Abstract
The Hfq protein, which shares sequence and structural homology with the Sm and Lsm proteins, binds to various RNAs, primarily recognizing AU-rich single-stranded regions. In this paper, we study the ability of the Escherichia coli Hfq protein to bind to a polyadenylated fragment of rpsO mRNA. Hfq exhibits a high specificity for a 100-nucleotide RNA harboring 18 3′-terminal A-residues. Structural analysis of the adenylated RNA–Hfq complex and gel shift assays revealed the presence of two Hfq binding sites. Hfq binds primarily to the poly(A) tail, and to a lesser extent a U-rich sequence in a single-stranded region located between two hairpin structures. The oligo(A) tail and the interhelical region are sensitive to 3′–5′ exoribonucleases and RNase E hydrolysis, respectively, in vivo. In vitro assays demonstrate that Hfq protects poly(A) tails from exonucleolytic degradation by both PNPase and RNase II. In addition, RNase E processing, which occurred close to the U-rich sequence, is impaired by the presence of Hfq. These data suggest that Hfq modulates the sensitivity of RNA to ribonucleases in the cell.
INTRODUCTION
The degradation of messenger RNA in Escherichia coli is carried out by a combination of endoribonucleolytic cleavages and 3′ to 5′ degradation, primarily by polynucleotide phosphorylase (PNPase) and RNase II. It is widely accepted that mRNA decay is initiated by a series of endonucleolytic cleavages catalysed by RNase E, or occasionally by RNase III, followed by processive exonucleolytic degradation of the message. The rpsO mRNA, which encodes ribosomal protein S15, has been used as a model for the study of mRNA decay. An RNase E cleavage located 10 nucleotides downstream of the coding sequence (M2 site) initiates the mRNA decay by removing the 3′ stabilizing terminal hairpin that protects the mRNA against the attack of exonucleases (1). The body of the messenger is then further degraded by the combined actions of poly(A) polymerase I (PAP I), polynucleotide phosphorylase (PNPase) and RNase II (2,3). Poly(A) tails were detected at the termini of nascent terminated mRNA, and at the end of decay intermediates produced by endonucleolytic and exonucleolytic digestion (4). Poly(A) tails promote decay of rpsO mRNA and of other structured RNAs by providing an unstructured 3′ end, to which 3′ exonucleases bind [for a review see (5)]. The degradation is controlled by RNA binding proteins (6), RNA helicases (7), ribosomes (8,9) and secondary structures (10–12).
The pleiotropic phenotype of hfq– strains shows that the Hfq protein acts on different pathways of E.coli metabolism (13,14). Hfq has been reported to be involved in the regulation of iron content (15,16), the expression of the σS subunit of RNA polymerase upon entry into stationary phase, the response to hyperosmolarity and low temperature (17–19), mRNA stability at slow growth-rate (20) and stationary phase (21), and in controlling the activity of regulatory sRNAs under stress conditions (see below). Hfq is known to stabilize some RNAs (22) and to target others for degradation (15,21,23). In the case of the ompA messenger, Hfq interferes with ribosome binding and unmasks an RNase E cleavage site, destabilizing the transcript (12). On the other hand, Hfq stimulates processivity of poly(A) polymerase I (24). We have shown that Hfq affects the position, frequency and length of oligo(A) tails at the 3′ end of rpsO mRNA in vivo (25). However, the failure to detect a direct interaction between Hfq and poly(A) polymerase I (M.Folichon, V.Arluison and E.Hajnsdorf, unpublished results) led us to the notion that the formation of a poly(A)–Hfq complex may stimulate poly(A) elongation by PAP I.
Hfq affects gene expression either by interacting directly with mRNA and/or by promoting annealing between regulatory RNAs that it directs to target messengers. The propensity of Hfq to bind single-stranded A–U-rich sequences (26) is exemplified by the direct interactions between Hfq and the OxyS, DsrA and Spot42 sRNAs (22,27,28). Hfq somehow facilitates the annealing of these riboregulators with their target mRNAs, namely the rpoS, fhlA and galK mRNAs, respectively. On the other hand, in the case of Qβ RNA, Hfq facilitates the access of replicase, presumably by melting its 3′ end (29,30). Similarly, Hfq binding weakens base-pairing in stem loops of OxyS sRNA (31).
In an attempt to understand the role of Hfq in the polyadenylation of prokaryotic mRNA, we have compared the interaction between Hfq and a polyadenylated or a non-polyadenylated mRNA fragment. This fragment corresponds to the 3′ end of the rpsO transcript, which has been shown to be polyadenylated both in vivo and in vitro (4,24,32). Measurements of dissociation constants show that Hfq exhibits a high affinity for rpsO mRNA and binds more steadily to molecules harbouring poly(A) tails. Consistently, Hfq binds preferentially to poly(A) tails and interacts with an internal site located between the two hairpins of the polyadenylated RNA at higher concentrations. Moreover, we show that the poly(A) tail and the internal binding site are protected by Hfq from ribonucleases, namely PNPase, RNase II and RNase E, which are known to trigger the degradation of this mRNA in the cell (2,32,33). Our findings raise the possibility that the high affinity of Hfq for polyadenylated RNA may play a general role in RNA maturation and degradation.
MATERIALS AND METHODS
Protein purification
Hfq was overproduced from strain BL21 (DE3) transformed with pTE607 plasmid (kindly provided by T. Elliott). Cells from induced cultures were resuspended at 4°C in 20 ml buffer containing 20 mmol/l Tris–HCl (pH 7.8), 500 mmol/l NaCl, 10% glycerol (v/v) and 0.1% Triton X-100 (v/v). The suspension was passed through a French press (1200 bar, 20 000 psi) and submitted to centrifugation (30 min at 15 000 g). Imidazole–HCl (pH 7.8) was added to the supernatant to a final concentration of 1 mmol/l. The resultant suspension was applied to a 1 ml Ni2+-NTA column (Qiagen). The resin was then sequentially washed with ∼15 column volumes of: (i) 20 mmol/l Tris–HCl (pH 7.8) buffer containing 0.3 mol/l NaCl and 20 mmol/l imidazole; and (ii) 50 mmol/l sodium phosphate (pH 6.0) buffer containing 0.3 mol/l NaCl. Hfq was eluted with buffer containing 50 mmol/l sodium phosphate (pH 6.0), 0.3 mol/l NaCl and 250 mmol/l imidazole. Fractions containing Hfq were determined by SDS–PAGE analysis, pooled and heated to 80°C for 15 min. Insoluble material was removed by centrifugation and the supernatant was dialyzed against the buffer containing 50 mmol/l Tris–HCl (pH 7.5), 1 mmol/l EDTA, 50 mmol/l NH4Cl, 5% glycerol (v/v) and 0.1% Triton X-100 (v/v). The protein was kept at 4°C.
