Abstract
Small DNA repeat tracts are located throughout the human genome. The tracts are unstable, and expansions of certain repeat sequences cause neuromuscular disease. DNA expansions appear to be associated with lagging-strand DNA synthesis and DNA repair. At some sites of repeat expansion, e.g. the myotonic dystrophy type 2 (DM2) tetranucleotide repeat expansion site, more than one repeat tract with similar sequences lie side by side. Only one of the DM2 repeat tracts, however, is found to expand. Thus, DNA base sequence is a possible factor in repeat tract expansion. Here we determined the expansion potential, during DNA replication by human DNA polymerase β, of several tetranucleotide repeat tracts in which the repeat units varied by one or more bases. The results show that subtle changes, such as switching T for C in a tetranucleotide repeat, can have dramatic consequences on the ability of the nascent-strand repeat tract to expand during DNA replication. We also determined the relative stabilities of self-annealed 100mer repeats by melting-curve analysis. The relative stabilities did not correlate with the relative potentials of the analogous repeats for expansion during DNA replication, suggesting that hairpin formation is not required for expansion during DNA replication.
INTRODUCTION
DNA repeat tracts, composed of multiple tandem repeats of identical sequence, are scattered throughout the human genome and vary in size and sequence (1). The repeats, the smallest of which are called microsatellites, are major sources of genomic instability, since they are subject to high frequencies of expansion and contraction mutations. Much attention has been focused on expansions of the trinucleotide repeats because of their association with at least 16 neuromuscular diseases, including myotonic dystrophy, Friedreich’s ataxia and Huntington’s disease (2). Expansions of other repeats have also been shown to cause disease, including the tetranucleotide repeat CCTG, responsible for myotonic dystrophy type 2 (DM2) (3), and the pentanucleotide repeat ATTCT, responsible for spinocerebellar ataxia type 10 (4). The mutations responsible for the diseases map to the repeat tracts alone, suggesting that properties inherent in the repeat tracts are responsible for the expansions. In addition, accumulating evidence suggests that the expansions are associated with lagging-strand DNA replication (5–12) and DNA repair involving DNA synthesis (13–19).
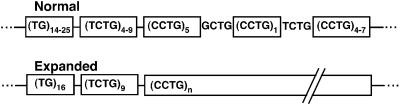
Some repeat loci are complicated, with different DNA sequence repeat tracts juxtaposed or near each other. For example, DM2 has been mapped to a mutation on chromosome 3q21. The mutation is an expansion of a CCTG repeat located in intron 1 of the zinc finger protein 9 (ZNF9) gene (3). The normal DM2 allele has a relatively complicated repeat structure (Fig. 1). The CCTG repeat lies adjacent to a TCTG repeat of similar size. The CCTG repeat is interrupted once or twice by single repeats of TCTG and GCTG. Only the CCTG repeat tract has been found to expand in DM2. TCTG expansion would be predicted to also cause DM2 based on its location and the presence of the CTG sequence within both the CCTG and TCTG repeats: recognition of the transcribed repeat RNA by CUG-binding protein is believed to play a major role in manifesting the DM1 phenotypes (20). Reasons for the apparent bias for expansion of CCTG repeats over TCTG repeats of similar size include the possible loss of interruptions to produce a large CCTG pre-mutation repeat tract (20), the possible larger potential of CCTG repeat tracts compared with same size TCTG repeat tracts to expand, or a combination of both possibilities. To determine if changes as subtle as a C to T in a tetranucleotide sequence could affect the potential of otherwise identical repeats to expand, we constructed a series of tetranucleotide repeat tracts for use as replication substrates. Determination of the relative abilities of these repeats to expand during replication with human DNA polymerase β (pol β) demonstrated significant effects of sequence on repeat expansion.
Figure 1.
Schematic of normal and DM2 expanded alleles in intron 1 of the ZNF9 gene. Adapted from Liquori et al. (3).
