Abstract
The causative agent of severe acute respiratory syndrome (SARS) is a previously unidentified coronavirus, SARS-CoV. The RNA-dependent RNA polymerase (RdRp) of SARS-CoV plays a pivotal role in viral replication and is a potential target for anti-SARS therapy. There is a lack of structural or biochemical data on any coronavirus polymerase. To provide insights into the structure and function of SARS-CoV RdRp, we have located its conserved motifs that are shared by all RdRps, and built a three-dimensional model of the catalytic domain. The structural model permits us to discuss the potential functional roles of the conserved motifs and residues in replication and their potential interactions with inhibitors of related enzymes. We predict important structural attributes of potential anti-SARS-CoV RdRp nucleotide analog inhibitors: hydrogen-bonding capability for the 2′ and 3′ groups of the sugar ring and C3′ endo sugar puckering, and the absence of a hydrophobic binding pocket for non-nucleoside analog inhibitors similar to those observed in hepatitis C virus RdRp and human immunodeficiency virus type 1 reverse transcriptase. We propose that the clinically observed resistance of SARS to ribavirin is probably due to perturbation of the conserved motif A that controls rNTP binding and fidelity of polymerization. Our results suggest that designing anti-SARS therapies can benefit from successful experiences in design of other antiviral drugs. This work should also provide guidance for future biochemical experiments.
INTRODUCTION
Severe acute respiratory syndrome (SARS) is a new viral disease that has spread to 32 countries and has resulted in more than 800 deaths from respiratory distress syndrome (1–3). The causative agent of SARS is a previously unidentified coronavirus, SARS-CoV (4–6), which is closely related to group II coronaviruses that include human virus OC43 and mouse hepatitis virus (7). Treatment of SARS with antiviral agents such as ribavirin and corticosteroids has not achieved satisfactory results (8). Furthermore, there is not yet a vaccine available for protection against SARS.
Coronaviruses are a group of enveloped positive strand RNA viruses. The viral genome of SARS-CoV is a single-stranded RNA of 29 727 nucleotides (9–11). By analogy with other coronaviruses, SARS-CoV gene expression is predicted to involve complex transcriptional and translational events (12). The 5′ two-thirds of the genome encode the replicase gene (∼21 kb) that is expressed by two very large open reading frames (ORFs), 1a and 1b. Expression of SARS-CoV proteins is expected to start with translation of two polyproteins, pp1a and pp1ab, with predicted lengths of 4328 and 7023 amino acids, respectively. pp1ab is the result of a translational frameshifting event at the end of ORF1a. These polyproteins undergo co-translational proteolytic processing into at least four key enzymes: an RNA-dependent RNA polymerase (RdRp), a picornavirus 3C-like proteinase, a papain-like proteinase and a helicase.
SARS-CoV RdRp is the essential enzyme in a replicase complex that is expected to contain additional viral and cellular proteins. The replicase complex is primarily used to transcribe: (i) full-length negative and positive strand RNAs; (ii) a 3′-co-terminal set of nested subgenomic mRNAs that have a common 5′ ‘leader’ sequence derived from the 5′ end of the genome; and (iii) subgenomic negative strand RNAs with common 5′ ends and leader complementary sequences at their 3′ ends (11,12).
Sequence comparisons and mutagenesis studies of RdRps from a wide range of RNA viruses have identified several conserved sequence motifs that are important for biological functions (13–19). Four of these conserved motifs exist in all polymerases (apart from polymerase β and multisubunit DNA-dependent RNA polymerases) and reside in their catalytic domain. Crystal structures of RdRps from five different RNA viruses have also been reported, including poliovirus (PV) (20), hepatitis C virus (HCV) (21–24), rabbit hemorrhagic disease virus (RHDV) (25), reovirus (RV) (26) and bacteriophage φ6 (φ6) (27). Those studies have revealed key aspects of the structural biology of RdRps and confirmed the hypothesis that RdRps share a common architecture and mechanism of polymerase catalysis (13).
Given the crucial role of RdRp in the virus life cycle and the success obtained with polymerase inhibitors in the treatment of viral infections, including human immunodeficiency virus type 1 (HIV-1), human hepatitis B virus (HBV), HCV and herpes virus, SARS-CoV RdRp is an attractive target for development of anti-SARS drugs. Yet there are no structural and very limited biochemical data on coronavirus polymerases.
To understand the structural basis of SARS-CoV RdRp enzymatic activity and potential drug susceptibility, we compared the sequence of SARS-CoV polymerase with those of PV, HCV, RHDV, RV, φ6 and HIV-1 polymerases whose crystal structures are known. Based on sequence comparisons, we have located the conserved sequence motifs that are shared in all RdRps and built a three-dimensional model of the catalytic domain. We also describe the potential roles of specific residues in the polymerization mechanism and in recognition of potential inhibitors. Structural analysis of SARS-CoV RdRp is likely to aid the development of anti-SARS agents and provide guidance in the design of future biochemical experiments.
MATERIALS AND METHODS
Sequence alignments
The sequence of SARS-CoV RdRp (932 amino acid residues; strain CUHK; NCBI accession no. AAP13566) was aligned with that of representatives of the other three groups of coronaviruses, and of five viral RdRps whose crystal structures are known. The coronaviruses used in sequence alignment are: group I, human coronavirus 229E (HcoV-229E; NCBI accession no. NC_002645); group II, murine hepatitis virus (MHV; NCBI accession no. NC_001846); and group III, avian infectious bronchitis virus (AIBV; NCBI accession no. NC_001451). The five viral RdRps are: PV, RHDV, HCV, RV and φ6 polymerases. HIV-1 reverse transcriptase (RT), which is both an RNA-dependent and DNA-dependent DNA polymerase, is also included in the sequence comparison because of the availability of abundant structural and functional data.
Sequence alignments of SARS-CoV RdRp with other coronavirus RdRps were obtained using program CLUSTAL-W (28). The SARS-CoV RdRp shares very high amino acid sequence identity with other coronavirus RdRps, but has very low sequence similarity to other viral RdRps and RTs of known structures. Therefore, any conventional method of sequence alignment is not helpful. We carried out the sequence comparison of SARS-CoV RdRp with other viral RdRps and HIV-1 RT of known crystal structures primarily based on manual alignments. The crystal structures of HCV, PV, RHDV, RV, φ6 and HIV-1 polymerases were used as guides in the sequence alignments to identify the conserved sequence motifs of SARS-CoV RdRp.
Comparison of the crystal structures of HCV, PV, RHDV, RV, φ6 and HIV-1 polymerases allowed us to align both the structures and primary sequences and identify the consensus sequences of the conserved motifs that are shared in all RdRps and RTs (motifs A–G). These consensus sequences were used as reference points to locate the conserved motifs in SARS-CoV RdRp. Comparison of the sequences of SARS-CoV and other coronarivus RdRps with those of HCV, PV, RHDV, RV, φ6 and HIV-1 polymerases allowed us to initially identify motifs A, B and C in SARS-CoV RdRp. For motif A, we searched for two strictly conserved aspartates separated by four residues. Motif B was expected to contain a strictly conserved ‘XSG’ sequence followed by a conserved threonine and a conserved asparagine in a long α-helix. For motif C, we searched for a conserved ‘XDD’ sequence. There are three ‘XDD’ sequences in SARS-CoV RdRp. We chose the first ‘XDD’ sequence as motif C because: (i) it is strictly conserved in all coronavirus RdRps (‘SDD’ in all); (ii) it is located between two predicted β-strands; and (iii) there are an appropriate number of residues at the C-terminus to accommodate conserved motifs D and E and the thumb subdomain. The locations of these three motifs facilitated the identification of other conserved motifs. The consensus ‘SXG’ sequence of motif G was identified based on the sequence alignment of SARS-CoV RdRp with PV and RHDV RdRps. Motif F was located by identifying several conserved positively charged, basic residues (K or R) based on the sequence alignment of SARS-CoV RdRp with other viral RdRps in the region between motifs G and A. Based on structural comparison of all viral RdRps and RTs of known structures, motif D appears to contain a hydrophilic residue (R/K/E/Q) in the middle of an α-helix, a polar residue (R/D/E/Y) at the C-terminus of the α-helix, and an aromatic residue (F/Y/W) in the following turn. We assigned motif D based on the alignment of a predicted α-helix after motif C in SARS-CoV RdRp with that in other viral RdRp structures, as well as the positions of a hydrophilic residue (K) and a residue containing a polar group (Y) in the C-terminus of the predicted α-helix and an aromatic residue (F/Y/W) in the following turn. Finally, motif E was recognized based on the position of a conserved aromatic residue and the sequence similarity (XCS) with HCV RdRp at the turn of the conserved hairpin structure following motif D. Subsequently, these conserved motifs were used as landmarks to guide further sequence and secondary structural element alignments. In less conserved regions or regions containing insertions or deletions, we adjusted the alignments manually by taking account of appropriate alignment of the predicted secondary structure of SARS-CoV RdRp with the secondary structures of other viral RdRps, and the properties of amino acids (hydrophobic or hydrophilic character).
