Abstract
Background
Alpha (α)-hemolysin is a pore forming cytolysin and serves as a virulence factor in intestinal and extraintestinal pathogenic strains of E. coli. It was suggested that the genes encoding α-hemolysin (hlyCABD) which can be found on the chromosome and plasmid, were acquired through horizontal gene transfer. Plasmid-encoded α-hly is associated with certain enterotoxigenic (ETEC), shigatoxigenic (STEC) and enteropathogenic E. coli (EPEC) strains. In uropathogenic E. coli (UPEC), the α-hly genes are located on chromosomal pathogenicity islands. Previous work suggested that plasmid and chromosomally encoded α-hly may have evolved independently. This was explored in our study.
Results
We have investigated 11 α-hly plasmids from animal and human ETEC, STEC and EPEC strains. The size of α-hly plasmids ranges from 48-157 kb and eight plasmids are conjugative. The regulatory gene (hlyR) located upstream of the hlyCABD gene operon and an IS911 element located downstream of hlyD are conserved. Chromosomally-encoded α-hly operons lack the hlyR and IS911 elements. The DNA sequence of hlyC and hlyA divided the plasmid- and chromosomally-encoded α-hemolysins into two clusters. The plasmid-encoded α-hly genes could be further divided into three groups based on the insertion of IS1 and IS2 in the regulatory region upstream of the α-hly operon. Transcription of the hlyA gene was higher than the housekeeping icdA gene in all strains (rq 4.8 to 143.2). Nucleotide sequence analysis of a chromosomally located α-hly determinant in Enterobacter cloacae strain indicates that it originates from an E. coli α-hly plasmid.
Conclusion
Our data indicate that plasmids encoding α-hly in E. coli descended from a common ancestor independent of the plasmid size and the origin of the strains. Conjugative plasmids could contribute to the spread of the α-hly determinant to Enterobacter cloacae. The presence of IS-elements flanking the plasmid-encoded α-hly indicate that they might be mobile genetic elements.
Background
Two major types of calcium dependent, pore forming cytolysins of the repeats in toxin (RTX)-family, called alpha-(α) and EHEC-hemolysin (enterohemolysin) were described in strains of Escherichia coli [1,2]. Both types of hemolysins are encoded by polycistronic operons consisting of four genes arranged in the order of hlyCABD [3,4]. The product of the hlyC gene is involved in activation of the hemolytic toxin the product of the hlyA gene. The gene products of hlyB and hlyD together with TolC are involved in secretion of the hemolysin through the bacterial cell wall [5].
EHEC-hemolysin is encoded on non-conjugative plasmids in strains of enterohemorrhagic E. coli (EHEC) that cause hemorrhagic diseases in humans [6,7]. In contrast, α-hemolysin is frequently associated with human uropathogenic E. coli (UPEC) strains [8] and with enterotoxigenic (ETEC), shigatoxigenic (STEC) and enteropathogenic E. coli (EPEC) strains that cause diarrhea and edema disease in animals [9-12].
In UPEC the α-hly genes are found on large chromosomal pathogenicity islands (PAI) [13,14]. The UPEC O4 (J96) and O6 (536) strains carry each two α-hly operons located on different PAIs [15,16], which contain divers junctions and adjacent sequences. This suggests that these loci have evolved independently [16,17]. Genetic analysis of chromosomal α-hly operons revealed differences in 5' flanking sequences and toxin expression [18-20].
Plasmid-encoded α-hly genes were found associated with EPEC O26 strains [21], as well as with ETEC and Shiga toxin 2e (Stx2e) producing STEC strains [9,10,22]. α-hly plasmids of E. coli were found to differ widely in size, incompatibility groups and conjugational transfer ability [10,20,21,23]. So far, only two plasmid α-hly operons were completely sequenced. The first is located on the 48 kb non-conjugative plasmid pHly152 from a murine E. coli strain [24]. The other is located on the 157 kb conjugative plasmid pEO5 of a human EPEC O26 strain [21]. Interestingly, despite the differences between pHly152 and pEO5, the DNA sequence of their α-hly operons are 99.2% similar while the sequence of the upstream regulatory hlyR region is 98.8% similar [21]. Importantly, the plasmid-inherited α-hly are less similar (96.0-96.4%) to the chromosomally inherited α-hlyCABD located on PAI I [GenBank AJ488511] and PAI II [GenBank AJ494981] of the E. coli strain 536 [18,21]. Moreover, chromosomally and plasmid-inherited α-hly operons also differ also for their 5' regulatory hlyR region. These findings suggest that the plasmid and chromosomal α-hly operons have evolved in parallel.
Studies on hemolysins of other bacterial species revealed similarities between the E. coli α-hemolysin genes and the Enterobacter, Proteus, Morganella and Mannheimia operons [25,26]. Codon usages base composition studies suggested that the α-hlyCABD genes of E. coli were originated from Proteus, Morganella or Mannheimia species [25,27]. Transposon-like structures found in the neighborhood of plasmid pHly152 and pEO5 encoded α-hly operons suggest that these were acquired by horizontal gene transfer [20,21].
The fact that the α-hlyCABD genes and their adjacent regions on pHly152 and pEO5 were highly similar to each other prompted us to investigate the genetic relationship between plasmid and chromosomal inherited α-hly operons in more strains of E. coli and in Enterobacter cloacae. Our results indicate that plasmid α-hly operons are highly similar regardless of differences in the plasmid backbone sequences, bacterial host and their source, suggesting that they have evolved from a common origin.
Results
Characterization of α-hly plasmids in E. coli strains
Nucleotide sequence analysis of the pEO5 α-hlyCABD operon and its 5' region revealed high similarity to the corresponding DNA segment of the α-hly plasmid pHly152 [21]. Since pEO5 and pHly152 differ in their origin, size and conjugative transfer, we investigated if plasmid α-hly operons have a common origin and evolved independently of chromosomal α-hlyCABD genes in E. coli.
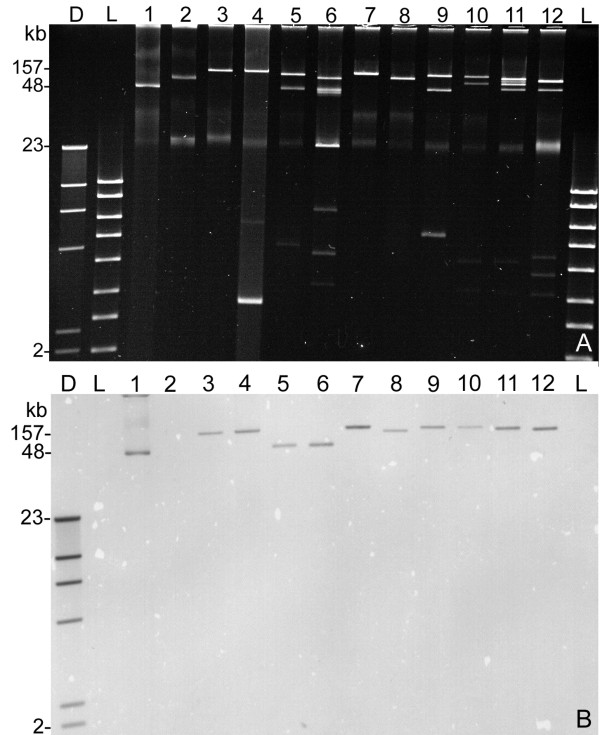
In order to explore the genetic relationship between plasmid α-hly genes we investigated five α-hly plasmids originating from canine ETEC strains and four plasmids of porcine ETEC and STEC strains (Table 1). α-hemolysin plasmids were detected by DNA-hybridization of Southern blotted plasmid DNA as described in Material and Methods (Fig. 1). The size of α-hly plasmids from dogs, pigs, mouse, cattle and human origin varied between 48 kb to 157 kb and other than pEO13, pEO14 and pEO860 all other plasmids were found transferable by conjugation (Table 1). Plasmid profile analysis has shown that the α-hly-plasmids are frequently found together with other large plasmids (Fig. 1).