MATERIALS AND METHODS
Synthetic oligodeoxyribonucleotides
Oligodeoxyribonucleotides (oligomers) were synthesized by the Lineberger Nucleic Acids Core Facility at UNC. Primers for the expansion assays were 12mer repeats of the sequences d(CAGN)3 and d(NCTG)3, where N is A, G, C or T. Templates for the expansion assays were 64mers of the sequences 5′-TTGCATTCGCAT(CAGN)10GCAATATAATCT and 5′-TTGCATTCGCAT(NCTG)10GCAATATAATCT. The templates contained a three-carbon spacer (Glen Research) at the 3′ terminus that cannot be removed or extended by DNA polymerase (not shown). A ‘random’ sequence primer (5′-GGATCAGGTCGC) and template (5′-ACTGTGTCTGTCAGGCTATCGATAGACAGTACTGCATACAGAGCGACCTGATCC) pair was used as a control. Templates were gel purified before use. Oligomers for determination of melting curves and mobilities by native gel electrophoresis were 100mers composed of one of the repeat sequences d(CAGN)25 or d(NCTG)25, where N was either A, C, T or G. The eight 100mers were gel purified by 12% denaturing PAGE containing 7.5 M urea.
DNA repeat expansion assays
Reactions (20 µl) contained 0.6 µM annealed DNA template/primer, 50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 20 mM NaCl, 2.5% glycerol, 2 mM dithiothreitol (DTT), 0.043 mCi of [α-32P]dCTP and 32 nM h-pol β [purified as previously described (21) using a construct generously provided by Dr Sam Wilson]. Assays were performed both as time course experiments and single 60 min time point reactions. For the time course assays, 20 µl reactions were aliquoted from a large reaction mix. The single time point (60 min) reactions were mixed individually using small amounts of reagents. All reaction mixes were kept on ice and started by incubation at 37°C for the indicated amounts of time. Reactions were stopped by the addition of EDTA to a final concentration of 115 mM. Excess [α-32P]dATP was removed by passing each reaction through one G-25 MicroSpin column (Amersham Biosciences). Reaction products were analyzed using 12% denaturing PAGE containing 7.5 M urea. Results were visualized with a Storm 840 PhosphorImager and were quantitated using ImageQuant 5.0 (Molecular Dynamics).
Melting temperature determination
Gel-purified d(CAGN)25 and d(NCTG)25 oligomers were self-annealed by boiling for 3 min and slow cooling to room temperature in a buffer composed of 50 mM sodium cacodylate pH 8.0, 10 mM MgCl2 and 20 mM NaCl in a final volume of 300 µl. This treatment allowed us to study hairpin formation under equilibrium conditions and thus enabled application of standard thermodynamic analysis. The OD (λ = 260 nm) was between 0.074 and 0.38 for each annealed oligo, at 25°C in a 0.1 cm path length cuvette. UV absorption readings were taken at the University of North Carolina Macromolecular Interactions Facility on a PiStar-CDF spectrophotometer (Applied Photophysics) at 260 nm with a temperature range from 2 to 98°C, raised in 1°C increments. The temperature was held constant at each new increment until the new temperature (T) achieved equilibrium as measured by a constant temperature reading (T ± 0.2°C) for 30 s. First derivative plots of the resulting melting curves were generated using GraFit 5.0 (Erithacus Software Ltd) and Excel 2000 (Microsoft Corp.).
Determination of ΔHo and ΔGo from DNA melting curves
At T = Tm, the amounts of ordered and disordered structures are equal, ΔG = 0 and ΔS = ΔH/Tm. The slope of the melting curve in the region surrounding the Tm can be used to calculate ΔHo using a variation of the van’t Hoff formula derived by Marky and Breslauer (22). This method, in which the melting curves are normalized to have respective low temperature and high temperature plateaus of 0 and 1, has been used to determine the stability of hairpin structures formed by CAG and CTG triplet repeats (23). The melting curves were normalized using the formula Atn = 1 – (Ah – At)/(Ah – Al), where Atn is the normalized absorbance at temperature T, Ah is the average absorbance of the high temperature absorbance plateau, Al is the average absorbance of the low temperature absorbance plateau and At is the absorbance at T. The normalized slope was found by obtaining the first derivative (with respect to T) of the normalized absorbance at the Tm. The formula ΔHo = (2 + 2n)R(Tm)2σ was used to calculate the van’t Hoff value of the standard enthalpy change (ΔHo), where the molecularity (n) = 1, R = 1.987 cal/mol-K, and σ is the normalized slope. The standard free energy change (ΔGo) at 37°C for each repeat oligomer was calculated using the formula ΔGo = ΔHo(Tm – T)/Tm, with T = 310 K.