The secondary structure prediction of SARS-CoV RdRp was performed using the program PHD (29,30). This program uses cascading neural network algorithms that have been trained on several hundred non-homologous protein structures. The accuracy of PHD has been reported to be on average >70% as judged by the Q3 index for globular proteins. Q3 and SOV are two widely used accuracy indices to evaluate algorithms of protein secondary structure prediction (31,32). To evaluate the reliability and accuracy of the PHD program, we carried out positive controls. We applied the program to the sequences of RHDV, HCV, RV and φ6 polymerases whose structures are known and were used in our study for structure comparisons (PV polymerase was not used in the test because its structure is less complete). The estimated accuracy of the predictions was judged by comparing the predicted secondary structures with the actual secondary structures extracted from the published structural results. For all four proteins, the overall Q3 index was 70–75% and the overall SOV index was 67–72.5% (data not shown). The accuracy of prediction for α-helices was higher than the average values (the Q3 index of 71–82% and the SOV index of 69–87.5%, respectively). The accuracy of prediction for β-strands was relatively low (the Q3 index of 43–62% and the SOV index of 44–65%, respectively). The accuracy of prediction for conserved motifs depends on their specific secondary structure: motifs consisting of α-helices were predicted with high reliability; motifs forming β-strands and random coils less accurately. Therefore, PHD was used in the secondary structure prediction of SARS-CoV RdRp, and the resulting secondary structure prediction should be considered as reasonably reliable, especially for the helical regions.
Homology modeling
Initial models of SARS-CoV RdRp were obtained using the MODELLER program (33) that generates three-dimensional structures based on amino acid sequence alignments of a molecule with one or more template structures. We used as template structures in the homology modeling the structures of HCV (PDB code 1QUV), PV (PDB code 1RDR), RHDV (PDB code 1KHV), RV (PDB codes 1N35 and 1N1H), φ6 (PDB codes 1HI0 and 1HI1) and HIV-1 (PDB codes 1RTD and 2HMI) polymerases. The scaffold of SARS-CoV RdRp was based on the crystal structure of RHDV polymerase. Other models derived from the crystal structures of HCV, PV, RV and φ6 polymerases were used as additional guides in building the molecular model of SARS-CoV RdRp. We built manually the less conserved regions and regions containing insertions and deletions using the graphics program O (34), consulting reference databases of known main chain and side chain conformations and preferred side chain rotamers. Buried side chains were manually adjusted to avoid steric conflict or to have favorable interactions with neighboring residues. The region of SARS-CoV RdRp residues 712–751 has no equivalent in other RdRps of known structure. Therefore, this region was not built in the current model. The N-terminal region (residues 376–388) and the C-terminal region (residues 891–932) are also omitted in the model because of the lack of consensus template structures for these regions. The final model was energy minimized using the molecular dynamics simulation procedure in program MODELLER. The quality and stereochemistry of the model were evaluated using the program PROCHECK (35). The main chain conformations for 99.3% of amino acid residues were within the favored or allowed regions of the Ramachandran plot, and the overall G factor was –0.23, indicating that the molecular geometry of the model is of good quality. Secondary structure assignments for the final model agree well with the secondary structure predicted from the sequence using the PHD program.
Models of the RNA–RNA template–primer and rNTP were built based on the structures of RV polymerase (26), φ6 polymerase (27) and HIV-1 RT (36,37) in their complexes with nucleic acid and NTP or dNTP substrates. The corresponding structures of these complexes were superimposed onto the structural model of SARS-CoV RdRp based on structural alignment of the palm subdomains. An A-form RNA template–primer duplex could be docked into the nucleic acid-binding cleft with only minor steric conflicts with structural elements of the protein.
RESULTS AND DISCUSSION
Sequence comparisons
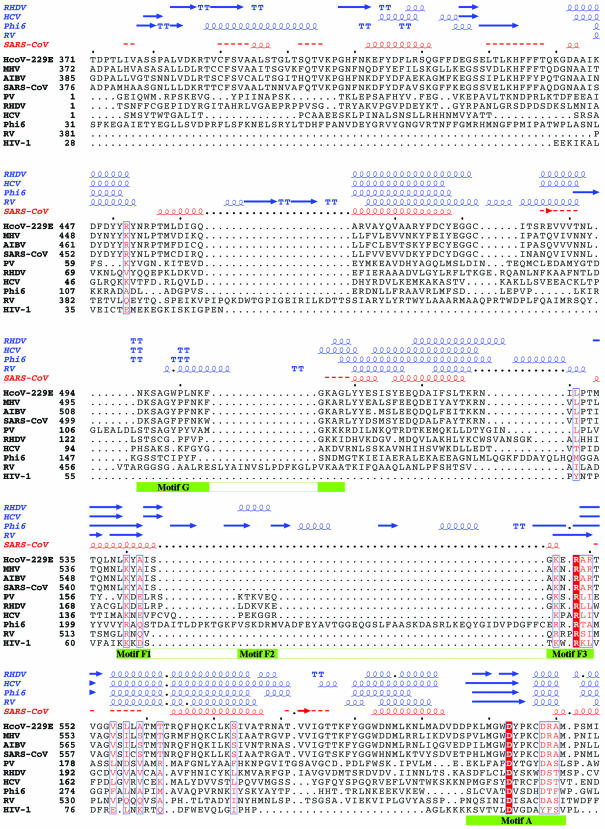
SARS-CoV RdRp is predicted to contain 932 amino acids. The N-terminal portion of SARS-CoV and other coronavirus RdRps is large and has no counterpart in other positive strand RNA virus RdRps of known structures. Thus, we consider this N-terminal portion of SARS-CoV RdRp (residues 1–375) as an N-terminal domain (NTD) and the C-terminal portion (residues 376–932) that is equivalent to other polymerases as the polymerase catalytic domain. Sequence alignments and comparisons indicate that SARS-CoV RdRp has a high sequence identity with other coronavirus RdRps (∼62–73%). However, it shares <10% sequence identity with other viral RdRps and RTs, including PV, HCV, RHDV, RV and φ6 RdRps and HIV-1 RT whose structures are known (Fig. 1). Normally, such a low level of homology would not permit reliable sequence alignment and homology modeling. However, we applied a stepwise protocol that relied on manual identification of key conserved motifs and used them as landmarks to guide subsequent alignment of primary sequence. Crucial for the sequence alignments were also the prediction of the secondary structure of SARS-CoV RdRp and the appropriate alignment of the predicted secondary structure elements of SARS-CoV RdRp with the secondary structures of PV, HCV, RHDV, RV and φ6 RdRps (as observed in the corresponding crystal structures of these enzymes) (Fig. 1). Prediction tests on the secondary structures of RHDV, HCV, RV and φ6 RdRps using the PHD program give overall accuracy indices Q3 >70% and SOV >67%. Nevertheless, it should be noted that the sequence alignment in the fingers and thumb subdomains is less reliable due to the low sequence similarity between SARS-CoV RdRp and other viral RdRps, and large structural variations among the known RdRp structures.