Table 1.
Relevant properties of strains carrying plasmid and chromosomally encoded α-hly determinants
| PCR products with primers pairsa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| strain | Serotypeb | Origin, referenced | hly-plasmid (kb) | Plasmid group |
1f/r (678 bp) |
32f/r (671 bp) |
44f/r (685 bp) |
99f/r (650 bp) |
72f/r (695 bp) |
81f/r (773 bp) |
| C4115 | O26:[H11] | human, EPEC [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| TPE422 | Or:H48 | E. coli K12 (pEO5) [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| CB9866 | O26:[H11] | cattle, EPEC [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| CB1027 | O26:[H11] | human, EPEC [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| CB1030 | O26:[H11] | human, EPEC [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| IP187 | O26:[H11] | human, EPEC [21] | pEO5 (157) | 1 | + | + | + | + | - | - |
| 84/2195 | Ont:H10 | dog [10] | pEO9 (146) | 1 | + | + | + | + | - | - |
| 84-R | O121:H46 | dog [10] | pEO13 (97) | 1 | + | + | + | + | - | - |
| 374 | Or:H48 | mouse [24] | pHly152 (48) | 2 | + | e) | + | + | - | - |
| 84-3208 | O42:H37 | dog, ETEC[10] | pEO11 (48) | 2 | + | e) | + | + | - | - |
| 84-2573 | O70:NM | dog, ETEC [10] | pEO12 (48) | 2 | + | e) | + | + | - | - |
| CB853 | O138:H14 | pig, STEC [29] | pEO853 (145) | 3 | + | f) | g) | + | - | - |
| CB855 | O138:NM | pig, STEC [29] | pEO855 (140) | 3 | + | f) | g) | + | - | - |
| CB857 | O157:NM | pig, ETEC [42] | pEO857 (97) | 3 | + | f) | g) | + | - | - |
| CB860 | O149:H10 | pig, ETEC [42] | pEO860 (48) | single | + | + | g) | + | - | - |
| 84-2S | O75:H2 | dog [10] | pEO14 (97) | single | - | - | - | - | - | - |
| 536h | O6:K15:H31 | human UPEC [20] | - | n.a | - | - | - | - | + | + |
| 536-14 | O6:K15:H31 | PAI I deletion mutant of 536 [20] | - | n.a | - | - | - | - | + | - |
| 695/83 | O126:H27 | human [19] | - | n.a | - | - | - | - | - | i) |
| J96h | O4:K6 | human UPEC [46] | - | n.a | - | - | - | - | + | j) |
| KK6-16 | E. cloacae | human [26] | - | n.a | k) | - | - | - | - | - |
a) primer pairs and size of the PCR products obtained with strains TPE422 (pEO5) (primers 1f/r, 32f/r and 44f/r) and 536 (primers 81f/r and 72f/r) (see Table 2).
+ = a PCR product of the same size as obtained with strains TPE422 (pEO5) or 536, respectively.
- = no PCR product obtained
PCR products with other sizes than obtained with the reference strains are indicated for their length in bp.
b) O:H serotype, Or = rough LPS, Ont = O-antigen not typable, NM = non motile
c) α = alpha-hemolytic (α-hly genes), e-hly = enterohemolytic (EHEC-hly genes)
d) All strains were from feces if not otherwise indicated, ETEC = enterotoxigenic E. coli, STEC = shigatoxigenic E. coli, UPEC = uropathogenic E. Coli
e) 2000 bp PCR product (2007 bp as calculated from the nucleotide sequence of pEO11 [GenBank FM210249]
f) 2900 bp PCR product (2868 bp as calculated from the nucleotide sequence of pEO860) [GenBank FM210351]
g) 1500 bp PCR products
h) strain carries two α-hly determinants in the chromosome
i) 950 bp PCR product
j) 860 bp PCR product
k) 778 bp PCR product (calculated from nucleotide sequencing of KK6-16 [GenBank FM210352])
Figure 1.
Detection of plasmid encoded α-hly genes in E. coli strains. A) Agarose gel (0.7%) with plasmid preparations obtained from E. coli strains. Lanes: D = digoxigenin-labelled molecular weight standard II (Roche); L = molecular weight standard hyperladder I (Bioline); 1 = 374 (phly152); 2 = TPE1313 (pO157); 3 = TPE422 (pEO5); 4 = TPE1030 (pEO5); 5 = 84-3208 (pEO11); 6 = 84-2573 (pEO12); 7 = 84-2195 (pEO9); 8 = 84-2 S (pEO14); 9 = 84-R (pEO13); 10 = CB853 (pEO853); 11 = CB855 (pEO855); 12 = CB857 (pEO857). B) Southern hybridization patterns of plasmid DNA from lanes 1-12 with the α-hlyA specific digoxigenin labelled gene probe generated with primers 10f/r from plasmid pEO5 DNA. The size of hybridizing α-hly plasmids varies from 48 (lane 1) to 157 kb (lane 3).
In addition, we investigated four E. coli and an E. cloacae strain with chromosomal α-hly operons (Table 1). A BLAST search using pEO5 [GeneBank FM180012] and phly152 [GeneBank M14107] sequences between hlyR and hlyC and downstream of hlyD revealed no similarity with sequences of chromosomal α-hly genes in strains CFT073 [GeneBank AE014075], UTI89 [CP000243] and 536 [CP000247].
Analysis of the plasmid and chromosomal upstream α-hly operons
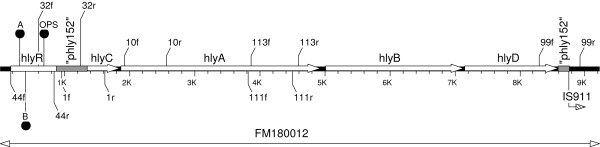
Based on the pEO5 DNA sequence (Fig. 2) we developed specific primers for amplification of fragments within the hlyR, and hlyR - hlyC regions (Table 2). In addition, we developed specific PCRs for the upstream hlyC sequences of the chromosomal α-hemolysin operons in PAI I and PAI II of strain 536 [15] (Table 2). We performed PCR analysis of all strains carrying plasmid and chromosomal α-hly operons; strains carrying α-hly-plasmids pEO5 and pHly152 and 536 served as positive controls. The results are summarized in Table 1.
Figure 2.
Map of the α-hly region of plasmid pEO5 (FM180012). The positions of PCR-primers used for investigation of strains with plasmid and chromosomally inherited α-hly genes are indicated as leaders carrying the primer designations (Table 2). Regulatory sequences inside the hlyR gene (A, B and OPS) are shown as filled ballons. "phly152" is a stretch of non-coding DNA showing strong homology to corresponding regions in the α-hly plasmid pHly152.