Native gel analysis
Gel-purified d(CAGN)25 and d(NCTG)25 oligomers were end labeled using T4 polynucleotide kinase (NEB). The kinase was heat killed at 65°C for 20 min, and excess [γ-32P]ATP was removed by passing the end-labeled products through two G-25 MicroSpin columns (Amersham Biosciences). After labeling, 1000–1500 c.p.m. of each oligomer was placed in replication buffer (consisting of 50 mM Tris–HCl pH 8.0, 20 mM NaCl and 10 mM MgCl2), boiled for 3 min and slowly cooled to room temperature. The oligomers were analyzed by 8% native PAGE.
RESULTS
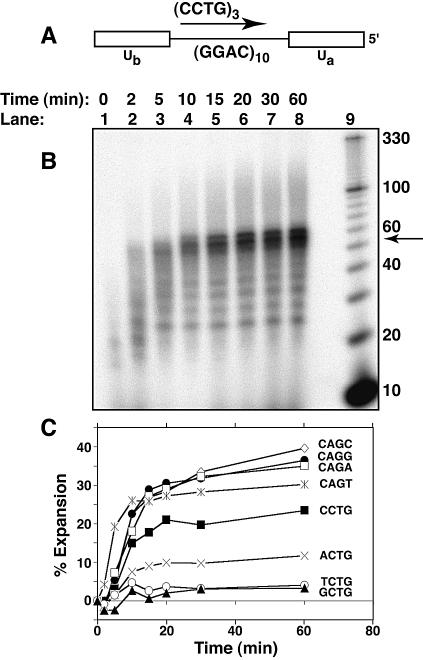
We determined the abilities of the various repeat-sequence template primers to expand during DNA replication. A template–primer construct was used with human DNA pol β to generate tetranucleotide expansion (Fig. 2A). This combination of substrate and polymerase generates observable amounts of triplet repeat expansion (24,25) and was therefore used to analyze the effects of DNA sequence on repeat expansion. The template contained 10 tandem repeats bounded on both sides by 12-base unique sequences. In addition, the 3′ end of the template was blocked to prevent extension of the template strand. Priming was from within the repeat tract, which gives much more expansion than priming from an upstream unique sequence (24, and B.L.Ruggiero and M.D.Topal, unpublished results). Percentage expansion was quantitated by measuring the density of the expanded region (≥56 nt), subtracting the appropriate background density and dividing by the total lane density (also corrected for background density). Tetranucleotide expansion was clearly seen to increase with reaction time (Fig. 2B and C). All of the expansion determinations as a function of time (Fig. 2C) were repeated at least twice. The overall shapes of the curves and especially the leveled-off regions of the curves were reproducible to ±4% expansion.
Figure 2.
Expansion of DNA tetranucleotide repeats during DNA replication in vitro. Reaction products were resolved on a denaturing 12% polyacrylamide gel containing 7.5 M urea. (A) Schematic of annealed primer and template used for DNA synthesis in vitro. Ua and Ub represent unique DNA sequences flanking the repeat tract in the template strand. (B) Expansion of the CCTG repeat-containing nascent strand. Lanes 1–8, replication with a (CCTG)3 primer by h-polβ for 0–60 min, as indicated. Lane 9, 10 bp ladder. The arrow indicates the size of the fully replicated template (52 nt); all higher bands are due to repeat expansion. (C) Comparison of tetranucleotide repeat expansions of varying sequences. Expansion is plotted as the percentage intensity of the expanded region (defined as the amount of labeled products >52 nt divided by the total amount of labeled products in the lane) versus reaction time. Sequences shown indicate the sequence of the nascent-strand repeat.
The time dependencies of expansion for the tetranucleotide template–primers were analyzed on separate gels. Therefore, the relative accuracies of the expansion measurements were checked by measuring expansions of all the different template–primers on the same gel at a single time point (60 min) (Fig. 3A). The percentage expansion for each repeat template–primer was quantitated (Fig. 3B) by measuring the density of labeled products above the predicted full length of 52 nt and dividing by the total density in the lane as described earlier. The determinations were done twice and the average and variance around the mean is shown by the error bars in Figure 3B.
Figure 3.