Figure 1.
Sequence alignment of the RdRp of SARS-CoV with those of representatives of the other three classes of coronaviruses and of five RNA viruses with known crystal structures. The representative coronaviruses are: group I, human coronavirus 229E (HcoV-229E; NCBI accession no. NC_002645); group II, murine hepatitis virus (MHV; NCBI accession no. NC_001846); and group III, avian infectious bronchitis virus (AIBV; NCBI accession no. NC_001451). The five RNA viruses are poliovirus 1 strain Mahoney (PV; PDB code 1RDR), rabbit hemorrhagic disease virus (RHDV; PDB code 1KHV), hepatitis C virus (HCV; PDB code 1QUV), reovirus (RV; PDB codes 1N35 and 1N1H) and bacteriophage φ6 (Phi6; PDB codes 1HI0 and 1HI1). HIV-1 RT (HIV-1; PDB code 1RTD), a widely studied RNA-dependent and DNA-dependent polymerase, is also included in the comparison. The sequence in the palm subdomain and regions containing the conserved motifs (highlighted with green bars) can be aligned confidently among different viral RdRps and HIV-1 RT. However, the sequence in the fingers and thumb subdomains is less conserved between SARS-CoV RdRp and other viral RdRps, and the structure in those subdomains also varies substantially among the known RdRp structures. Thus, the sequence alignment and the structural model in these regions are less reliable. Invariant residues are highlighted in a shaded red box, and conserved residues are in red. The secondary structures of RHDV, HCV, RV and φ6 polymerases extracted from the corresponding structures and the predicted secondary structure of SARS-CoV RdRp are shown above the sequence alignment. α-Helices are shown as spirals and β-strands as arrows. The alignment was drawn with ESPript (66).
Structural model of SARS-CoV RdRp
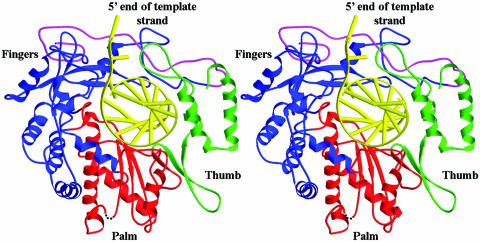
After identification of the conserved sequence motifs and establishment of reliable sequence alignments, we built a three-dimensional homology model of the catalytic domain of SARS-CoV RdRp based on the crystal structures of HCV, PV, RHDV, RV, φ6 and HIV-1 polymerases. By analogy with other polymerases, the catalytic domain of SARS-CoV RdRp consists of fingers, palm and thumb subdomains that form an encircled nucleic acid-binding tunnel (Fig. 2). Analysis of the structural model permits us to discuss the potential functional roles of the conserved motifs and specific residues in polymerization (Fig. 3 and Table 1).
Figure 2.
Ribbon diagram of the homology model of SARS-CoV RdRp with a docked RNA template–primer. α-Helices are shown as spirals and β-strands as arrows. The subdomains of the catalytic domain are colored as the N-terminal portion of the fingers subdomain (376–424) in magenta, the base of the fingers (residues 425–584 and 626–679) in blue, palm (residues 585–625 and 680–807) in red, and thumb (residues 808–932) in green.
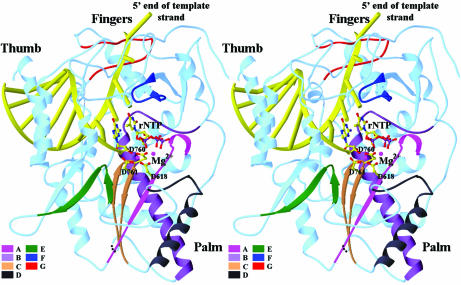
Figure 3.
Stereoview of the polymerase active site and the rNTP-binding site. The conserved sequence motifs (A–G) are highlighted. A docked rNTP substrate is shown as a ball-and-stick model. The catalytic active site is defined by the three conserved aspartates, Asp618, Asp760 and Asp761 (shown with side chains) that are coordinated with two divalent metal ions (shown as magenta spheres).
Table 1. Conserved motifs of SARS-CoV RdRp and their potential functions.
| Motif | Sequence | Possible functions | References |
|---|---|---|---|
| A | 612 PHLMGWDYPKCDRAM | Asp618: metal ion chelation | (24,26,27,36,40) |
| Asp623: recognition of rNTP sugar ring | (24,26,27) | ||
| B | 678 GGTSSGDATTAYANSVFNICQAVTANVNALLST | Ser682 and Thr687: recognition of template–primer | (21,25–27) |
| Ser682, Thr687 and Asn691: help sugar selection of rNTP | (20,21,23–26) | ||
| C | 753 FSMMILSDDAVVCYN | Asp760 and Asp761: metal ion chelation | (24,26,27,36,40) |
| Ser759: binding of 3′-primer terminus or priming nucleotide | (36,37) | ||
| D | 771 AAQGLVASIKNFKAVLYYQNNVFMSE | Stabilize the core structure; may also help position Asp618 | |
| E | 810 HEFCSQHTMLV | Control the flexibility of the thumb | (36,37,45) |
| Cys813 and Ser814: positioning of priming nucleotide | (26,27,37,45) | ||
| F | 544 LKYAISAKNRARTVAGV | Lys545, Lys551 and Arg553: rNTP binding and positioning of template overhang | (24,26,27,36) |
| G | 499 DKSAGFPFNKWGK | Positioning of template overhang | (26,27) |
The conserved motifs of SARS-CoV RdRp are assigned based on manual sequence alignments and structural comparisons with other viral RdRps of known structures. The highlighted residues are highly conserved in most viral RdRps. The potential functions of these motifs and specific residues are proposed for SARS-CoV RdRp based on comparisons with other RdRps whose structures and functions are known.
N-terminal domain
SARS-CoV and other coronavirus RdRps contain an NTD (approximately residues 1–375 in SARS-CoV RdRp) that is expected to form at least one protein domain. There is no equivalent structural domain in other positive strand RNA virus RdRps with known structures. The double-stranded RNA RV RdRp contains an NTD that is comparable in size with that of SARS-CoV RdRp. However, the two NTDs share a very low sequence similarity (<12% identity) and contain no conserved sequence motif, making it difficult to perform a reliable sequence alignment and build a meaningful homology model of the NTD of SARS-CoV RdRp. The functional implications of this domain are unclear. The NTD of RV polymerase bridges the fingers and thumb subdomains on one side of the catalytic cleft and participates in the formation of a channel through which the incoming nucleotide is likely to diffuse into the active site during polymerization (26). It is plausible that some of the coronavirus-specific replicase and transcription activities map to this domain. For example, this domain may be involved in interactions with the leader or intragenic sequences during transcription of the characteristic nested mRNAs of coronaviruses and/or in protein–protein interactions with the viral helicase or other viral and/or host proteins involved in coronavirus replication.
Fingers subdomain
The sequence and structure of the fingers subdomain are less conserved than those of the palm subdomain among different viral RdRps. In all known RdRp structures, the fingers subdomain is composed of two polypeptide segments, an N-terminal segment and a segment spanning motifs A and B of the palm subdomain. The ‘base’ of the fingers is mainly α-helical and the ‘tip’ of the fingers consists primarily of β-strands and random coils. The SARS-CoV RdRp fingers subdomain spans approximately from residues 376 to 584 and 626 to 679 and is also predicted to consist of α-helices in the base and β-strands and coils in the tip (Figs 1 and 2). Despite the high sequence variability among the fingers subdomains of different viral RdRps, there are two conserved sequence motifs (F and G) shared by all RdRps that play important functional roles in the mechanism of polymerization (Figs 1 and 3).