Table 2.
Specific PCRs for identification of plasmid and chromosomally inherited α-hly determinants
| DNA-target (position in sequence) | GenBank Accession | Primer | nucleotide sequence (5' - 3') | Tm (°C) | PCR product bp |
|---|---|---|---|---|---|
| hlyA (1915-1936) (2560-2580) | FM180012 | 10f 10r |
GCTGCAAATAAATTGCACTCAG CCCTGCACCGATATTATCAAG |
53.1 | 666 |
| "pHly152" (953-974) &hlyC (1612-1630) | FM180012 | 1f 1r |
GTAGTTCAAAAGACAACTCGTG ATCCCCGAAAGGAGCAATC |
50.6 | 678 |
| hlyR (597-618) & "pHly152" (1246-1267) | FM180012 | 32f 32r |
GTCTTGCCGTACAATAATTTCC TCCGTTTAATGTCATAACTCGC |
56.5 | 671a |
| hlyR (167-188) (830-851) | FM180012 | 44f 44 |
ATTCCAAGCGAAGTCCATCCCC CATAAAGCATGATGCCACCACG |
66.5 | 685a |
| hlyA (3817-3839) (4497-4518) | FM180012 | 111f 111r |
GATGGCACAAAAGCAACCGAAG TTCTCGCTTGAAGGCCACATCC |
55.4 | 702 |
| hlyA (3865-3883) (4592-4613) | FM180012 | 113f 113r |
CTTGGTGGCGATGTTAAGG GACTCTTTTTCAAACCAGTTCC |
53.5 | 749 |
| hlyD (8297-8319) & IS911 (8925-8946) | FM180012 | 99f 99r |
GCAGAATGCCATCATTAAAGTG CCATGTAGCTCAAGTATCTGAC |
53.8 | 650 |
| PAI I (536) (44506-44524) &hlyC (45278-45299) | AJ488511 | 81f 81r |
CCTGTGACACTTCTCTTGC CCCAAGAACCTCTAATGGATTG |
52.3 | 773a |
| PAI II (536) (31974-31995) &hlyC (32650-32668) | AJ494981 | 72f 72r |
CCCAACTACAATATGCAACAGG CGCCAATAGAGTTGCCTTC |
51.9 | 695 |
a) PCR products of different lengths were obtained with these primers depending on the DNA template (see Table 1)
Primers 1f/r are specific for the upstream hlyC region in pEO5 and yielded a PCR product of 678 bp (Fig. 2). PCR products of the same size were obtained with all strains carrying α-hly plasmids, except 84/S (pEO14); restriction enzyme analysis revealed all the fragments had a similar HinfI profile (data not shown). Primers 1f/r gave no products using E. coli strains carrying chromosomally encoded α-hly as template with the exception of the E. cloacae strain KK6-16 which yielded a PCR product; DNA sequencing revealed a 778 bp fragment [GenBank FM210352, position 72-849] (Table 1).
Primers 32f/r spanning the region between hlyR and the "phly152" segment amplify a 671 bp product in pEO5 [GenBank FM180012, position 597-1267] (Fig. 2). A PCR product of the same size was obtained with pEO5 and derivative plasmids as well as with plasmids pEO9 [GenBank FM210248 position 427-1097], pEO13 and pEO860 (Table 1, Fig. 3). Primers 32f/r yielded PCR products of 2007 bp with pEO11, [GenBank FM210249, position 392-2398), pHly152 and pEO12, and 2784 bp PCR products with pEO853 [GenBank FM10347 position 399-3182], pEO855 and pEO857 (Table 1). All amplicons of a given size (671 bp, 2007 bp and 2784 bp), yielded a similar HinfI restriction pattern (data not shown). Strains with chromosomally encoded α-hemolysin gave no products in the 32f/r PCR, as well as strain 84/2 S carrying plasmid pEO14 (Table 1).
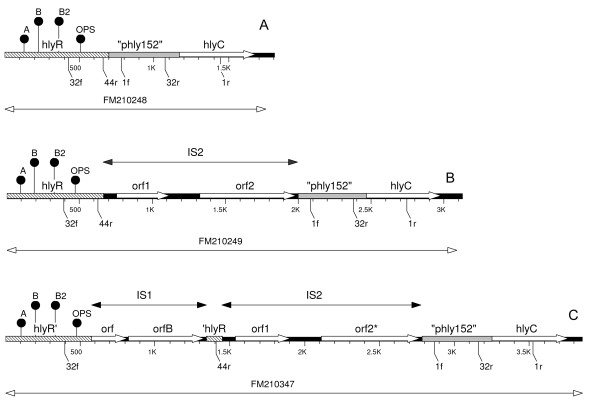
Figure 3.
Map of the hlyR-hlyC region of representative plasmids of groups 1, 2 and 3. Genetic map of the corresponding regions from hlyR to hlyC of α-hly determinants from plasmids representing groups 1-3. A) pEO9, (strain 84-2195) B) pEO11, (84-3208); and C) pEO853 (CB853). The positions of PCR-primers used for identification and nucleotide sequencing are indicated as leaders carrying the primer designations (Table 2). Regulatory sequences inside the hlyR gene (A, B and OPS) are shown as filled ballons. "phly152" is a stretch of non-coding DNA showing strong homology to corresponding regions in the α-hly plasmid pHly152.
Primer pair 44f/r (Fig. 2) is specific for hlyR and amplified related sequences in all E. coli carrying α-hly plasmids except pEO14. The hlyR PCR product was 685 bp (pEO5) [GeneBank FM180012, position 167-851] for all plasmids except for pEO853, pEO855, pEO857 and pEO860 which generated amplicons of about 1400 bp (Table 1). The 685 bp and 1400 bp size PCR products yielded similar HinfI restriction profiles, respectively. Strains with chromosomally inherited α-hly genes were negative for hlyR sequences (Table 1).
None of the strains with α-hly plasmids, or the E. cloacae strain KK6-16 yielded PCR products with primer pairs 81f/r and 72f/r, that are specific for PAI I and PAI II α-hemolysins (Table 2) [17]. All strains with chromosomal α-hly except KK6-16 produced PCR products with one or both of these primer pairs (Table 1). Taken together, the PCR typing indicated that all plasmid α-hly except pEO14 were similar for the regulatory regions located upstream of the hly-genes which differed from the chromosomal α-hly operons.
Genetic analysis of the region between hlyR and hlyC in α-hly plasmids
A 464 bp DNA segment that carries a promoter (pHhlyL) for expression of α-hly-genes is located directly upstream of the hlyC gene in plasmid pHly152 [24] [GenBank M14107]. A 466 bp region with 98.9% sequence homology was found upstream of hlyC in pEO5 [21]. The "phly152" region is not present in E. coli strains containing chromosomal α-hly genes [20] (this work). Sequences highly homologous to a large part of the "phly152" region were found in all α-hly plasmids investigated here, except pEO14. Comparison of the complete 466 bp "phly152" DNA stretch of plasmids pEO5 [GenBank FM180012], pEO9 [FM210248], pEO853 [FM210347], pEO11 [FM210249] and pEO860 [FM210351] revealed similarities from 97.9% to 100%. Interestingly, a 427 bp fragment with 93% similarity to the "phly152" segment was found upstream of hlyC in the E. cloacae strain KK6-16 [GenBank FM210352, position 1-427].