RNase II was overproduced from BL21 (DE3) strain transformed with plasmid GC100, (kindly provided by G. Mackie) and purified as described previously (34).
Protein concentrations were determined by using the Bradford protein assay with bovine serum albumin as a standard. Hfq concentration was calculated on the basis of the monomer form.
PNPase was a gift from C. Portier and F. Allemand, and RNase E, purified as described previously (35), was a gift from A.J. Carpousis.
RNA preparation and labelling
The 97RNA, referred to as the ΔrpsO mRNA, corresponds to the last 97 nucleotides of the rpsO transcript, with a GGG sequence at the 5′ end. This RNA and its polyadenylated derivative were synthesized by T7 RNA polymerase as described previously (24). An extended polyadenylated ΔrpsO mRNA was transcribed using a PCR-amplified template, using the following primers: 5′-TAATACGACTCACTATAGGGAGACGTAGCACGTTACACC (upstream primer) and 5′-GATCCCGGGATCCACCACCAT18GAAAAAAGGGGCCACTCAGG (downstream primer). Underlined nucleotides correspond to the T7 RNA polymerase promoter. Transcription reactions were carried out either as described previously, yielding uniformly labelled RNA with high specific activity (24), or according to Ambion’s MEGAshortscript protocol and with [α-32P]UTP as tracer, except that 20 mmol/l guanosine was added to allow direct 5′-labelling. Radiolabelled RNAs were gel purified, eluted and resuspended in water. The percentage of radioactivity incorporated was determined and used to calculate RNA concentrations. 5′ end labelling was performed with [γ-32P]ATP and T4 polynucleotide kinase. Labelled RNAs were separated from unincorporated nucleotides using a ProbQuant G-50 Micro column (Pharmacia Biotech).
Heterogeneous poly(A) (Sigma) was separated on denaturating polyacrylamide gels. Bands were located by comparing with radioactive RNA markers. The gel was cut into horizontal strips and poly(A) RNA was eluted. Precipitated poly(A) was dissolved in water. Its length was determined by analysing 5′-end-labelled aliquots on sequencing gel along with RNA markers of known lengths and alkaline hydrolyzed poly(A). Due to the resolution of the preparative gel, the poly(A) size fractions contained different size distributions. Values are estimated mean values of each fraction. Poly(A) concentration was determined by spectrophotometry with the extinction coefficient taken from the Pharmacia catalogue.
Gel shift assays
Radiolabelled RNAs (5 pmol/l) were titrated against Hfq protein (15 pmol/l to 2 nmol/l, expressed as monomers) in 50 µl buffer containing 10 mmol/l Tris–HCl (pH 8), 1 mmol/l EDTA, 80 mmol/l NaCl, 1% glycerol (v/v), 0.01% dodecyl maltoside (v/v) at 37°C for 30 min. Reactions were loaded onto running native gels (27) and quantified using a PhosphorImager (Molecular Dynamics). The apparent dissociation constant (Kd) was determined for Hfq–rpsO RNA complexes, assuming that complex formation obeys a simple bimolecular equilibrium and the concentration of the labelled RNA is negligible compared to Hfq concentration. In practice, the reactions were performed with protein in excess over RNA, and thus the free Hfq concentration changed little during RNA binding. Therefore, the results were plotted using the equation:
Q = [Hfq]/(Kd + [Hfq])
Q represents the fraction of labelled RNA bound to Hfq. The apparent Kd values were calculated from at least three independent experiments. The percentage of free and complexed RNAs in each sample were deduced by counting each band and normalizing to the total radioactivity in each lane.
Chemical and enzymatic probing
Hfq-polyadenylated ΔrpsO mRNA complex formation was carried out with 105 c.p.m. of 5′-labelled RNA in the presence of 1 µg E.coli tRNA in 50 mmol/l HEPES–NaOH (pH 7.7), 1 mmol/l Mg–acetate, 80 mmol/l sodium acetate for 15 min at 37°C. The chemical reaction was performed in 10 µl in the presence of 16 mmol/l lead acetate (Merck) at 20°C for 5 min. The reactions were stopped by adjusting the sample to 50 mmol/l EDTA. RNase V1 (Pharmacia Biotech) (5 × 10–5 U) and RNase T2 (Sigma and Ambion) (0.02 U) were used for enzymatic hydrolysis. Incubation controls in the absence of the probes were carried out in parallel. All samples were extracted with phenol/chloroform and precipitated with ethanol. Pellets were counted, resuspended in 8 mol/l urea, 0.025% bromophenol blue (w/v) and 0.025% xylene cyanol blue (w/v), to load identical amounts of radioactivity on the sequencing gel. Cleavage positions were identified by running RNase T1 (Ambion) (indicated on the right side of panel C) and alkaline ladders of the same 5′-end-labelled RNA, performed under denaturing conditions, in parallel (36).
Assays for RNase II activity were performed in a 10 µl reaction volume containing 105 c.p.m. 5′-labeled RNA (∼100 fmol) and 1 µg of E.coli tRNA in reaction buffer containing 50 mmol/l HEPES–NaOH (pH 7.7), 1 mmol/l Mg–acetate, 80 mmol/l sodium acetate, in the absence and in the presence of 1 µmol/l Hfq, and samples were incubated at 37°C for 15 min. Next, 0.2 pmol RNase II was added, and samples were withdrawn at various times and quenched by phenol extraction. After precipitation, samples were resuspended in 8 mol/l urea, 0.1% xylene cyanol (w/v) and 0.1% bromophenol blue (w/v). Normalized amounts of RNA were separated on 8% sequencing gels and visualized using a PhosphorImager (Molecular Dynamics).