Similarly to HCV and RHDV RdRps, the fingers subdomain of SARS-CoV RdRp contains an N-terminal portion (residues 405–444) that forms a long loop emanating from the fingertip that bridges the fingers and thumb subdomains (Fig. 2). In the PV RdRp structure, the equivalent region is disordered, and in the φ6 RdRp structure it has a different structural fold but also bridges the fingers and thumb subdomains (27). In RV polymerase, however, the bridging of the two subdomains is accomplished by the NTD (26). As a result of these interactions, all RdRps form an encircled nucleic acid-binding ‘tunnel’ that can accommodate binding and translocation of a nucleic acid without major conformational changes of the enzymes. This is different from HIV-1 RT and other DNA polymerases that form a U-shaped DNA-binding cleft due to the lack of the fingers–thumb subdomain interaction and require large-scale subdomain movements to accommodate the template–primer and dNTP substrates. The interaction of these subdomains is believed to ensure coordinated movement and help modulate initiation, elongation and termination of RNA synthesis by contributing to high processivity of viral replication (26,27). The N-terminal region of the fingers subdomain is also suggested to be involved in recognition of nucleotide substrate, protein–protein interactions and oligomerization of the polymerase (20,38,39).
Motif F. Motif F contains several conserved positively charged, basic residues (K or R) and has been proposed to consist of three submotifs, F1, F2 and F3 (19). Submotif F2 does not appear to be present in SARS-CoV, RV and HIV-1 polymerases (Fig. 1). Motif F forms part of a ‘β-strand, loop and β-strand’ structure that, similarly to the N-terminal loop (see above), also extends from the fingers to interact with the thumb. The size of the loop, however, varies in different polymerases. In φ6 polymerase, motif F is ∼60 residues longer than in other polymerases because of two insertions between submotifs F1 and F2 (15 residues), and submotifs F2 and F3 (40 residues), respectively (19). In SARS-CoV RdRp, motif F contains several highly conserved basic residues, including Lys545, corresponding to submotif F1, and Lys551 and Arg553, corresponding to submotif F3 (Figs 1 and 3; Table 1).
In HIV-1 RT, the structural element containing motif F rotates inwards towards the polymerase active site upon binding of dNTP, allowing the three conserved residues (Lys65, Lys70 and Arg72) to interact with the triphosphate of the incoming dNTP (36). In HCV, RHDV, RV and φ6 RdRps, this structural element adopts a closed conformation and has (or is proposed to have) no major conformational change upon rNTP binding (22–24,26,27). Though the three conserved basic residues are separated by a varying size of residues in the primary sequence, they are structurally close to each other and interact with the incoming rNTP and the template overhang. In the structural model of SARS-CoV RdRp, residues of motif F are also predicted to form part of the rNTP-binding pocket and help position the template overhang (Fig. 3 and Table 1).
Motif G. Motif G consists of a conserved SXGXP sequence possibly followed by a conserved basic residue in many RdRps (18). The same motif can be found in SARS-CoV (corresponding to Ser501, Gly503, Pro505 and Lys511), PV and RHDV RdRps (Fig. 1). These residues are less conserved in HCV, RV and φ6 polymerases and do not exist in HIV-1 RT. The segment containing motif G forms a ‘loop and α-helix’ in most RdRp structures, except RV RdRp which has a 16-residue insertion between the loop and the α-helix (Figs 1 and 3). In the structures of RV and φ6 polymerases, residues of motif G contact the nucleic acid at its 5′ template overhang and form part of the channel for the template strand (26,27). In the structural model of SARS-CoV RdRp, residues of motif G are also predicted to be involved in positioning of the 5′ template strand (Fig. 3 and Table 1).
Palm subdomain
The palm subdomain of SARS-CoV RdRp (residues 585–625 and 680–807) forms the catalytic core of polymerase and contains the four highly conserved sequence motifs (A–D) found in all polymerases and a fifth motif (E) unique to RdRps and RTs (14). The core structure of the palm subdomain is well conserved across all classes of polymerases and is primarily comprised of a central three-stranded β-sheet flanked by two α-helices on one side and a β-sheet and an α-helix on the other (Figs 2 and 3). Residues forming the catalytic active site are found within motifs A and C.
Motif A. As in all viral RdRps, motif A of SARS-CoV RdRp contains two highly conserved aspartic acid residues separated by four residues (Asp618 and Asp623) (Fig. 1 and Table 1). Motif A is composed of a ‘β-strand and short α-helix’ structure. The β-strand of motif A, together with the β-strands formed by motif C, forms the central β-sheet (Fig. 3). The first aspartate (Asp618) is located near the end of the β-strand and, together with the two strictly conserved aspartates in motif C (Asp760 and Asp761), forms the catalytic center of SARS-CoV RdRp. Structural studies of other polymerases indicate that the corresponding three aspartates are involved in binding divalent metal ions required for catalysis (24,26,27,36,40). Similar to other polymerases, mutation of any of those aspartates in SARS-CoV RdRps is expected to abrogate polymerase activity. The second aspartate (Asp623) is located in the short α-helix. In the structures of HCV, RV and φ6 polymerases, the corresponding residues (Asp225, Asp590 and Asp329, respectively) form a hydrogen bond with the 2′-OH group of the incoming NTP and appear to be involved in sugar selection (24,26,27). The same interaction has been proposed in PV polymerase (20,22,23,41). The equivalent residues in HIV-1 RT and MMLV RT are Tyr115 and Phe155, respectively. These bulky hydrophobic residues form a steric gate that prevents binding of rNTPs because of their 2′-OH group (36,37,42,43). Asp623 of motif A in SARS-CoV RdRp is expected to also be involved in sugar selection (Table 1).
Motif B. Motif B of SARS-CoV RdRp forms a ‘loop and α-helix’ structure and contains several highly conserved residues (Ser682, Gly683, Thr687 and Asn691) that appear to participate in recognition of the correct nucleic acid and selection of the correct substrate (Figs 1 and 3). As in other RdRp structures, the N-terminal loop of motif B contains three conserved residues (Ser682, Gly683 and Thr687) that appear to interact with the nucleotide that base-pairs with the incoming rNTP (21,25–27). The equivalent residues in HIV-1 RT (Gln151, Gly152 and Ser156) are also involved in positioning the template nucleotide that base-pairs with the incoming dNTP (36,37,42). The α-helical part of motif B, together with an α-helix formed by motif D, packs beneath the central β-sheet (Fig. 3). The conserved asparagine on this α-helix (corresponding to Asn691 in SARS-CoV, Asn291 in HCV, Asn317 in RHDV, Asn297 in PV and His691 in RV, respectively; φ6 RdRp has a glycine at this position) is proposed to contribute to the specificity of RdRp for rNTP versus dNTPs via a hydrogen-bonding interaction with the second conserved aspartate of motif A which in turn hydrogen-bonds to the 2′-OH of rNTP (20,21,23–26). The equivalent residue in HIV-1 RT (Phe160) forms hydrophobic interactions with the side chain of Tyr115 of motif A, which in turn determines the substrate specificity of RT by preventing binding of rNTPs through steric conflicts with their 2′-OH. In SARS-CoV RdRp, Asn691 of motif B appears to interact with Asp623 of motif A through a hydrogen bond and, by analogy, is likely to have similar function (Table 1).
Motif C. SARS-CoV and other coronavirus RdRps contain the highly conserved XSDD motif C (Leu758–Ser759–Asp760–Asp761) at the polymerase active site. This motif forms a ‘β-strand, turn and β-strand’ hairpin structure in all types of polymerases; the two conserved aspartates are located at the turn (Fig. 3). The first two residues of motif C show some degree of variation. The first position has an invariant leucine in all coronavirus RdRps which has no apparent functional role in the molecular model of SARS-CoV RdRp. This position is occupied by a tyrosine in PV (Tyr326), RHDV (Tyr352) and HIV-1 (Tyr183) polymerases. In the HIV-1 RT structure, the phenoxyl group of Tyr183 forms hydrogen bonds with the nucleotide bases of both template and primer strands and is suggested to be involved in positioning the template–primer (37). The equivalent residue (Gln732) in RV polymerase does not interact with nucleic acid. Instead, it has contacts with the sugar ring of the NTP substrate (26). The corresponding residue (Lys451) in φ6 polymerase has no contacts with either nucleic acid or rNTP substrate (27).