Sequences specific for hlyR [GenBank X07565], a regulatory region located about 2000 bp upstream of the α-hly determinant in pHly152 [28] were present in all α-hly plasmids except pEO14. The hlyR regions of five representative plasmids (pHly152, pEO5, pEO9, pEO11 and pEO853) were analyzed and compared to each other (Fig. 3). Short DNA sequences that were reported to be involved in regulation of α-hly expression located inside hlyR, i.e regulatory sequences A and B [28] and the "operon polarity suppressor (ops) [18], were identified in the corresponding hlyR region of the five plasmids. A clustal analysis performed with a 565 bp segment of the hlyR region beginning with the regulatory sequence A to the end of the hlyR region revealed 98.8 to 100% similarity between these five plasmids.
IS1 and IS2 elements are present in the α hly-upstream region of plasmid hly
According to the length of PCR products obtained for the region located between hlyR and hlyC genes three groups of α-hly plasmids were established (Table 1). "Group 1" is represented by pEO5, its homologues from other E. coli O26 strains and by pEO9 and pEO13. "Group 2" is represented by pHly152, pEO11 and pEO12. "Group 3" is formed by plasmids pEO853, pEO855 and pEO857 from porcine strains. Two strains with α-hly plasmids pEO14 and pEO860 showed individual patterns by PCR-typing (Table 1). In order to explore the differences between the major groups of α-hly-plasmids we determined the nucleotide sequence of the region located between hlyR and hlyC of three representative plasmids, namely pEO9 [GenBank FM210248], pEO11 [FM210249] and pEO853 [FM210347] (Fig. 3).
Major differences between the α-hly plasmids in the region between hlyR and hlyC caused by insertion of IS1 and IS2. While "group 1" plasmids (pEO5, pEO9 and pEO13) carry no IS elements all "group 2" plasmids (phly152, pEO11 and pEO12) carry an IS2 element inserted directly downstream of the 3' end of hlyR (5' CCTGG 3') in pEO11. A 326 bp part of the IS2 element was previously described in pHly152 [GenBank M14107] [24], it is 99.4% identical to the corresponding IS2 element of pEO11. The IS2 elements in pEO11 and pHly152 are inserted at the DNA same site and are both flanked by the duplicated 5' CCTGG 3' DNA sequence.
Plasmids belonging to "group 3", which were all from pig strains (pEO853, pEO855 and pEO857), carry two IS elements in the region between hlyR to hlyC. In pEO853, the 786 bp IS1 is inserted immediately downstream of the hlyR internal sequence 5' AACAAAATT 3'. This 9 bp DNA stretch is repeated at the right hand end of the inserted IS1 and followed by the 94 bp residual 3' end of the hlyR region (Fig. 3). The IS2 element of pEO853 is 99.8% similar to that of pEO11 and inserted at the same position as in "group 2" plasmids pEO11 and pHly152.
Investigation of hlyR-hlyC region of STEC strains of porcine origin
We used the primers specific for the region between hlyR to hlyC (Table 1) to investigate 26 α-hemolysin/stx2e STEC strains from diseased pigs or pork meat [29]. PCR products were obtained from all. According to the length of the amplicons generated with primers 1f/r, 32f/r and 44f/r all but one strain showed patterns indicating the presence of a "group 2" or "group 3" plasmid with IS-elements in the region between hlyR and hlyC (Table 3). The PCR-profiles were closely associated with serotypes of strains causing edema disease in pigs (O138:H14, O139:H1 and O141:H4) suggesting that α-hly plasmids are conserved in these strains.
Table 3.
Detection of α-hly plasmid specific sequences in porcine STEC strains.
| Size of PCR products with primersa | ||||||
|---|---|---|---|---|---|---|
| Serotype | No. strains | Plasmid group | 1f+1r | 32f + 32r | 44 f + 44r | 99 f + 99r |
| O138:H14b | 4 | 3 | 678 | 2900 | 1500 | 650 |
| O139:H1b | 9 | 3 | 678 | 2900 | 1500 | 650 |
| O139:H1b | 1 | single | 678 | 2000 | 1500 | 650 |
| O141:H4b | 7 | 3 | 678 | 2900 | 1500 | 650 |
| O141:H4b | 3 | 2 | 678 | 2000 | 685 | 650 |
| O2:H32c | 1 | 2 | 678 | 2000 | 685 | 650 |
| O36:H19c | 1 | 2 | 678 | 2000 | 685 | 650 |
a) size of the PCR products (bp) with primers 1f/r, 32f/r, 44f/r and 99f/r as described in Tables 1+ 2. Digestion of PCR products by HinfI resulted in identical patterns between strains corresponding to the size of the PCR products obtained (Table 1)
b) Strains isolated from feces of diseased pigs [29]
c) Strains isolated from pork meat [29]
Sequence analysis of the hlyC gene of plasmid and chromosomal α-hemolysin
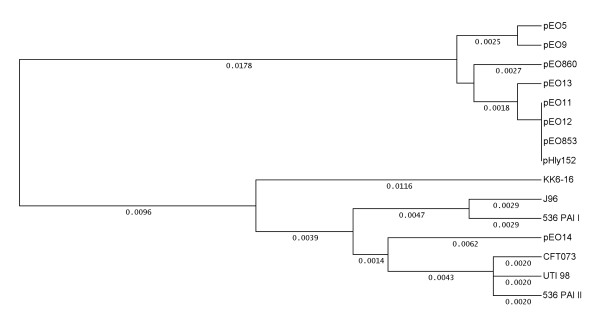
The fact that α-hly plasmids were similar for the regulatory sequences upstream of the α-hly operon prompted us to analyze the coding sequence of seven plasmid hlyC genes, namely pEO9 [GenBank FM210248], pEO860 [FM210351], pEO13 [FM210348], pEO14 [FM210350], pEO11 [FM210249], pEO853 [FM210347], and pEO12 [FM210349] (Table 1). We used Clustal W analysis to compare the DNA sequences of the plasmid hlyC genes and the chromosomal hlyC genes of strain 536, PAI [GenBank AJ488511] and PAI II [AJ494981] UTI98 [CP000243], CFT073 [AE014075], J96 [M14107] and that of the E. cloacae strain KK6-16 [FM210352]. All plasmid hlyC sequences, except that of pEO14, showed 99.2 to 100% nucleotide sequence homology to each other and were grouped into one cluster (Fig. 4). A second cluster (98.5% to 99.6% similarity) was formed by the chromosomal and pEO14 hlyC genes (Fig. 4). The hlyC gene encoded by pEO14 was most similar to that of PAI II from strain 536 (99.2% homology). The hlyC genes of all other α-hly plasmids showed 94.9-95.9% homology to chromosomal hlyC genes of E. coli. The amino acid (aa) sequences of hlyC translation products revealed five aa-exchanges (positions 3, 5, 40, 51, and 160) in the 170 aa-sequence that were closely associated with the origin (plasmid or chromosome) of the E. coli hlyC genes (data not shown).
Figure 4.