PNPase reactions were as for RNase II reactions, except that the buffer was supplemented with 10 mmol/l K2HPO4 and 2 pmol PNPase were added to initiate the enzymatic assay. The formation of a Hfq-polyadenylated ΔrpsO mRNA complex under these buffer conditions was monitored by band shift assay.
RNase E digestions were carried out in buffer containing 100 mmol/l NaCl, 10 mmol/l Tris–HCl (pH 7.5), 1% glycerol (v/v), 1 mmol/l MgCl2, 0.1% Triton (v/v), 0.2 mmol/l DTT, 1 µg of E.coli tRNA and 50 ng of a degradosome preparation for 5 min at 37°C.
RESULTS
Equilibrium dissociation constant measurement of complex formation by mobility shift assay
Polyadenylated and non-polyadenylated RNAs were synthesized to examine the contribution of a poly(A) tail to the binding of Hfq to RNA. We used an RNA fragment corresponding to the 3′ end of the rpsO transcript, which was shown to be polyadenylated both in vivo and in vitro (4,24,32). An 18-A tail was added to the 3′ end of this RNA for two reasons. First, it corresponds to the minimum size of the oligo(A) tails at the 3′ end of this fragment (ranging from 20 to 35 As) required to detect Hfq stimulation of poly(A) tail elongation by poly(A) polymerase I (24). Secondly, it fits the size of the tails found at the end of total RNA, which can be up to 50 nucleotides in length (37,38). The RNA, referred to as ΔrpsO mRNA, corresponds to the last 97 nucleotides of the rpsO primary transcript, and is predicted to contain two secondary structures separated by a single-stranded region (see Fig. 2 below). This RNA and its adenylated derivative containing an 18-residue poly(A) tail [ΔrpsO-(A)18] were incubated with increasing quantities of Hfq and of His6-Hfq. Overexpressed His6-Hfq had the same binding capacity as overexpressed native Hfq (data not shown), indicating that the histidine tag at the C-terminus of the protein does not perturb the RNA binding capacity of the protein. The tagged protein was used throughout this study, and is referred to as Hfq.
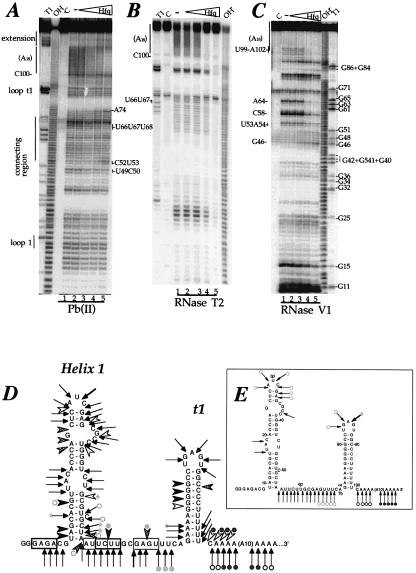
Figure 2.
Enzymatic and chemical probing of polyadenylated ΔrpsO-(A)18 mRNA, free and complexed to Hfq. (A) Chemical probing of ΔrpsO-(A)18 with Pb(II). (B) Enzymatic probing with RNase T2. (C) Enzymatic probing with RNase V1. 5′-end-labelled polyadenylated ΔrpsO-(A)18 mRNA with (A) or without a UGGUGGUGGAUCCCGGGAUC 3′-extension [In (B) and (C) the extension is not shown on the secondary structure]. Pb probing was also performed on the RNA lacking the 3′ extension. The extension did not alter the cleavage pattern (results not shown). RNase V1, free (lanes C, 1) or bound with increasing amounts of Hfq protein: 10 nmol/l (lanes 2), 100 nmol/l (lanes 3), 1 µmol/l (lanes 4) and 10 µmol/l (lanes 5). Samples were treated with RNase T2, RNase V1 or Pb(II), under conditions where statistically less than one cleavage per molecule takes place (lanes 1–5). Lanes T1 and OH– correspond to the sequencing products generated by RNase T1 cleavages of the same RNA (G-specific cleavage) and an alkaline hydrolysis ladder under denaturating conditions, respectively. (D and E) Summary of the probing results of three independent experiments, represented on the secondary structure of polyadenylated ΔrpsO mRNA. (D) Enzymatic cleavages by RNase V1: strong (filled arrowheads), moderate (grey arrowheads) and low (open arrowheads). Pb2+ cleavages: strong (bold-tailed arrows), moderate (thin-tailed arrows) and low (dashed-tailed arrows). Enhancement of cleavage is represented with an asterisk. Potential base-pairings that may occur between GAGA6 and UCUU60, and AUUC58 and GAGU66, are boxed in black and grey, respectively. (E) Enzymatic cleavages by RNase T2: strong (bold-tailed arrows) and moderate (thin-tailed arrows). Positions that became protected in the complex are denoted with filled circles for strong protection and open circles for weaker protection. Dark grey and light grey symbols reflect modification of cleavage intensity observed at low and high Hfq concentrations, respectively. Higher Hfq concentrations are required in footprinting experiments than in gel retardation assays because of tRNA addition.
