 3, 1034–1039. [DOI] [PubMed] [Google Scholar]
- Marabini R., Masegosa,I.M., San Martin,M.C., Marco,S., Fernandez,J.J., de la Fraga,L.G., Vaquerizo,C. and Carazo,J.M. (1996) Xmipp: An image processing package for electron microscopy. J. Struct. Biol., 116, 237–240. [DOI] [PubMed] [Google Scholar]
- Marabini R., Herman,G.T. and Carazo,J.M. (1998) 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements (blobs). Ultramicroscopy, 72, 53–65. [DOI] [PubMed] [Google Scholar]
- Marco S., Chagoyen,M., de la Fraga,L.G., Carazo,J.M. and Carrascosa,J.L. (1996) A variant of the ‘random approximation’ of the reference-free alignment algorithm. Ultramicroscopy, 66, 5–10. [Google Scholar]
- Mastrangelo I.A., Hough,P.V., Wall,J.S., Dodson,M., Dean,F.B. and Hurwitz,J. (1989) ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature, 338, 658–662. [DOI] [PubMed] [Google Scholar]
- Mastrangelo I.A., Bezanilla,M., Hansma,P.K., Hough,P.V. and Hansma,H.G. (1994) Structures of large T antigen at the origin of SV40 DNA replication by atomic force microscopy. Biophys. J., 66, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Montano A., Donate,L.E., Valle,M., Barcena,M., Pascual-Marqui,R.D., Carazo,J.M. (2001) A novel neural network technique for analysis and classification of EM single-particle images. J. Struct. Biol., 133, 233–245. [DOI] [PubMed] [Google Scholar]
- Radermacher M. (1994) Three-dimensional reconstruction from random projections: orientational alignment via Radon transforms. Ultramicroscopy, 53, 121–136. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht,T., Verschoor,A. and Frank,J. (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc., 146, 113–136. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Ruiz,T., Wieczorek,H. and Gruber,G. (2001) The structure of the V(1)-ATPase determined by three-dimensional electron microscopy of single particles. J. Struct. Biol., 135, 26–37. [DOI] [PubMed] [Google Scholar]
- San Martin M.C., Gruss,C. and Carazo,J.M. (1997) Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol., 268, 15–20. [DOI] [PubMed] [Google Scholar]
- Saxton W.O. and Baumeister,W. (1982) The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc., 127, 127–138. [DOI] [PubMed] [Google Scholar]
- Seinsoth S., Uhlmann-Schiffler,H. and Stahl,H. (2003) Bidirectional DNA unwinding by a ternary complex of T antigen, nucleolin and topoisomerase I. EMBO Rep., 4, 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta D.J. and Borowiec,J.A. (1994) Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J., 13, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V. and Lane,D.P. (1985) An immunoaffinity purification procedure for SV40 large T antigen. Virology, 144, 88–100. [DOI] [PubMed] [Google Scholar]
- Simmons D.T. (2000) SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res., 55, 75–134. [DOI] [PubMed] [Google Scholar]
- Squire P.G. and Himmel,M.E. (1979) Hydrodynamics and protein hydration. Arch. Biochem. Biophys., 196, 165–177. [DOI] [PubMed] [Google Scholar]
- Stahl H., Droge,P. and Knippers,R. (1986) DNA helicase activity of SV40 large tumor antigen. EMBO J., 5, 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Uhlmann-Schiffler H., Seinsoth,S. and Stahl,H. (2002) Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res., 30, 3192–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Gruss,C., Halmer,L., Carazo,J.M. and Donate,L.E. (2000) Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol., 20, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLoock M.S., Alexandrov,A., Yu,X., Cozzarelli,N.R. and Egelman,E.H. (2002) SV40 Large T antigen hexamer structure, domain organization and DNA-induced conformational changes. Curr. Biol., 12, 472–476. [DOI] [PubMed] [Google Scholar]
- Wessel R., Schweizer,J. and Stahl,H. (1992) Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol., 66, 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]