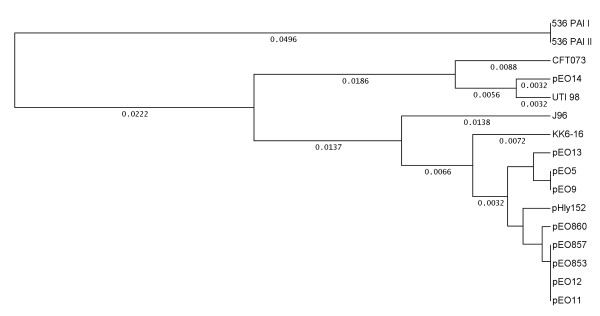
Genetic relationship between plasmid and chromosomally inherited hlyC genes. Clustal analysis of the coding sequence of the hlyC gene (513 bp) of strains 84-3208 (pEO11) [GenBank FM210249], 84-2 S (pEO14] [FM210350], 84-R (pEO13) [FM210348], 84-2195 (pEO9) [FM210248], C4115 (pEO5) [FM180012], CB860 (pEO860) [FM210351], CB853 (pEO853) [FM210347], 84-2573 (pEO12) [FM210349], KK6-16 [FM210352], 536 PAI I [AJ488511], 536 PAI II [AJ494981], CFT073 [AE014075], UTI98 [CP000243] and J96 [M10133]. UPGMA was used as tree building method and distances calculated according to Tajima and Nei 1984 [45].
The hlyC gene of the E. cloacae strain KK6-16 was more distant for its nucleotide and aa sequence from both the E. coli plasmid and chromosomal hlyC gene clusters and most similar to chromosomal PAI I, PAI II (98.2%) and pEO13 (97.2%) hlyC genes (Fig. 4).
Comparison of nucleotide sequences of plasmid and chromosomal α-hlyA genes
Comparing the nucleotide sequences of hlyA revealed significant differences between chromosomal and plasmid genes. By BLAST search, a 755 bp stretch of the hlyA gene (position 3000-4095 in the pHly152 sequence, GenBank M14105) was found only 91% similar to the corresponding regions of the chromosomal hlyA genes from PAI I [AJ488511] and PAI II [AJ494981].
In order to explore the differences between plasmid and chromosomal hlyA genes we have developed PCR primers (111f/r and 113f/r from GenBank FM180012, Table 2) for amplification of this DNA region. The nucleotide sequence of the corresponding 633 bp PCR products from strains with α-hly plasmids and from E. cloacae strain KK6-16 was determined. The results are presented in Fig. 5. Except for pEO14, all plasmid encoded hlyA internal sequences were very similar to each other with a maximum difference of 1.4% (pHly152 and pEO13). In contrast, chromosomal hlyA genes showed differences of up to 9.5% when compared to each other (J96 compared to 536 both PAI I and PAI II). The 211 aa HlyA translation products showed aa-exchanges at positions 58 and 78 that were associated with the E. coli plasmid or chromosomal origin of the genes (data not shown).
Figure 5.
Genetic relationship between plasmid and chromosomally inherited hlyA genes. Clustal analysis of 633 bp internal hlyA sequence of strains 84-3208 (pEO11) [GenBank FN673696], 84-2 S (pEO14) [FN673697], 84-R (pEO13) [FN673698], 84-2195 (pEO9) [FN673699], C4115 (pEO5) [FM180012], CB860 (pEO860) [FN673700], CB853 (pEO853) [FN673701], CB857 (pEO857) [FN673702], 84-2573 (pEO12) [FN673703], KK6-16 [FN673704], 536 PAI I [AJ488511], 536 PAI II [AJ494981], CFT073 [AE014075], UTI98[CP000243] and J96 [M10133]. UPGMA was used as tree building method and distances calculated according to Tajima and Nei 1984 [45].
The nucleotide sequence of the hlyA region on plasmid pEO14 was found closely related to the chromosomal hlyA gene of strain UTI98 (0.6% difference), and showed 5-6% sequence differences to all other α-hly-plasmids. Interestingly, the E. cloacae hlyA gene sequence was found 99% similar to that of plasmids pEO5 and pEO9 and more distantly related to the E. coli chromosomal hlyA genes (2.6 to 10.4% differences).
IS911 is present downstream of hlyD in strains carrying α-hly plasmids
It was suggested that the hlyCABD operons were spread in E. coli by mobile genetic elements [20] and a truncated IS911 segment of 254 bp was found located closely and downstream of the hlyD gene in plasmid pEO5 [21]. In order to investigate the other α-hly plasmids for the presence of this element we developed PCR-primers (99f/r) encompassing a 650 bp stretch of DNA starting inside hlyD and ending inside the IS911 sequence. All α-hly plasmids except pEO14 yielded a PCR product. None of the strains carrying chromosomal α-hly genes reacted positive with this PCR (Table 1). The nucleotide sequence of the 579 bp amplicons from nine α-hly plasmids (strains CB860 [GenBank FN678780], CB857 [FN678781], CB853 [FN678782], 84-3208 [FN678783], 84-2573 [FN678784], 374 [FN678785], 84-R [FN678786], 84-2195 [FN678787] and CB855 [FN678788] were compared by Clustal W analysis. The sequences were 99.5-100% identical, indicating that the 3' part of the hlyD gene and downstream IS911 sequences are conserved among plasmid α-hly determinants. In addition, all 26 STEC strains from pigs or pork meat that carried α-hly-plasmids (Table 3) yielded 650 bp products with primers 99f/r, that showed similar HinfI digestion profiles (257, 222 and 171 bp) to those of the sequenced plasmids [FN678782-88] indicating that the hlyD-IS911 region is conserved in these strains.
Transcriptional analysis of plasmid and chromosomal α-hlyA genes
We investigated if the presence of IS elements in the regulatory region upstream hlyC has an affect on transcription of the α-hlyA gene. Phenotypically, all strains with α-hly plasmids showed large and clear zones of hemolysis on blood agar plates similar to that found with strains carrying chromosomally inherited α-hly genes. An exception was made for strains 536-14 (the PAI I deletion mutant of strain 536) and the wildtype strain 695/83 (Table 1), which generated small, turbid zones of hemolysis on blood agar plates [19].
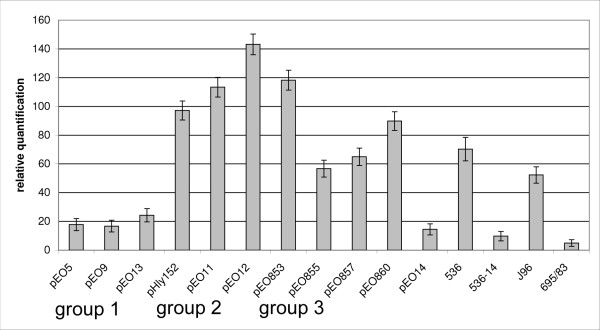
We compared the transcriptional activity of 15 E. coli strains carrying plasmid and chromosomal α-hly operons by analyzing the mRNA transcription level of the α-hlyA gene in a relative quantification (rq) assay by Real-Time PCR. The E. coli icdA housekeeping gene was used as a standard (Fig. 6). Transcription of the hlyA gene was higher than icdA in all strains (rq 4.8 to 143.2). Relatively low hlyA transcription rates (rq 4.8 and 9.7) were found with poor hemolysin producing strains 536-14 and 695/83. Strains carrying "group 1" α-hly plasmids (pEO5, pEO9 and pEO13) as well as pEO14 showed significantly (95% confidence intervals) lower transcription rates (rq 14.4 -24.3) compared to "group 2" and "group 3" strains with IS elements inserted upstream hlyC (rq 56.7 to 143.2). Significant differences in hlyA transcription rates were found between individual strains carrying "group 2" and "group 3" plasmids but they could not be clearly assigned to one of two groups. Except for pEO12 and pEO853, all "group 2" and "group 3" strains showed hlyA transcription rates that were not significantly different from those of strains 536 and J96, the latter carry each two chromosomally inherited α-hly genes [16,17].