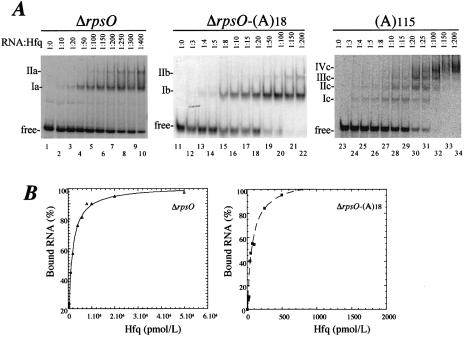
ΔrpsO or ΔrpsO-(A)18 RNAs were incubated with increasing concentrations of Hfq protein. The formation of complexes was analysed by mobility shift assay. Figure 1 shows that Hfq binds both non-polyadenylated and polyadenylated RNA. In both cases, two complexes were observed (referred to as Ia and IIa, and Ib and IIb in Fig. 1A), the second ones (IIa and IIb) forming at higher protein concentrations. The equilibrium dissociation constants (Kd) were deduced from the strongest binding site (Ia and Ib) for both ΔrpsO and ΔrpsO-(A)18 mRNAs, assuming a bimolecular model. The apparent binding affinity of Hfq for the adenylated RNA (Kd of 90 ± 16 pM) is 15 times higher than that for the non-polyadenylated RNA (1.4 ± 0.1 nM). It is likely that the binding of the protein to the poly(A) tail accounted for this difference. To address this point, we incubated Hfq with poly(A) RNA under the same conditions. We performed the titration experiment with poly(A)115, which has the same length as the polyadenylated RNA, ΔrpsO-(A)18. The mobility shift assay revealed the appearance of distinct intermediary complexes, which presumably correspond to the sequential filling of binding sites as the protein concentration is increased (Fig. 1A). These data suggest that Hfq binds non-cooperatively to the poly(A)115. A similar Hfq concentration is required to detect binding to either poly(A)115 or ΔrpsO-(A)18 RNA (Fig. 1A, lanes 13–14 and 24–25, respectively). Thus the formation of complex Ib with ΔrpsO-(A)18 is likely to be due to the binding of the protein to the polyadenylated extremity of the RNA. This result also implies that the ΔrpsO mRNA does not affect the binding of Hfq to its poly(A) tail and that a tail of 18 As is sufficient for efficient Hfq binding (24). Moreover, the fact that similar Hfq concentrations were required for the formation of complex Ia with non-polyadenylated ΔrpsO RNA (Fig. 1A, lane 7) and complex IIb with polyadenylated ΔrpsO-(A)18 RNA (Fig. 1A, lane 22), suggests that this latter complex corresponds to the interaction of the protein with an internal region of ΔrpsO RNA. We cannot determine, however, if complex IIa on Figure 1A results from sequential binding of Hfq to two sites on ΔrpsO RNA or if protein–protein interactions account for the appearance of complex IIa (39).
Figure 1.
Affinity of Hfq for ΔrpsO mRNA. (A) Non-polyadenylated ΔrpsO mRNA, polyadenylated ΔrpsO mRNA fragments [ΔrpsO-(A)18] and poly(A)115 were incubated with various concentrations of Hfq. Five pmol/l of 5′-end-labelled non-polyadenylated ΔrpsO mRNA (lanes 1–10), uniformly labelled polyadenylated ΔrpsO mRNA (lanes 11–22) and 5′ labelled poly(A)115 (lanes 23–34) were mixed with increasing concentrations of Hfq, ranging from 15 to 2000 pmol/l. RNA:Hfq molar ratios are indicated at the top. Complexes were separated on native polyacrylamide gels as described in Materials and methods. (B) The data were plotted using KALEIDAGRAPH 3.0.4 (Abelbeck Software, Reading, PA) and the generated curves were fitted by non-linear least squares regression assuming a bimolecular model such that the Kd values represent the protein concentration at half-maximal RNA binding. The data show one representative experiment of three performed to calculate the Kd value.
Enzymatic and chemical probing of polyadenylated RNA complexed to Hfq
Chemical and enzymatic probing was used to determine where Hfq binds on the polyadenylated RNA and whether Hfq binding causes conformational changes in the polyadenylated ΔrpsO mRNA. The polyadenylated RNA, free or bound to Hfq, was submitted to partial enzymatic hydrolysis with RNase V1 (specific for double-stranded regions) and RNase T2 (specific for unpaired nucleotides with a preference for adenines). The RNA was also subjected to Pb(II)-induced cleavages, which has proven to be exquisitely sensitive to subtle structural variations and which induces cleavages, mainly in interhelical regions, loops and bulged nucleotides. The experimental results and a schematic view of the secondary structure of the RNA are shown in Figure 2.
The data correlate well with the existence of the two hairpin structures (helix 1 and the terminator stem–loop, t1) connected by a single-stranded interhelical region (connecting region shown in Fig. 2A, D and E). Both helices are cleaved by RNase V1, whereas the single-stranded and loop regions are cleaved by both RNase T2 and Pb(II). The presence of several Pb(II) cleavages in helix I (C26–C28/G32–G34) and in helix II (A72–A73) suggests that these two regions are metastable. Finally, the presence of concomittant RNase V1 cleavages (at positions 53–54, 58 and 64) and single-stranded specific cleavages in region 53–60 suggests the existence of an equilibrium between unpaired and paired residues, resulting in the formation of alternative conformers. Indeed potential base pairings may occur either between residues GAGA6 and UCUU60 or between residues AUUC58 and GAGU66.
The addition of increasing concentrations of Hfq induced a strong reduction in the number of cleavages in the poly(A) tail region by Pb(II) and RNase T2 hydrolysis. Moreover, the RNase V1 cleavages at position 99–102 were strongly reduced by Hfq binding. These data indicate that the poly(A) tail is the major binding site of Hfq. Higher Hfq concentrations caused a significant reduction in Pb(II) cleavages in the U-rich sequence U66–U68. Hfq, concomitantly, prevented cleavages at positions U53–A54, C58 and A64 by RNase V1. Since Pb(II) cleavages were not affected by Hfq binding in region 55–60, a more open conformation of residues C52–U60 may be favoured. Subtle rearrangements induced by Hfq binding are also supported by the fact that weak, but reproducibly enhanced, cleavages by RNase V1 (at G46) and Pb(II) (at C52 and U53) were observed at the bottom of helix 1.
In summary, the results suggest that Hfq binds strongly to the poly(A) tail and to a lesser extent to the U-rich interhelical region, and induces subtle changes in the structure of the polyadenylated RNA.