The second position of motif C has a serine in all coronavirus and φ6 RdRps, but a glycine in PV, HCV, RHDV and RV RdRps (Fig. 1). Other residues have been observed at this position in RTs and DNA polymerases (44). In the molecular model of SARS-CoV RdRp, Ser759 appears to help position the 3′-primer terminus and/or priming nucleotide (Table 1). The equivalent residue in φ6 polymerase (Ser452) forms a hydrogen bond with the 3′-OH of the priming nucleotide (27). The corresponding residue in HIV-1 RT (Met184) also helps position the 3′ end of the primer strand and the incoming dNTP through hydrophobic interactions with the deoxyribose ring (36,37).
Together with Asp618 of motif A, the two conserved aspartates of motif C (Asp760 and Asp761) form the polymerase active site of SARS-CoV RdRp. The first aspartate (Asp760) is strictly conserved in all polymerases and is coordinated with the metal ions during catalysis (24,26,27,36,40). The corresponding residue in HIV-1 RT (Asp185) can also form a hydrogen bond with the primer terminal 3′-OH, suggesting that it might activate the 3′-OH of the primer strand for nucleophilic attack on the incoming dNTP α-phosphate (36,37,42). The second aspartate (Asp761) is strictly conserved in all RdRps and RTs, but can be replaced by glutamate in several DNA polymerases (44). This residue does not interact with metal ions in most RdRp and RT structures, except in the structures of HCV and RV RdRps (24,26). It may help position the side chains of the other two aspartates and the 3′-primer terminus by interacting with the 3′-terminal phosphate of the primer strand (36,37).
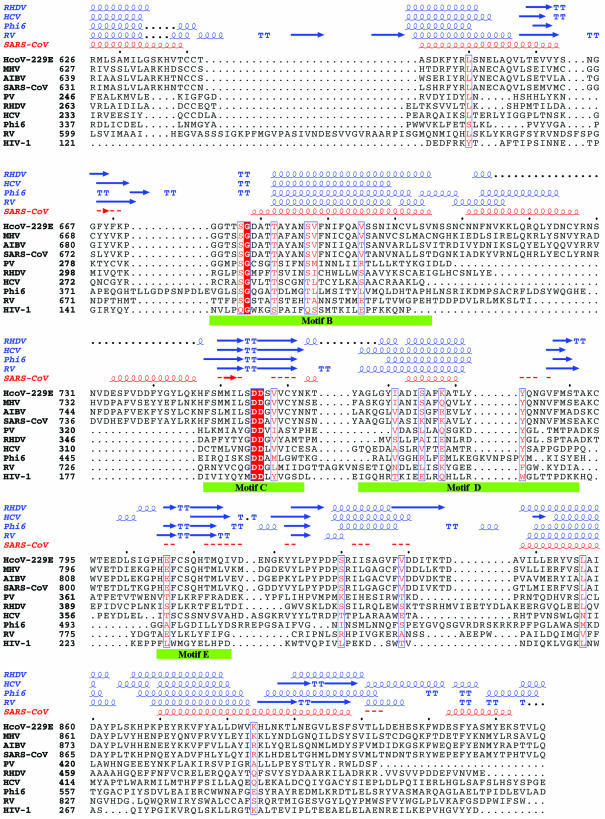
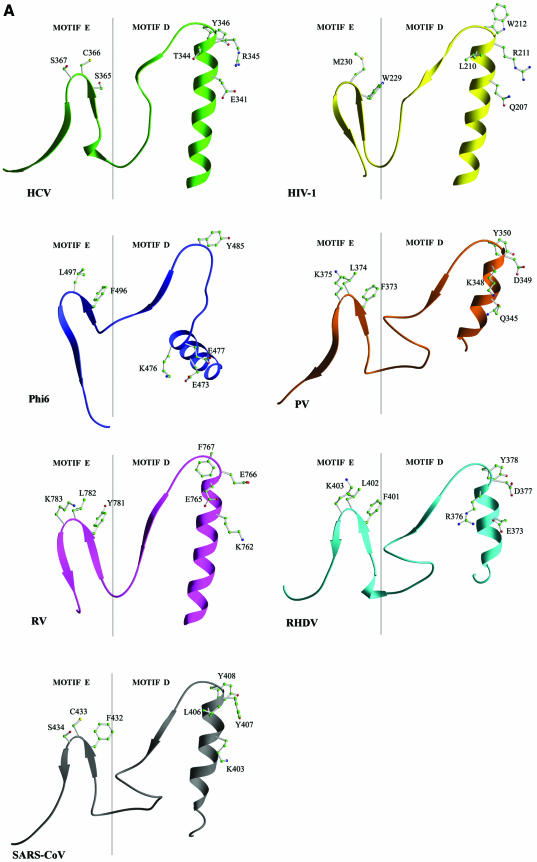
Motif D. Although the primary sequence of motif D is not well conserved, this motif always forms an ‘α-helix, turn and short β-strand’ in all known RdRp and RT structures except φ6 RdRp which contains a seven-residue insertion between the α-helix and the turn (Fig. 4). The α-helix of this motif flanks the central β-sheet containing the catalytic aspartates (Fig. 3). The C-terminal β-strand forms an antiparallel β-sheet with the β-strand of motif A. Structural comparisons indicate that motif D appears to contain a hydrophilic residue in the middle of the α-helix and a polar residue at the C-terminus of the α-helix followed by an aromatic residue at the turn (Lys783, Tyr787 and Tyr788 in SARS-CoV; Glu341, Arg345 and Tyr346 in HCV; Gln345, Asp349 and Tyr350 in PV; Glu373, Asp377 and Tyr378 in RHDV; Lys762, Glu766 and Phe767 in RV; Glu473, Glu477 and Tyr485 in φ6; and Lys207, Arg211 and Trp212 in HIV-1 polymerases, respectively) (Figs 1 and 4). The exact functional role(s) of motif D is not yet clear. It is likely that motif D is involved in stabilizing the core structure of the catalytic domain and in helping position motif A in all viral RdRps, including SARS-CoV RdRp (Table 1).
Figure 4.
Structural comparison of HCV, PV, RHDV, RV, φ6 and SARS-CoV RdRps and HIV-1 RT in the regions containing motifs D and E. (A) Ribbon presentation of motifs D and E in the structures of HCV, PV, RHDV, RV, φ6 and HIV-1 polymerases, and in the structural model of SARS-CoV polymerase. (B) Superposition of the regions containing motifs D and E in different viral RdRp and RT structures [the color coding is the same as in (A)].
Motif E. Motif E is present only in RdRps and RTs, and its primary sequence is not well conserved (14) (Fig. 1). However, motif E has a conserved ‘β-strand, turn and β-strand’ structure that is part of a three-stranded antiparallel β-sheet in all known RdRp and RT structures (Fig. 4). It is located at the junction of the palm and thumb subdomains and is suggested to control the flexibility of the thumb during DNA polymerization (36,37,45). Structural comparison of all viral RdRps and HIV-1 RT studied in this work reveals a possible consensus sequence at the turn that consists of an aromatic residue (F/Y/W) followed by a hydrophobic residue (L/C/M) and a polar residue (S/K) in most RdRps and RTs (Figs 1 and 4). The structural element containing motif E has been designated as the ‘primer grip’ in HIV-1 RT because the residues at the turn (Met230 and Gly231) help position the primer strand at the polymerase active site (37,45). Residues of the primer grip have also been implicated in processivity and fidelity of polymerization (46). The equivalent residues of motif E in RV polymerase (Leu782 and Lys783) and φ6 polymerase (Leu497 and Gly498) interact with the phosphate of the priming rNTP in the initiation complex (26,27). In the molecular model of SARS-CoV RdRp, motif E corresponds to residues 810–820, and the residues at the turn (Cys813 and Ser814) help position the primer strand at the polymerase active site and are likely to contribute to the fidelity of processive polymerization (Table 1).