Figure 6.
Relative quantification of the hlyA gene transcription in E. coli strains encoding plasmid and chromosomally inherited α-hly determinants. Strains and plasmids as well as plasmid groups are listed in Table 1. Means and standard deviations from two separate experiments performed in duplicate are shown.
Discussion
We have recently determined the nucleotide sequence of the pEO5 α-hly genes, which are commonly occurring in EPEC O26 strains from humans and animals [21]. Surprisingly, the α-hly genes were 99.2% similar to that of pHly152 which originates from a murine E. coli strain. Moreover, plasmid encoded α-hly genes from pHly152 and pEO5 showed close similarity in the regulatory region upstream of hlyC and thus differed from chromosomally inherited α-hly determinants [21]. These findings may indicate that plasmid encoded α-hemolysins have evolved from one source and separately from the chromosomal hemolysin operons. In order to explore this possibility we compared plasmid α-hly from unrelated E. coli strains of human, mouse, canine and porcine origin for similarities the regulatory and structural genes and their adjacent sequences.
Plasmid encoded α-hly determinants were found similar to each other in their genes (hlyR, hlyC, hlyA and hlyD) as well as in the adjacent sequences upstream and downstream of the α-hly-operon. Plasmid encoded hlyC and hlyA genes showed typical alterations in the nucleotide and in the amino acid sequence compared to their chromosomally encoded homologues. Moreover, chromosomally encoded α-hly genes were found different for the regions encompassing the α-hly-operon. The finding that chromosomal hlyC and hlyA genes clustered separately and showed greater sequence diversity compared to the plasmid homologues suggests that plasmid α-hly-genes have emerged more recently in E. coli and thus accumulated fewer changes compared to the chromosomal α-hly genes.
It was previously suggested that α-hly genes were acquired by strains of E. coli by horizontal gene transfer [25,27,30]. This hypothesis is supported by the location of chromosomally encoded hemolysin genes on pathogenicity islands [13,14,16,17] and the flanking of plasmid encoded α-hly genes by transposable elements [20,21]. A truncated IS911 element located downstream of the hlyD gene was found in all α-hly plasmids investigated in our study indicating that the plasmid encoded α-hly determinants may have descended from a common progenitor [31]. We do not know much about the genetic similarity between the α-hly plasmids investigated in this study, except that they show differences in size (48-157 kb) and conjugation ability. Further investigation of plasmid backbone sequences could reveal if they have descended from a common progenitor. At present, we cannot exclude that the α-hly determinant was transposed independently to different plasmids in E. coli.
Interestingly, plasmid pEO14 differed largely from all other α-hly-plasmids investigated in this study. The nucleotide sequence analysis of its α-hly genes and the adjacent sequences revealed close similarity to chromosomal α-hly genes. Because the α-hly genes present on plasmid pEO14 shows all features of chromosomal α-hly operon it is likely that it was generated by recombination between a plasmid and chromosomal α-hly loci in E. coli. A similar event might have been involved in generation of a truncated α-hly segment in plasmid Vir68 that has been analyzed for its complete nucleotide sequence [GenBank CP001162].
The chromosomally located α-hly genes of the E. cloacae strain KK6-16 showed similarities to E. coli plasmid encoded α-hly determinants. The nucleotide sequence analysis of the 552 bp upstream region of the α-hly operon of KK6-16 was identical to that of the α-hly plasmid pANN681 [GeneBank L01627] [32] and the "pHly152" region found in all α-hly plasmids except pEO14 is present in KK6-16. Since α-hly is not common in strains of Enterobacter species [26], it seems likely that strain KK6-16 acquired the α-hly genes by conjugation from E. coli. Similar findings have been made for plasmids encoding antimicrobial resistance [33,34]. However, we have not investigated this possibility.
Interestingly, the hlyC and hlyA sequences of the KK6-16 showed characteristic features which made it difficult to assign its α-hly determinant to the group of plasmid- or chromosomally inherited α-hly genes (Figs. 4+5). It is possible that characteristic alterations found in the KK6-16 α-hly sequence are due to E. cloacae as a different bacterial host species.
Multiple copies of IS1 and IS2 were frequently found in genetically unrelated strains of E. coli. IS1 and IS2 were found to be non-randomly scattered in the genomes of wild-type E. coli strains [35-37]. IS-elements are involved in chromosomal rearrangements, integration of F-plasmids and transposition of genes [38] and thus could have been involved in the generation of E. coli α-hly plasmids. Activation of downstream genes by presence of IS1 and IS2 elements in E. coli has been reported [39] and this could explain the relatively high hlyA transcription rates in plasmids carrying IS2 or IS1 and IS2. However, we have not tested this possibility experimentally and other factors such as plasmid copy numbers and differences between the E. coli host strains could have an influence on the transcription rates.
α-hemolysin plasmids are frequently found in STEC strains producing Stx2e, agents of edema disease in pigs [40], and in ETEC strains producing heat-stable enterotoxin causing diarrhea in dogs [10]. The α-hly plasmid pEO5 is closely associated with EPEC O26 strains as diarrheal agents of human infants and calves [21,41]. In contrast, E. coli strains carrying chromosomal α-hly are associated with UPEC which are characterized by other virulence attributes and serotypes than ETEC, EPEC and STEC strains [13,14,16,17]. The association of α-hly plasmids with intestinal and of chromosomal α-hly determinants with extraintestinal strains points to a separate evolution in these two major groups of pathogenic E. coli.
Conclusion
Our results indicate that the α-hly genes present on plasmids in ETEC, STEC and EPEC strains have a common origin. The presence of IS-sequences flanking the plasmid α-hly genes suggest that these were introduced in E. coli by horizontal gene transfer. Plasmids were shown to play a role in the spread of α-hly determinant to Enterobacter cloacae. Chromosomally α-hly genes present in UPEC are genetically more diverse and seem to have evolved separately from the plasmid α-hly genes.
Methods
Bacteria
The bacterial strains used in this work are listed in Table 1. Strain C4115, the source of the plasmid pEO5, the E. coli strain 374 carrying the α-hly-plasmid pHly152 as well as other EPEC O26 strains were described previously [21]. The relevant characteristics of strains with chromosomally located α-hemolysin determinants are listed elsewhere [10,18,19]. The α-hemolytic E. cloacae strain KK6-16 as well as the canine and porcine ETEC and STEC strains carrying α-hly plasmids were described previously [10,26,29,42]. The EHEC-hemolysin plasmid pO157 carrying strain TPE1313 was used as negative control is described elsewhere [21]. Mating of bacteria with E. coli K-12 recipient strains and isolation of α-hemolytic transconjugants was performed as described by Burgos et al. 2009 [21]. Phenotypes corresponding to E. coli α-hemolysin were analyzed on washed sheep blood agar [43].