Hfq binding protects poly(A) from RNase II- and PNPase-mediated decay
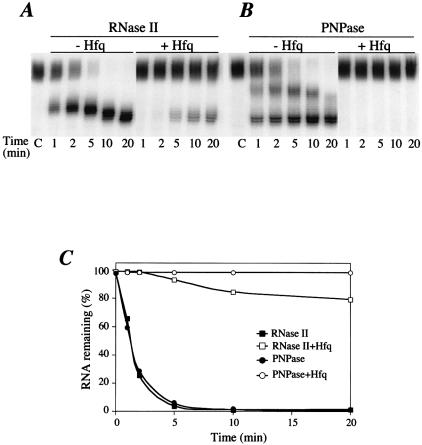
We next used RNase II, an exoribonuclease of 3′ to 5′ polarity that degrades single-stranded polyribonucleotides, to examine the effect of Hfq binding to the poly(A) tail. The rpsO mRNA fragment used as a substrate (see above) contained a tail of 18 As downstream of the stable terminator stem–loop, which protects the upstream RNA from attack by exonucleases. We confirm here that degradation of this transcript by RNase II is arrested by the terminator structure and show that Hfq protects the poly(A) tail from digestion by RNase II.
RNase II degraded the polyadenylated substrate in a two-step process. An initial rapid shortening from the poly(A) tail yielded a set of stalled intermediates (Fig. 3A, first three lanes) followed by a slower progressive shortening of the intermediates. These data suggest that the first step is processive, while the second step is distributive, i.e. RNase II dissociates from its substrate after the removal of each nucleotide. Carrying out the RNase II digestion with five times less enzyme confirms this two-step reaction (data not shown). The processive step of RNase II degradation generates intermediates with 3′ tails of approximately eight A-residues 3′ to the stable hairpin structure ending at C100 (Fig. 3A), in agreement with Coburn et al. (40). Then, RNase II generates stable intermediates distributively, with 3′ ends close to the base of the stem–loop structure. A comparison with a non-polyadenylated rpsO substrate used as a marker indicated that RNase II hydrolysis of the ΔrpsO-(A)18 RNA stopped three A-residues downstream of the secondary structure under these conditions (data not shown). In the presence of 1 µmol/l Hfq, 80% of the substrate was protected against RNase II degradation (Fig. 3A and C). We have calculated that Hfq concentration ranges from 8 to 20 µmol/l, depending on physiological conditions (41). This suggests that Hfq binding protects poly(A) tailed RNA by preventing the access of RNase II to the 3′ extremity of the RNA. Surprisingly, the few molecules that are still attacked by RNase II are shortened up to residues A4 and A5 of the oligo(A) tail, suggesting that processive degradation stops closer to the t1 hairpin. This is in agreement with the footprinting experiments, which suggest that Hfq binding weakens base-pairing at the bottom of the hairpin.
Figure 3.
Hfq impedes the poly(A) shortening activity of PNPase and RNase II. Digestion of 5′-labelled polyadenylated ΔrpsO mRNA in the absence (–Hfq) and presence of 1 µmol/l Hfq (+Hfq) was performed as described in Materials and methods. Aliquots were removed at 1, 2, 5, 10 and 20 min digestion. ‘C’ indicates the non-digested sample. (A) Digestion with 0.2 pmol RNase II and (B) with 2 pmol PNPase. We chose conditions in which each enzyme exhibits comparable exoribonuclease activity and in which the activity was limited. Lanes with sequence ladders and non-polyadenylated mRNA are not shown. (C) Quantification of experiments shown in (A) and (B).
We have shown previously that PNPase degradation of polyadenylated rpsO transcript is arrested three to five adenine residues downstream of the transcription terminator site in vivo (42). In vitro assays, using the polyadenylated substrate described above, also revealed a two-step degradation process by PNPase. The first step is characterized by the appearance of an intermediate, seven residues shorter than the initial substrate. This intermediate was further degraded to the final stalling site of PNPase, one or two adenines downstream of C100 (Fig. 3B). The addition of purified Hfq dramatically impeded PNPase digestion of poly(A) tails (Fig. 3B and C). Hfq may either mask the 3′ end of the RNA or prevent interactions between the enzyme and its RNA binding site.
Hfq binding decreases RNase E cleavage efficiency
Our data indicate that Hfq also binds to the 14-nucleotide-long sequence lying between the two hairpins of the polyadenylated ΔrpsO mRNA fragment, albeit with a weaker binding affinity than for the poly(A) sequence. In particular, the GCGAGUUU sequence, containing the M2 RNase E cleavage site (underlined) (1), is protected by Hfq from cleavage by RNase T2 and Pb(II). We performed in vitro processing experiments with a degradosome preparation containing RNase E to determine the effect of Hfq binding to this region. We omitted phosphate from the buffer to prevent exonucleolytic degradation of the RNA by PNPase present in the degradosome.
We first showed that most of the full-length RNA was efficiently cleaved using different concentrations of the degradosome, and concomitantly a shorter RNA fragment corresponding to the M2 RNase E cleavage site accumulated. Interestingly, the addition of increasing amounts of Hfq (10 nmol/l, 100 nmol/l, 1 µmol/l and 10 µmol/l) progressively reduced the processing of the polyadenylated ΔrpsO mRNA by RNase E from 89 to 20% (Fig. 4). This was also accompanied by a lesser amount of the RNase E main cleavage product as Hfq concentration increased. Furthermore, the non-adenylated ΔrpsO RNA was protected by Hfq binding against RNase E hydrolysis, suggesting that binding of Hfq to the poly(A) tail does not contribute to the inhibition of RNase E cleavage (results not shown). Altogether, these result suggest that Hfq binding to the connecting region inhibits RNase E cleavage by masking the GUUU sequence of the cleavage site. However, the Hfq binding effect was less pronounced on RNase E cleavage efficiency than on RNase II or PNPase hydrolysis. This may result from the fact that binding of Hfq to the GUUU sequence is weaker than for the poly(A) tail.
Figure 4.