Thumb subdomain
The C-terminal portion of SARS-CoV RdRp (residues 808–932) contains only the thumb subdomain. The sequence of the thumb subdomain is less conserved in all polymerases. The thumb subdomain of SARS-CoV RdRp is similar in size to that of PV and RHDV RdRps, but considerably smaller than that of HCV, RV and φ6 RdRps. It is likely to assume a similar α-helical structure to that seen in the PV and RHDV RdRp structures (Figs 1 and 2).
Structural studies of HIV-1 RT and other polymerases indicate that the thumb subdomain has great flexibility that is essential for nucleic acid binding and polymerization and appears to function as part of a translocation track during polymerization (36,37,45). However, due to the inflexibility of the nucleic acid-binding cleft in RdRps, both the fingers and thumb subdomains are expected to have only modest conformational changes upon nucleic acid binding (22,23, 25–27). The thumb subdomain of SARS-CoV RdRp is predicted to have a relatively unobstructed nucleic acid-binding cleft that can accommodate double-stranded RNA. This is in contrast to HCV RdRp that has part of the nucleic acid-binding cleft obstructed by a β-hairpin that is proposed to ensure replication of the 3′ portion of the genome during initiation (47).
Implications for design of anti-SARS therapeutics
Two major classes of antiviral agents that target polymerases have been identified: nucleoside analog and non-nucleoside analog inhibitors [see review by De Clercq (48)]. There are several reports on inhibition of flavivirus RdRps with nucleoside and non-nucleoside analog inhibitors. However, there are no data on inhibition of coronavirus polymerase by any inhibitors. We review here the available biochemical data on inhibition of other related RdRps, most notably HCV polymerase, by antiviral agents in the context of the SARS-CoV RdRp model and discuss their inhibitory potential for and interactions with SARS-CoV polymerase.
Potential nucleoside analog inhibitors of SARS-CoV polymerase
Nucleoside analogs are analogs of dNTPs or rNTPs that lack the 3′-OH group. These inhibitors directly compete with nucleotide substrates for binding to the polymerase active site and lead to chain termination once they are incorporated into the elongating chain of the nucleic acid. Nucleoside inhibitors have been widely used in the treatment of HIV-1, HBV, HCV and herpes virus infections.
It has been reported that dNTPs lacking the 3′-OH group (3′-dNTPs) (with cytidine as the preferred nucleobase) can function as chain terminators and inhibit in vitro recombinant HCV polymerase (Kis ranging from 0.6 to 25 µM). However, the inhibition in cell culture is considerably less efficient (49,50). Those results suggest that the 3′-OH of nucleotide analogs is not required for their incorporation; however, a polar group may be required at the 3′ position of the sugar ring to facilitate the activation of the prodrug and nucleoside metabolism. Removal of the 2′-OH resulted in elimination of the inhibitory activity, indicating that the known chain terminators of DNA polymerases, dideoxynucleotides, are not recognized by HCV polymerase (50). Other antivirals that also lack a 2′-OH, such as AZT, do not inhibit RdRps (49,51), consistent with a requirement for a 2′-OH group.
In the molecular model of SARS-CoV RdRp, the 2′-OH group of a docked canonical rNTP (and presumably of a nucleotide analog) interacts with Asp623 of motif A and Asn691 of motif B. The 3′-OH of the rNTP also forms a hydrogen bond with Asp623. Hence, potential nucleoside analog inhibitors of SARS-CoV RdRp should contain groups at the 2′ and 3′ positions that are capable of making hydrogen-bonding interactions with the neighboring Asp623 and Asn691. Also, analysis of the molecular model of SARS-CoV RdRp also suggests that potential nucleoside inhibitors should have the C3′ endo sugar puckering conformation to maintain its ability for making a hydrogen bond at the 3′ position and to avoid steric conflicts at the 2′ position.
Nucleoside analogs with sugar ring modifications, 2′-C-methyladenosine and 2′-O-methylcytidine, are effective HCV polymerase inhibitors (IC50s ∼2 and 4 µM, respectively) (52). The 2′-methyl group of the first inhibitor projects from the opposite side of the 2′-OH group of the sugar ring and interacts with the conserved Arg158 of motif F in the fingers subdomain of HCV RdRp. Similar interactions are expected with Arg553 of SARS-CoV polymerase. The second compound has a 2′-methoxy group instead of the 2′-OH of rNTP. This group would be proximal to Asp225 of motif A and Asn291 of motif B in HCV polymerase. Despite some structural differences in the active sites of HCV and SARS-CoV polymerases, our molecular model suggests that 2′-C-methyadenosine and 2′-O-methylcytidine may be potential inhibitors of SARS-CoV polymerase.
Addition of a 5-methyl group to a pyrimidine base is detrimental to the potency of 3′-dNTPs as inhibitors of HCV polymerase (50). The 5-methyl group is in the major groove of the base pair between the incoming rNTP and the templating base. This area is proximal to the conserved elements of the fingers subdomain (motifs G and F). Due to structural variation in this area between the SARS-CoV and HCV polymerases, this modification of the nucleobase may have a different effect on the inhibition of SARS-CoV polymerase.
Design of nucleoside analogs that are non-chain terminators can also be pursued as a possibility for anti-SARS therapy. Biochemical data show that some analogs may possess promiscuous base-pairing properties and, once misincorporated, they cause errors in viral replication and induce mutations in the viral genome (53). Recently, it has been shown that ribavirin monophosphate is incorporated into viral genomes during RNA synthesis and causes mutations because of its ability to pair with both uracil and cytosine (54–56). Ribavirin can increase the error frequency in both PV and HCV replication and reduces the fitness of viruses to the point of extinction.
A possible mechanism for the natural resistance of SARS-CoV to ribavirin
The only nucleoside analog that has been used therapeutically against HCV infection is ribavirin, but its efficacy is limited by the emergence of drug-resistant mutations. SARS-CoV has natural resistance to ribavirin (8). Ribavirin resistance of PV polymerase can be caused by a single amino acid change, G64S, in an unresolved portion of the fingers subdomain (56). Sequence alignment and structural comparison indicate that the structural segment containing the equivalent residue in HCV, RHDV, RV and φ6 polymerases forms an α-helix that interacts with the short α-helix of motif A, suggesting that Gly64 of PV polymerase is located in the vicinity of the conserved Asp238 of motif A. This aspartate has been suggested to help bind the incoming nucleotide (20,26, 27,41). The equivalent residue in HIV-1 RT (Tyr115) has also been shown to affect the fidelity of the enzyme (57). Based on the present structural analysis, we propose that ribavirin resistance in PV polymerase caused by the G64S mutation is due to a change in the enzyme’s fidelity through the repositioning of the structurally conserved α-helix of motif A. In the molecular model of SARS-CoV polymerase, the three-dimensional arrangement of the corresponding structural elements is conserved. However, there are variations in the interactions between the two α-helices that may modulate differently the enzyme’s fidelity and susceptibility to mutagens such as ribavirin and account for the clinically natural resistance of SARS-CoV towards ribavirin.
It is possible that additional structural elements that can affect the fidelity of polymerization may also contribute to the low susceptibility of SARS-CoV to ribavirin. Such structural elements may involve motifs F and G of the fingers subdomain that may ‘proof-read’ errors in the major groove of the nucleic acid. Similarly, motif C of the palm subdomain may also contribute to the fidelity of SARS-CoV polymerase by proof-reading mismatches in the minor groove, by analogy to what we have previously observed in HIV-1 RT (37). In addition, the ‘primer grip’ of motif E that is expected to control the positioning of the elongating primer strand may also contribute to the enzyme’s fidelity and susceptibility to ribavirin and other mutagens.
Non-nucleoside inhibitors
Non-nucleoside inhibitors of polymerases have been known to be effective therapeutics with great specificity against HIV-1, and are currently under development as anti-HCV drugs (58–60). In both cases, the inhibitors are hydrophobic in nature and act kinetically in a non-competitive manner with respect to dNTP or rNTP substrates, which is consistent with inhibitor binding at a site different from the nucleotide substrate. In HIV-1 RT, the inhibitors bind at a hydrophobic pocket that is proximal to, but distinct from the polymerase active site and is located at the palm–thumb subdomain interface. Binding of these inhibitors causes restriction on the movement of the thumb, conformational changes of the residues at the polymerase active site, and displacement of the ‘primer grip’ (61–65). The HCV polymerase non-nucleoside inhibitors bind to a hydrophobic pocket on the surface of the thumb subdomain and have an allosteric effect that interferes with the conformational change of the thumb (59).