Isolation of DNA, RNA and cDNA
Total DNA of bacteria was isolated as described [29]. Purified plasmid DNA of bacteria that was used for restriction digestion, DNA-hybridization, PCR and nucleotide sequencing was isolated with the large construct kit following the instructions of the producer (Qiagen, Hilden, Germany). Analysis of total plasmid profiles of E. coli strains was performed as described previously [44]. Total RNA was isolated from 20 ml of exponentially growing aerated cultures (3-5 × 108 bacteria/ml) of bacteria in L-Broth with the RNeasy minikit (Qiagen). Isolation of RNA and preparation of cDNA was performed as described previously [29].
DNA hybridization
Southern blot hybridization of plasmid DNA and labeling of gene probes with Digoxigenin-11-dUTP was performed as described [21]. Dig-labeled molecular markers (Dig Roche) were used for size determination of hybridizing DNA fragments. For identification of α-hly plasmids in Southern blotted gels a 666 bp PCR product of the α-hlyA gene generated with primers 10f/r (Table 2) was used as internal DNA probe for detection of α-hly specific sequences [21]. Plasmids pHly152, pO157 and pEO5 served as reference plasmids for size determination of α-hly plasmids [21] (Fig. 1).
Nucleotide sequencing of α-hemolysin and associated sequences
Nucleotide sequence analysis of the α-hly determinants and adjacent sequences was performed as described [21]. PCR products were purified and used for sequencing applying the dye terminator chemistry (PE Applied Biosystems, Darmstadt, Germany) and separated on an automated DNA sequencer (ABI PRISM® 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA). The sequences were analyzed using the Lasergene software (DNASTAR, Madison,WI) and Accelrys Gene v2.5 software.
Development of specific PCRs for plasmid- and chromosomally inherited α-hly determinants and their associated sequences
Primer pairs specific for α-hly-plasmid specific sequences hlyR (primers 44f/r), the region between hlyR and hlyC (primers 1f/r, 32f/r), hlyA (111f/r and 113f/r) and hlyD and downstream (99f/r) (Table 2) were developed with Accelrys software using the pEO5 sequence [GenBank FM180012]. Primer pairs for amplification of regions upstream of hlyC in chromosomally inherited α-hemolysins were developed from sequences of PAI I (81f/r) and PAI II (72f/r) from the uropathogenic E. coli strain 536 (Tables 1+2). Primers 10f/r served as positive control for general detection of plasmid and chromosomally inherited α-hly determinants. Primers and PCR conditions are listed in Table 2. PCR reactions were performed as described previously [29].
Transcriptional analysis of α-hlyA genes by qRT-PCR
Quantitative real time reverse transcription PCR (qRT-PCR) was performed with the Applied Biosystems 7500 real time PCR system (Applied Biosystems) with cDNA samples from bacteria (see above). Transcription rates of the α-hlyA gene were compared to those of the icdA housekeeping gene. Primers hlyA-f 5' ACCTTGTCAGGACGGCAGAT 3' and hlyA-r 5' CCGTGCCATTCTTTTCATCA 3' and the VIC labeled TaqMan MGB probe 5' ACTGGGAATTGAAGTCC 3' were used for amplification of the α-hlyA gene. The primers and the gene probe for detection of the icdA gene were described recently [29]. Real time PCR amplification were performed in an "icdA & α-hlyA" multiplex assay and were analyzed with the 7500 system SDS software version 1.4 as described [29].
GenBank accession numbers
The following nucleotide sequences derived from the α-hemolysin producing strains and α-hly plasmids from Table 1 were submitted to GenBank: strain 374 (pHly152) [GenBank FN678785]; 84-2195 (pEO9) [GenBank FM210248, FN673699, FN678787]; 84-3208 (pEO11) [GenBank FM210249, FN678787, FN673696]; CB853 (pEO853) [GenBank FM210347, FN678782, FN673701]; 84-R (pEO13) [GenBank FM210348, FN678786, FN673698]; 84-2573 (pEO12) [FM210349, FN678784, FN673703]; 84-2 S (pEO14) [GenBank FM210350, FN673697]; CB860 (pEO860) [GenBank FM210351, FN678780, FN673700]; CB855 (pEO855) [GenBank FN678788]; CB857 (pEO857) [GenBank (FN678781, FN673702] and strain KK6-16 [FM210352, FN673704].
Authors' contributions
LB took an integral part of project conception and both YB and LB in method development. YB took most part in the design and performance of the experimental procedures. Data analysis was performed by both researchers LB and YB as well as interpretation of results and preparation of the manuscript. All authors have read and approved the final manuscript
Contributor Information
Ylanna Burgos, Email: Ylana.Kelner-Burgos@bfr.bund.de.
Lothar Beutin, Email: lothar.beutin@bfr.bund.de.
Acknowledgements
Y. Burgos was partially supported from Brazil by "Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)", process of number 2006//53805-2. The authors are grateful to Eckhard Strauch (BfR, Berlin) for valuable discussions and suggestions and to Karin Pries for technical assistance.
References
- Welch RA. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- Menestrina G, Moser C, Pellet S, Welch R. Pore-formation by Escherichia coli hemolysin (HlyA) and other members of the RTX toxins family. Toxicology. 1994;87:249–267. doi: 10.1016/0300-483X(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Stanley P, Koronakis V, Hughes C. Acylation of Escherichia coli hemolysin: a unique protein lipidation mechanism underlying toxin function. Microbiol Mol Biol Rev. 1998;62:309–333. doi: 10.1128/mmbr.62.2.309-333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Kernbach C, Karch H. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:h7. Microbiology. 1996;142(Pt 4):907–914. doi: 10.1099/00221287-142-4-907. [DOI] [PubMed] [Google Scholar]
- Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Mol Membr Biol. 2005;22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Karch H, Beutin L. The large-sized plasmids of enterohemorrhagic Escherichia coli O157 strains encode hemolysins which are presumably members of the E. coli alpha-hemolysin family. FEMS Microbiol Lett. 1994;117:189–196. doi: 10.1111/j.1574-6968.1994.tb06763.x. [DOI] [PubMed] [Google Scholar]
- Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145(Pt 5):1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertschinger HU, Gyles CL. In: Escherichia coli in domestic animals and humans. Gyles CL, editor. Wallingford: CAB International; 1994. Oedema Disease of Pigs; pp. 193–219. [Google Scholar]
- Prada J, Baljer G, De Rycke J, Steinruck H, Zimmermann S, Stephan R. Characteristics of alpha-hemolytic strains of Escherichia coli isolated from dogs with gastroenteritis. Vet Microbiol. 1991;29:59–73. doi: 10.1016/0378-1135(91)90110-2. [DOI] [PubMed] [Google Scholar]
- Hampson DJ. In: Escherichia coli in Domestic animals and Humans. Gyles CL, editor. Wallingford: CAB International; 1994. Postweaning Escherichia coli Diarrhoea in Pigs; pp. 171–191. [Google Scholar]
- Beutin L. Escherichia coli as a pathogen in dogs and cats. Vet Res. 1999;30:285–298. [PubMed] [Google Scholar]