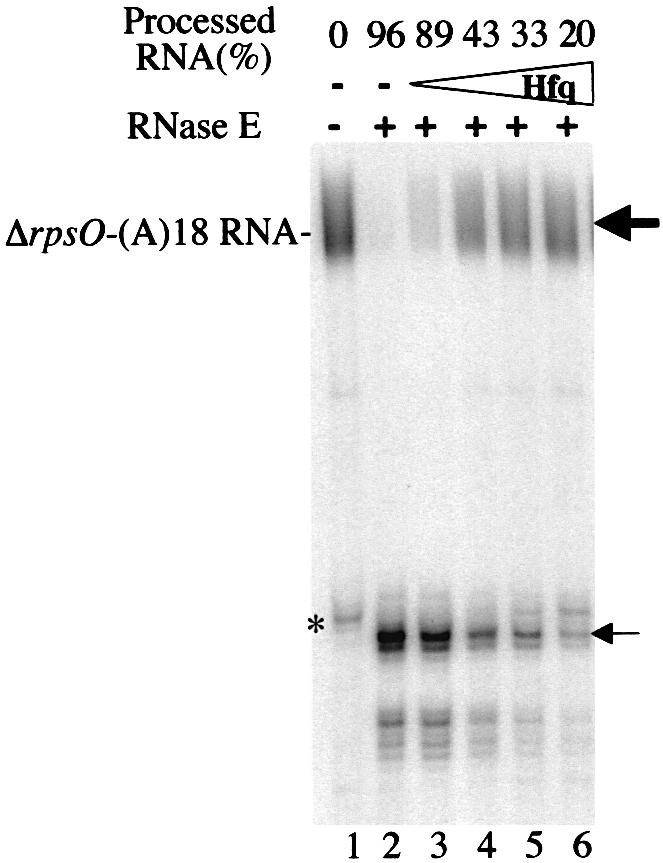
Hfq impedes the endonucleolytic activity of RNase E in the degradosome. 5′-end-labelled polyadenylated ΔrpsO-(A)18 mRNA (lanes 1) was incubated with 50 ng of the degradosome preparation in the absence (lane 2) and in the presence of 10 nmol/l (lane 3), 100 nmol/l (lane 4), 1 µmol/l (lane 5) and 10 µmol/l (lane 6) Hfq. The positions of 5′-labelled RNA and of the cleavage product by RNase E are indicated by the bold and normal arrows, respectively. Positions were determined relative to sequence ladders obtained by alkaline hydrolysis and RNase digestion, and to location of non-polyadenylated mRNA (not shown). The asterisk indicates a contaminant RNA of ΔrpsO-(A)18 RNA preparation, which is also cleaved by RNase E. The fraction of processed RNA is indicated at the top. This fraction was derived from the ratio of the amount of full-length labelled RNA divided by the total amount of radioactivity in each lane.
DISCUSSION
This report shows that Hfq forms stable complexes with rpsO RNAs and preferentially binds the poly(A) tail of polyadenylated mRNA. A second and weaker binding site is also detected in the single-stranded region just upstream of the transcription terminator hairpin. We further demonstrate that binding of these sites by the Hfq protein protects the polyadenylated rpsO RNA from the two 3′ to 5′ exoribonucleases, RNase II and PNPase, and from cleavage by RNase E.
Hfq binds tightly to the poly(A) tail of rpsO mRNA
Gel retardation assays demonstrated that Hfq exhibits a high affinity for polyadenylated RNA. The discrepancy with the lower Kd value previously reported (24) probably reflects variation in the proportion of active protein resulting from the purification method or from aggregation of the protein used in early experiments. The high affinity of the Hfq protein for the poly(A)-tailed ΔrpsO mRNA is of the same order of magnitude as that for poly(A)115 (Fig. 1A). The Kd values of Hfq bound to poly(A)-tailed RNA and non-polyadenylated RNA reported here are 90 pM (15 pM if a Hfq hexamer binds the RNA) and 1.4 nM (0.23 nM for the hexamer), respectively. The Kd for poly(A)-tailed rpsO RNA is also of the same order of magnitude as the value reported by de Haseth and Uhlenbeck (43) for (Ap)27 (6.25 pM for the hexamer) at a similar salt concentration. With RNA substrates containing significantly fewer adenosine residues, the equilibrium dissociation of both Staphylococcus aureus and E.coli Hfq proteins with AAAAAAG and A7, respectively, was only 30.5 nM (44) (V.Arluison, unpublished data). These data suggest that a critical number of adenine residues is required for high binding affinity. This is in agreement with earlier fluorescence titration experiments indicating a saturation point of 14 A-residues per Hfq hexamer (43). On the other hand, the Kd value we found for non-polyadenylated rpsO mRNA (0.23 nM for hexameric Hfq) differs from the values measured for Spot42 [20 nM (28)] and DsrA [1.2 µM (45)]. A comparison of known binding sites (Table 1) suggests Hfq does not recognize a strict RNA consensus. Our data are nevertheless consistent with earlier observations that Hfq binds single-stranded A–U-rich RNA regions (26), either adjacent to hairpin structures [Spot42 (28), ompA mRNA (12)] or surrounded by them [DsrA (45), OxyS (27), Spot42 (28)]. These RNA hairpin structures probably help to constrain the Hfq binding site in a single-stranded conformation.
Table 1. Comparison of Hfq binding sites with mRNAs, viral RNA and small regulatory RNAs.
We propose that two Hfq hexamers bind independently to ΔrpsO RNA-(A)18; the first strong binding occurs on the poly(A) moiety of the RNA, and the second weaker binding on the U-rich connecting region (Fig. 2). Moreover, the gel-shift assay performed with poly(A)115 (Fig. 1A) and previous experiments (43) suggest that Hfq hexamers can sequentially bind every 14 residues and cover long poly(A)-sequence-like ‘pearls on a string’. We cannot exclude, however, that Hfq hexamers exhibit several distinct binding sites for RNA. Two sites were indeed described for the Pyrococcus abyssi Sm protein (46), and the contribution of these sites differs depending on the RNA sequence. Mutational analysis of E.coli Hfq will be required to define the poly(A) binding site more precisely.