In the structural model of SARS-CoV RdRp, there is no hydrophobic pocket similar to that of HIV-1 RT near the polymerase active site. Besides, the thumb subdomain of SARS-CoV RdRp is considerably smaller than that of HCV RdRp and a substantial part of the non-nucleoside inhibitor-binding pocket of HCV does not exist in SARS-CoV RdRp. Thus, it is likely that the non-nucleoside inhibitors that can inhibit HCV or HIV-1 polymerase may not work for SARS-CoV polymerase. Nevertheless, different allosteric sites may exist in SARS-CoV polymerase that can be targeted for developing antivirals. Information on novel inhibitor-binding sites is likely to emerge as detailed structural data are available and/or new inhibitors of SARS-CoV polymerase are discovered through high-throughput drug screening efforts.
Conclusion
Although the SARS pandemic appears to be currently under control, the lack of effective therapeutics against a potentially devastating disease that could re-emerge at any time has triggered intensive research efforts to identify possible vaccine and chemotherapeutic strategies. Because of its pivotal role in viral replication, SARS-CoV polymerase is an excellent target for anti-SARS drugs. Despite substantial differences between the polymerases of SARS-CoV and other RNA viruses, we were able to build a three-dimensional homology model of the catalytic domain of SARS-CoV polymerase. In the absence of any biochemical and structural data on coronavirus polymerases, this model provides the first insights into the functional roles of conserved residues and motifs of this enzyme and a structural basis to evaluate potential interactions with inhibitors of related enzymes. This information should be helpful in designing anti-SARS agents and provide guidance for future biochemical experiments.
Protein Data Bank accession code
The full atomic coordinates of the catalytic domain of SARS-CoV polymerase have been deposited with the RCSB Protein Data Bank (entry 1O5S) for immediate release.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Zhen Xu, Manwu Zha, Yadong Yu, Yongcheng Lu, Ning Shen, Jinyue Zhao, Sheng Li, Peng Zhang, Baozheng Peng and Meilan Zhang for assistance and discussion. The research in J.D.’s laboratory was funded in part by the National Natural Science Foundation of China (NSFC) grants (30125011, 30170223 and 30130080), the 863 Hi-Tech Program grants (2001AA235071, 2001AA233021 and 2002BA711A13) and the Chinese Academy of Sciences grant (KSCX1-SW-17). E.A.’s laboratory was sponsored in part by NIH grants (AI 27690 and GM 66671). S.W.’s laboratory was supported by an NIH grant (AI 17418).
PDB code 1O5S
REFERENCES
- 1.Lee N., Hui,D., Wu,A., Chan,P., Cameron,P., Joynt,G.M., Ahuja,A., Yung,M.Y., Leung,C.B., To,K.F. et al. (2003) A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med., 348, 1986–1994. [DOI] [PubMed] [Google Scholar]
- 2.Poutanen S.M., Low,D.E., Henry,B., Finkelstein,S., Rose,D., Green,K., Tellier,R., Draker,R., Adachi,D., Ayers,M. et al. (2003) Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med., 348, 1995–2005. [DOI] [PubMed] [Google Scholar]
- 3.Tsang K.W., Ho,P.L., Ooi,G.C., Yee,W.K., Wang,T., Chan-Yeung,M., Lam,W.K., Seto,W.H., Yam,L.Y., Cheung,T.M. et al. (2003) A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med., 348, 1977–1985. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C., Gunther,S., Preiser,W., van der Werf,S., Brodt,H., Becker,S., Rabenau,H., Panning,M., Kolesnikova,L., Fouchier,R.A.M. et al. (2003) Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med., 348, 1967–1976. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek T.G., Erdman,D., Goldsmith,C.S., Zaki,S.R., Peret,T., Emery,S., Tong,S., Urbani,C., Comer,J.A., Lim,W. et al. (2003) A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med., 348, 1953–1966. [DOI] [PubMed] [Google Scholar]
- 6.Peiris J.S.M., Lai,S.T., Poon,L.L.M., Guan,Y., Yam,L.Y.C., Lim,W., Nicholls,J., Yee,W.K.S., Yan,W.W., Cheung,M.T. et al. (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snijder E.J., Bredenbeek,P.J., Dobbe,J.C., Thiel,V., Ziebuhr,J., Poon,L.L.M., Guan,Y., Rozanov,M., Spaan,W.J.M. and Gorbalenya,A.E. (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol., 331, 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cinatl J., Morgenstern,B., Bauer,G., Chandra,P., Rabenau,H. and Doerr,H.W. (2003) Glycyrrhizin, an active component of liquorice roots and replication of SARS-associated coronavirus. Lancet, 361, 2045–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rota P.A., Oberste,M.S., Monroe,S.S., Nix,W.A., Campagnoli,R., Icenogle,J.P., Penaranda,S., Bankamp,B., Maher,K., Chen,M. et al. (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science, 300, 1394–1399. [DOI] [PubMed] [Google Scholar]
- 10.Marra M.A., Jones,S.J.M., Astell,C.R., Holt,R.A., Brooks-Wilson,A., Butterfield,Y.S.N., Khattra,J., Asano,J.K., Barber,S.A., Chan,S.Y. et al. (2003) The genome sequence of the SARS-associated coronavirus. Science, 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
- 11.Thiel V., Ivanov,K.A., Putics,A.,T. Hertzig,T., Schelle,B., Bayer,S., Weissbrich,B., Snijder,E.J., Rabenau,H., Doerr,H.W. et al. (2003) Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol., 84, 2305–2315. [DOI] [PubMed] [Google Scholar]
- 12.Lai M.M.C. and Holmes,K.V. (2001) Coronaviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott Williams and Wilkins, Philadelphia, pp. 1163–1185. [Google Scholar]
- 13.Kamer G. and Argos,P. (1984) Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res., 12, 7269–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y. and Eickbush,T.H. (1990) Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J., 9, 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin E.V. (1991) The phylogeny of RNA-dependent RNA polymerases of positive-stranded RNA viruses. J. Gen. Virol., 72, 2197–2206. [DOI] [PubMed] [Google Scholar]
- 17.O'Reilly E.K. and Kao,C.C. (1998) Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology, 252, 287–303. [DOI] [PubMed] [Google Scholar]
- 18.Gorbalenya A.E., Pringle,F.M., Zeddam,J.-L., Luke,B.T., Cameron,C.E., Kalmakoff,J., Hanzlik,T.N., Gordon,K.H.J. and Ward,V.K. (2002) The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol., 324, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruenn J.A. (2003) A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res., 31, 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen J.L., Long,A.M. and Schultz,S.C. (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure, 5, 1109–1122. [DOI] [PubMed] [Google Scholar]
- 21.Ago H., Adachi,T., Yoshida,A., Yamamoto,M., Habuka,N., Yatsunami,K. and Miyano,M. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure, 7, 1417–1426. [DOI] [PubMed] [Google Scholar]
- 22.Bressanelli S., Tomei,L., Roussel,A., Incitti,I., Vitale,R.L., Mathieu,M., De Francesco,R. and Rey,F.A. (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA, 96, 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesburg C.A., Cable,M.B., Ferrari,E., Hong,Z., Mannarino,A.F. and Weber,P.C. (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol., 6, 937–943. [DOI] [PubMed] [Google Scholar]
- 24.Bressanelli S., Tomei,L., Rey,F.A. and De Francesco,R. (2002) Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol., 76, 3482–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng K.K., Cherney,M.M., Vazquez,A.L., Machin,A., Alonso,J.M., Parra,F. and James,M.N. (2002) Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem., 277, 1381–1387. [DOI] [PubMed] [Google Scholar]