- Guyer DM, Kao JS, Mobley HL. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and Catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JS, Stucker DM, Warren JW, Mobley HL. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Janke B, Piechaczek K, Nagy G, Ziebuhr W, Fischer G. Toxin genes on pathogenicity islands: impact for microbial evolution. Int J Med Microbiol. 2000;290:307–311. doi: 10.1016/S1438-4221(00)80028-4. [DOI] [PubMed] [Google Scholar]
- Swenson DL, Bukanov NO, Berg DE, Welch RA. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70:6365–6372. doi: 10.1128/IAI.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Altenhoefer A, Knapp O, Maier E, Dobrindt U, Blum-Oehler G. Both alpha-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect. 2006;8:2006–2012. doi: 10.1016/j.micinf.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Prada J, Zimmermann S, Stephan R, Beutin L. Restriction fragment length polymorphisms associated with alpha-hemolysin determinants are correlating with the expression of alpha-hemolysin in strains of Escherichia coli. Zentralbl Bakteriol. 1992;276:152–164. doi: 10.1016/s0934-8840(11)80002-2. [DOI] [PubMed] [Google Scholar]
- Knapp S, Then I, Wels W, Michel G, Tschape H, Hacker J. Analysis of the flanking regions from different haemolysin determinants of Escherichia coli. Mol Gen Genet. 1985;200:385–392. doi: 10.1007/BF00425721. [DOI] [PubMed] [Google Scholar]
- Burgos YK, Pries K, Pestana de Castro AF, Beutin L. Characterization of the alpha-haemolysin determinant from the human enteropathogenic Escherichia coli O26 plasmid pEO5. FEMS Microbiol Lett. 2009;292:194–202. doi: 10.1111/j.1574-6968.2009.01496.x. [DOI] [PubMed] [Google Scholar]
- Wu XY, Chapman T, Trott DJ, Bettelheim K, Do TN, Driesen S. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl Environ Microbiol. 2007;73:83–91. doi: 10.1128/AEM.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunig HM, Lebek G. Haemolytic activity and characteristics of plasmid and chromosomally borne hly genes isolated from E. coli of different origin. Zentralbl Bakteriol Mikrobiol Hyg [A] 1988;267:485–494. [PubMed] [Google Scholar]
- Hess J, Wels M, Vogel M, Goebel W. Nucleotide sequence of a plasmid-encoded hemolysin determinant and its caomparison with a corresponding chromosomal hemolysin sequence. FEMS Microbiol Lett. 1986;34:1–11. [Google Scholar]
- Strathdee CA, Lo RY. Extensive homology between the leukotoxin of Pasteurella haemolytica A1 and the alpha-hemolysin of Escherichia coli. Infect Immun. 1987;55:3233–3236. doi: 10.1128/iai.55.12.3233-3236.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada J, Beutin L. Detection of Escherichia coli alpha-haemolysin genes and their expression in a human faecal strain of Enterobacter cloacae. FEMS Microbiol Lett. 1991;63:111–114. doi: 10.1016/0378-1097(91)90537-k. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Cross M, Senior B, Koronakis E, Hughes C. The secreted hemolysins of Proteus mirabilis, Proteus vulgaris, and Morganella morganii are genetically related to each other and to the alpha-hemolysin of Escherichia coli. J Bacteriol. 1987;169:1509–1515. doi: 10.1128/jb.169.4.1509-1515.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Hess J, Then I, Juarez A, Goebel W. Characterization of a sequence (hlyR) which enhances synthesis and secretion of hemolysin in Escherichia coli. Mol Gen Genet. 1988;212:76–84. doi: 10.1007/BF00322447. [DOI] [PubMed] [Google Scholar]
- Beutin L, Kruger U, Krause G, Miko A, Martin A, Strauch E. Evaluation of major types of Shiga toxin 2e producing Escherichia coli present in food, pigs and in the environment as potential pathogens for humans. Appl Environ Microbiol. 2008. [DOI] [PMC free article] [PubMed]
- Strathdee CA, Lo RY. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J Bacteriol. 1989;171:916–928. doi: 10.1128/jb.171.2.916-928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen E, Rousseau P, Duval-Valentin G, Chandler M. Truncated forms of IS911 transposase downregulate transposition. Mol Microbiol. 2006;62:1102–1116. doi: 10.1111/j.1365-2958.2006.05424.x. [DOI] [PubMed] [Google Scholar]
- Frechon D, Le Cam E. Fur (ferric uptake regulation) protein interaction with target DNA: comparison of gel retardation, footprinting and electron microscopy analyses. Biochem Biophys Res Commun. 1994;201:346–355. doi: 10.1006/bbrc.1994.1708. [DOI] [PubMed] [Google Scholar]
- Khalaf NG, Eletreby MM, Hanson ND. Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae clinical isolates from Cairo, Egypt. BMC Infect Dis. 2009;9:84. doi: 10.1186/1471-2334-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I, Souli M, Koratzanis E, Koratzanis G, Chryssouli Z, Giamarellou H. Emerging bacterial pathogens: Escherichia coli, Enterobacter aerogenes and Proteus mirabilis clinical isolates harbouring the same transferable plasmid coding for metallo-beta-lactamase VIM-1 in Greece. J Antimicrob Chemother. 2007;59:578–579. doi: 10.1093/jac/dkl508. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Dykhuizen DE, Dubose RF, Green L, Mutangaduramhlanga T, Wolczyk DF. Distribution and Abundance of Insertion Sequences Among Natural Isolates of Escherichia-Coli. Genetics. 1987;115:51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Hartl DL. Nonrandom location of IS1 elements in the genomes of natural isolates of Escherichia coli. Mol Biol Evol. 1997;14:725–732. doi: 10.1093/oxfordjournals.molbev.a025812. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Leonard C, Chandler M. IS elements as constituents of bacterial genomes. Res Microbiol. 1999;150:675–687. doi: 10.1016/S0923-2508(99)00124-2. [DOI] [PubMed] [Google Scholar]
- Bachellier S, Gilson E, Hofnung M, Hill CW. In: Escherichia coli and Salmonella. 2. Neidhardt FC, Curtiss R III, Ingraham J, Lin ECC, Low KB, Megasanik WS, et al, editor. Vol. 2. Washington D.C.: ASM Press; 2010. Repeated Sequences; pp. 2012–2040. [Google Scholar]
- Galas DJ, Chandler M. In: Mobile DNA. Berg DE, Howe MM, editor. Washington D.C.: ASM; 1989. Bacterial Insertion Sequences; pp. 109–162. [Google Scholar]
- Jorgensen ST, Poulsen AL. Antibiotic resistance and Hly plasmids in serotypes of Escherichia coli associated with porcine enteric disease. Antimicrob Agents Chemother. 1976;9:6–10. doi: 10.1128/aac.9.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol Lett. 2005;249:335–342. doi: 10.1016/j.femsle.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Han W, Liu B, Cao B, Beutin L, Kruger U, Liu H. DNA Microarray-Based Identification of Serogroups and Virulence Gene Patterns of Escherichia coli Isolates Associated with Porcine Postweaning Diarrhea and Edema Disease. Appl Environ Microbiol. 2007;73:4082–4088. doi: 10.1128/AEM.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L, Zimmermann S, Gleier K. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J Clin Microbiol. 1996;34:2812–2814. doi: 10.1128/jcm.34.11.2812-2814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Welch RA, Hull R, Falkow S. Molecular cloning and physical characterization of a chromosomal hemolysin from Escherichia coli. Infect Immun. 1983;42:178–186. doi: 10.1128/iai.42.1.178-186.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]