The high affinity of Hfq for poly(A) and A–U-rich regions, implies that this abundant protein [between 2500 and 35 000 Hfq molecules per cell (47,48)] can be strongly associated with RNA fragments during exponential growth. However, Hfq has been shown to be a co-factor of the transiently expressed sRNA (22,27,28). This binding would imply either that new Hfq molecules are synthesized or that Hfq is released from short-lived RNAs, or possibly that Hfq is transferred from storage RNA to the transiently expressed sRNAs under particular physiological conditions. It is indeed conceivable that the capacity of Hfq to promote RNA–RNA interactions could allow the protein to be rapidly transferred from a storage RNA, for example poly(A) tails, to a newly synthesized small regulatory RNA, despite the high affinity of the Hfq for RNA. This hypothesis is presently under investigation.
Hfq modulates rpsO mRNA degradation
The footprinting data suggest that Hfq destabilizes the bottom of both helices in the ΔrpsO mRNA. This correlates well with the data showing that RNase II continues the processive degradation of nucleotides to the bottom of the t1 hairpin of the rpsO transcript in the presence of Hfq. These data are consistent with the proposed role of Hfq as an RNA chaperone (31,49) and with the idea that Hfq facilitates base pairing between small RNAs and their target mRNAs (28,31). It has also been reported that Hfq affects the orientation of helices of the DsrA sRNA rather than its secondary structure (45). In the case of ΔrpsO mRNA, we propose that weakening of base pairing at the bottom of the transcription terminator may facilitate the addition of A-residues at the 3′ end of the molecule by PAP I (24).
Our data also suggest that Hfq may play a role in the stabilization of RNAs. We showed here that at physiological concentrations, Hfq masks the M2 RNase E site, where RNase E initiates degradation of the rpsO transcript (1,33). Since RNase E and Hfq both recognize single-stranded A–U-rich regions of RNA, one can speculate that Hfq may bind the RNase E sites of other RNAs, therefore may modulate processing and degradation of the RNA (50). In contrast, Hfq may also cause degradation of ompA mRNA by competing with the binding of the ribosome to the A–U-rich translation initiation site of the ompA mRNA and as a result the mRNA is no longer protected from ribonucleases by translating ribosomes (12).
We proposed that Hfq also protects poly(A) tails from degradation by the exoribonucleases PNPase and RNase II. This may explain why poly(A) can be detected in the cell (51). However, there is an apparent paradox; on the one hand Hfq stimulates processive synthesis of poly(A) tails, which promote RNA degradation, and on the other hand Hfq protects these tails from PNPase and RNase II. This property raises the interesting possibility that Hfq prevents poly(A)-dependent degradation of full-length transcripts whose stability is mostly controlled by RNase E and favors poly(A)-dependent degradation of stably structured RNAs devoid of RNase E sites.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to P. Romby and C. Condon for careful critical reading of the manuscript, and S. Guillou for contribution to this work as an undergraduate student. We thank C. Portier and F. Allemand for providing PNPase, A.J. Carpousis for degradosome preparation, G. Mackie for the gift of plasmid GC100 and T. Elliott for the gift of pTE607. We are indebted to P. Romby for her help in performing probing experiments. This work was supported by Université Paris 7-Denis Diderot (plan quadriennal), the Centre National de la Recherche Scientifique (UPR 9073), the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires of the Ministère de l’Education Nationale de la Recherche et de la Technologie (MNRT). M. F. is the recipient of a grant from MNRT and V. A. is supported by a fellowship from Université Paris 7-Denis Diderot.
REFERENCES
- 1.Braun F., Hajnsdorf,E. and Régnier,P. (1996) Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol. Microbiol., 19, 997–1005. [DOI] [PubMed] [Google Scholar]
- 2.Hajnsdorf E., Steier,O., Coscoy,L., Teysset,L. and Régnier,P. (1994) Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams-rnb-pnp mutant. EMBO J., 13, 3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajnsdorf E. and Régnier,P. (1999) E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol., 286, 1033–1043. [DOI] [PubMed] [Google Scholar]
- 4.Haugel-Nielsen J., Hajnsdorf,E. and Regnier,P. (1996) The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J., 15, 3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn G.A. and Mackie,G.A. (1999) Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. [DOI] [PubMed] [Google Scholar]
- 6.Jerome L.J., van Biesen,T. and Frost,L.S. (1999) Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol., 285, 1457–1473. [DOI] [PubMed] [Google Scholar]
- 7.Iost I. and Dreyfus,M. (1994) mRNAs can be stabilized by DEAD-box proteins. Nature, 372, 193–196. [DOI] [PubMed] [Google Scholar]
- 8.Braun F., Le Derout,J. and Régnier,P. (1998) Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J., 17, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bechhofer D.H. and Zen,K.H. (1989) Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol., 171, 5803–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouvet P. and Belasco,J. (1992) RNA degradation in E. coli by endonuclease RNase E: control by 5′-terminal base pairing. Nature, 360, 488–491. [DOI] [PubMed] [Google Scholar]
- 11.Joyce S.A. and Dreyfus,M. (1998) In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol., 282, 241–254. [DOI] [PubMed] [Google Scholar]
- 12.Vytvytska O., Moll,I., Kaberdin,V.R., von Gabain,A. and Bläsi,U. (2000) HFq (HFI) stimulates ompA mRNA decay by interfering with ribosomes binding. Genes Dev., 14, 1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 13.Tsui H.-C.T., Leung,H.-C.E. and Winkler,M.E. (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol., 13, 35–49. [DOI] [PubMed] [Google Scholar]
- 14.Muffler A., Traulsen,D.D., Fischer,D., Lange,R. and Hengge-Aronis,R. (1997) The RNA-binding protein HF-1 plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol., 179, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wachi M., Takada,A. and Nagai,K. (1999) Overproduction of the outer-membrane proteins FepA and FhuE responsible for iron transport in Escherichia coli hfq::cat mutant. Biochem. Biophys. Res. Commun., 264, 525–529. [DOI] [PubMed] [Google Scholar]