- 26.Tao Y., Farsetta,D.L., Nibert,M.L. and Harrison,S.C. (2002) RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell, 111, 733–745. [DOI] [PubMed] [Google Scholar]
- 27.Butcher S.J., Grimes,J.M., Makeyev,E.V., Bamford,D.H. and Stuart,D.I. (2001) A mechanism for initiating RNA-dependent RNA polymerization. Nature, 410, 235–240. [DOI] [PubMed] [Google Scholar]
- 28.Higgins D., Thompson,J., Gibson,T., Thompson,J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rost B. and Sander,C. (1993) Prediction of protein structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 30.Rost B. (1996) PHD: predicting one-dimensional protein structure by use of sequence profiles and neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- 31.Schultz G.E. and Schirmer,R.H. (1979) Principles of Protein Structure. Springer-Verlag, New York, NY. [Google Scholar]
- 32.Rost B., Sander,C. and Schneider,R. (1994) Redefining the goals of protein secondary structure prediction. J. Mol. Biol., 235, 13–26. [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Potterton,L., Yuan,F., van Vlijmen,H. and Karplus,M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins, 23, 318–326. [DOI] [PubMed] [Google Scholar]
- 34.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 35.Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- 36.Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- 37.Ding J., Das,K., Hsiou,Y., Sarafianos,S.G., Clark,J.A.D., Jacobo-Molina,A., Tantillo,C., Hughes,S.H. and Arnold,E. (1998) Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with double-stranded DNA and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol., 284, 1095–1111. [DOI] [PubMed] [Google Scholar]
- 38.Hobson S.D., Rosenblum,E.S., Richards,O.C., Richmond,K., Kirkegaard,K. and Schultz,S.C. (2001) Oligomeric structures of poliovirus polymerase are important for function. EMBO J., 20, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathak H.B., Ghosh,S.K., Roberts,A.W., Sharma,S.D., Yoder,J.D., Arnold,J.J., Gohara,D.W., Barton,D.J., Paul,A.V. and Cameron,C.E. (2002) Structure–function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol): a surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J. Biol. Chem., 277, 31551–31562. [DOI] [PubMed] [Google Scholar]
- 40.Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gohara D.W., Crotty,S., Arnold,J.J., Yoder,J.D., Andino,R. and Cameron,C.E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical and biological analysis of conserved structural motifs A and B. J. Biol. Chem., 275, 25523–25532. [DOI] [PubMed] [Google Scholar]
- 42.Sarafianos S.G., Das,K., Ding,J., Boyer,P.L., Hughes,S.H. and Arnold,E. (1999) Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem. Biol., 6, R137–R146. [DOI] [PubMed] [Google Scholar]
- 43.Gao G., Orlova,M., Georgiadis,M.M., Hendrickson,W.A. and Goff,S.P. (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl Acad. Sci. USA, 94, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delarue M., Poch,O., Tordo,N., Moras,D. and Argos,P. (1990) An attempt to unify the structure of polymerases. Protein Eng., 6, 461–467. [DOI] [PubMed] [Google Scholar]
- 45.Jacobo-Molina A., Ding,J., Nanni,R.G., Clark,A.D.,Jr, Lu,X., Tantillo,C., Williams,R.L., Kamer,G., Ferris,A.L., Clark,P. et al. (1993) Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutierrez-Rivas M. and Menendez-Arias,L. (2001) A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res., 29, 4963–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong Z., Cameron,C.E., Walker,M.P., Castro,C., Yao,N., Lau,J.Y. and Zhong,W. (2001) A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology, 285, 6–11. [DOI] [PubMed] [Google Scholar]
- 48.De Clercq E. (2001) Molecular targets for antiviral agents. J. Pharmacol. Exp. Ther., 297, 1–10. [PubMed] [Google Scholar]
- 49.De Francesco R., Tomei,L., Altamura,S., Summa,V. and Migliaccio,G. (2003) Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res., 58, 1–16. [DOI] [PubMed] [Google Scholar]
- 50.Shim J., Larson,G., Lai,V., Naim,S. and Wu,J.Z. (2003) Canonical 3′-deoxyribonucleotides as a chain terminator for HCV NS5B RNA-dependent RNA polymerase. Antiviral Res., 58, 243–251. [DOI] [PubMed] [Google Scholar]
- 51.Ishii K., Tanaka,Y., Yap,C.-C., Aizaki,H., Matsuura,Y. and Miyamura,T. (1999) Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology, 29, 1227–1235. [DOI] [PubMed] [Google Scholar]
- 52.Carroll S.S., Tomassini,J.E., Bosserman,M., Getty,K., Stahlhut,M.W., Eldrup,A.B., Bhat,B., Hall,D., Simcoe,A.L., LaFemina,R. et al. (2003) Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem., 278, 11979–11984. [DOI] [PubMed] [Google Scholar]
- 53.Loeb L.A. and Mullins,J.I. (2000) Lethal mutagenesis of HIV by mutagenic ribonucleoside analogs. AIDS Res. Hum. Retroviruses, 16, 103. [DOI] [PubMed] [Google Scholar]
- 54.Crotty S., Maag,D., Arnold,J.J., Zhong,W., Lau,J.Y., Hong,Z.R.A. and Cameron,C.E. (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature Med., 6, 1375–1379. [DOI] [PubMed] [Google Scholar]
- 55.Lau J.Y., Tam,R.C., Liang,T.J. and Hong,Z. (2002) Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology, 35, 1002–1009. [DOI] [PubMed] [Google Scholar]
- 56.Pfeiffer J.K. and Kirkegaard,K. (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl Acad. Sci. USA, 100, 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin-Hernandez A., Domingo,E. and Menendez-Arias,L. (1996) Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J., 15, 4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 58.Dhanak D., Duffy,K.J., Johnston,V.K., Lin-Goerke,J., Darcy,M., Shaw,A.N., Gu,B., Silverman,C., Gates,A.T., Nonnemacher,M.R. et al. (2002) Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem., 277, 38322–38327. [DOI] [PubMed] [Google Scholar]
- 59.Love R.A., Parge,H.E., Yu,X., Hickey,M.J., Diehl,W., Gao,J., Wriggers,H., Ekker,A., Wang,L., Thomson,J.A. et al. (2003) Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol., 77, 7575–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan L., Reddy,T.J., Proulx,M., Das,S.K., Pereira,O., Wang,W., Siddiqui,A., Yannopoulos,C.G., Poisson,C., Turcotte,N. et al. (2003) Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem., 46, 9489–9495. [DOI] [PubMed] [Google Scholar]
- 61.Kohlstaedt L.A., Wang,J., Friedman,J.M., Rice,P.A. and Steitz,T.A. (1992) Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science, 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- 62.Ding J., Das,K., Tantillo,C., Zhang,W., Clark,A.D., Jr., Jessen,S., Lu,X., Hsiou,Y., Jacobo-Molina,A., Andries,K. et al. (1995) Structure of HIV-1 reverse transcriptase in a complex with the nonnucleoside inhibitor α-APA R 95845 at 2.8 Å resolution. Structure, 3, 365–379. [DOI] [PubMed] [Google Scholar]
- 63.Ren J., Esnouf,R., Garman,E., Somers,D., Ross,C., Kirby,I., Keeling,J., Darby,G., Jones,Y., Stuart,D. et al. (1995) High resolution structures of HIV-1 RT from four RT–inhibitor complexes. Nature Struct. Biol., 2, 293–302. [DOI] [PubMed] [Google Scholar]
- 64.Esnouf R., Ren,J., Ross,R., Jones,Y., Stammers,D. and Stuart,D. (1995) Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nature Struct. Biol., 2, 303–308. [DOI] [PubMed] [Google Scholar]
- 65.Das K., Ding,J., Hsiou,Y., Clark,J.A.D., Moereels,H., Koymans,L., Andries,K., Pauwels,R., Janssen,P.A.J., Boyer,P.L. et al. (1996) Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J. Mol. Biol., 264, 1085–1100. [DOI] [PubMed] [Google Scholar]
- 66.Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]