- 16.Massé E. and Gottesman,S. (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA, 99, 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sledjeski D.D., Gupta,A. and Gottesman,S. (1996) The small RNA, dsrA, is essential for the low temperature expression of rpoS during exponential growth in Escherichia coli.EMBO J., 15, 3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 18.Muffler A., Fischer,D. and Hengge-Aronis,R. (1996) The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev., 10, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 19.Brown L. and Elliott,T. (1996) Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires Host Factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol., 178, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vytvytska O., Jakobsen,J.S., Balcunate,G., Andersen,J.S., Baccarini,M. and von Gabain,A. (1998) Host-factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl Acad. Sci. USA, 95, 14118–14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada A., Wachi,M. and Nagai,K. (1999) Negative regulatory role of the Escherichia coli hfq gene in cell division. Biochem. Biophys. Res. Commun., 266, 579–583. [DOI] [PubMed] [Google Scholar]
- 22.Sledjeski D.D., Whitman,C. and Zhang,A. (2001) HFq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol., 183, 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsui H.-C.T., Feng,G. and Winkler,M.E. (1997) Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol., 179, 7476–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajnsdorf E. and Régnier,P. (2000) Host factor HFq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Derout J., Folichon,M., Briani,F., Dehò,G., Régnier,P. and Hajnsdorf,E. (2003) Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res., 31, 4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senear A.W. and Steitz,J.A. (1976) Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J. Biol. Chem., 251, 1902–1912. [PubMed] [Google Scholar]
- 27.Zhang A., Altuvia,S., Tiwari,A., Argaman,L., Hengge-Aronis,R. and Storz,G. (1998) The oxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-1) protein. EMBO J., 17, 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moller T., Franch,T., Hojrup,P., Keene,D., Bächinger,H.P., Brennan,R.G. and Valentin-Hansen,P. (2002) HFq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell, 9, 23–30. [DOI] [PubMed] [Google Scholar]
- 29.Schuppli D., Miranda,G., Tsui,H.-C.T., Winckler,M.E., Sogo,J.M. and Weber,H. (1997) Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 10239–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda G., Schuppli,D., Barrera,I., Hausherr,C., Sogo,J.M. and Weber,H. (1997) Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factors. J. Mol. Biol., 267, 1089–1103. [DOI] [PubMed] [Google Scholar]
- 31.Zhang A., Wassarman,K.M., Ortega,J., Steven,A.C. and Storz,G. (2002) The Sm-like HFq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell, 9, 11–22. [DOI] [PubMed] [Google Scholar]
- 32.Hajnsdorf E., Braun,F., Haugel-Nielsen,J. and Regnier,P. (1995) Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 3973–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Régnier P. and Hajnsdorf,E. (1991) Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol., 217, 283–292. [DOI] [PubMed] [Google Scholar]
- 34.Coburn G.A. and Mackie,G.A. (1996) Overexpression, purification and properties of Escherichia coli ribonuclease II. J. Biol. Chem., 271, 1048–1053. [DOI] [PubMed] [Google Scholar]
- 35.Carpousis A.J., van Houwe,G., Ehretsmann,C. and Krisch,H.M. (1994) Co-purification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell, 76, 889–900. [DOI] [PubMed] [Google Scholar]
- 36.Donis-Keller H., Maxam,A.M. and Gilbert,W. (1977) Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res., 4, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Hara E.B., Chekanova,J.A., Ingle,C.A., Kushner,Z.R., Peters,E. and Kushner,S.R. (1995) Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 1807–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohanty B.K. and Kushner,S.R. (2000) Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli.Mol. Microbiol., 6, 982–994. [DOI] [PubMed] [Google Scholar]
- 39.Arluison V., Derreumaux,P., Allemand,F., Folichon,M., Hajnsdorf,E. and Regnier,P. (2002) Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol., 320, 705–712. [DOI] [PubMed] [Google Scholar]
- 40.Coburn G.A. and Mackie,G.A. (1996) Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′ exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem., 271, 15776–15781. [DOI] [PubMed] [Google Scholar]
- 41.Azam T.A., Iwata,A., Nishimura,A., Ueda,S. and Ishihama,A. (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol., 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marujo P.E., Hajnsdorf,E., Le Derout,J., Andrade,R., Arraiano,C.M. and Régnier,P. (2000) RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli.RNA, 6, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Haseth P.L. and Uhlenbeck,O.C. (1980) Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry, 19, 6138–6146. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher M.A., Rearson,R.F., Moller,T., Valentin-Hansen,P. and Brennan,R.G. (2002) Structures of the pleiotropic translational regulator HFq and an HFq–RNA complex: a bacterial Sm-like protein. EMBO J., 21, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brescia C.C., Mikulecky,P.J., Feig,A.L. and Sledjeski,D.D. (2003) Identification of the HFq-binding site on DsrA RNA: HFq binds without altering DsrA secondary structure. RNA, 9, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thore S., Mayer,C., Sauter,C., Weeks,S. and Suck,D. (2003) Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA: common features of RNA-binding in Archae and Eukarya. J. Biol. Chem., 278, 1239–1247. [DOI] [PubMed] [Google Scholar]
- 47.Carmichael G.G., Weber,K., Niveleau,A. and Wahba,A.J. (1975) The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation and purifcation by polyadenylate-cellulose chromatography. J. Biol. Chem., 250, 3607–3612. [PubMed] [Google Scholar]
- 48.Azam T.A. and Ishihama,A. (1999) Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem., 274, 33105–33113. [DOI] [PubMed] [Google Scholar]
- 49.Moll I., Leitsch,D., Steinhauser,T. and Bläsi,U. (2003) RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep., 4, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massé E., Escorcia,F.E. and Gottesman,S. (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev., 17, 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohanty B.K. and Kushner,S.R. (1999) Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol., 34, 1094–1108. [DOI] [PubMed] [Google Scholar]