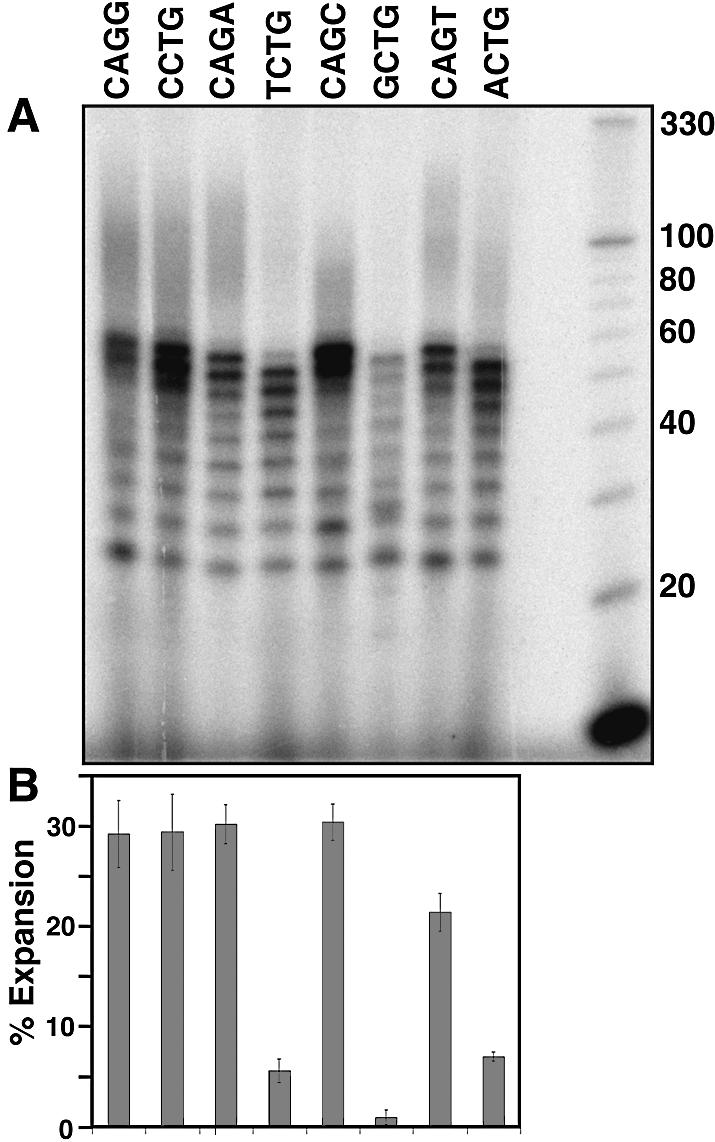
Comparison of expansion of the different repeat sequences at a single time point (60 min). (A) Gel analysis of the repeat expansion reaction products. Reaction products were resolved on a denaturing 12% polyacrylamide gel containing 7.5 M urea. Base sequences above each lane are the repeat sequences of the primers used in each expansion reaction. Repeat expansion reactions were carried out for 60 min at 37°C. (B) Bar graph summarizing quantitation of the expansion results. Bars correspond to the sequences shown in the top panel, and represent the percentage expansion (as defined in Fig. 2), and are the average of two determinations. The error bars indicate the variance between the two determinations.
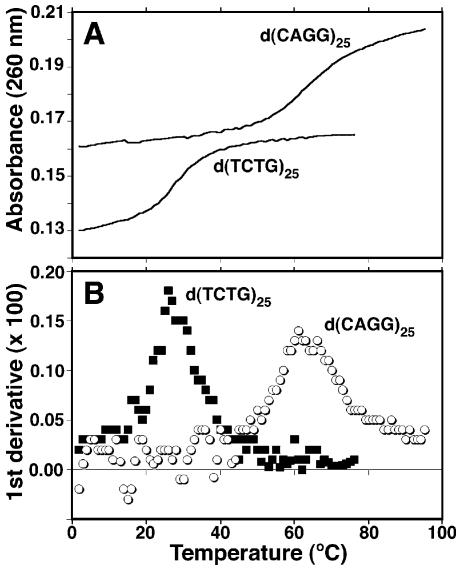
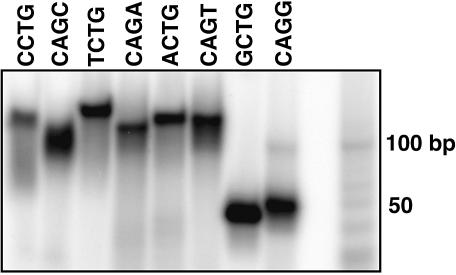
The ability of repeat tracts to self-anneal to give hairpin structures is predicted to be important for several different aspects of repeat expansion. These include hairpin formation in the template repeat that increases DNA slippage, and hairpin formation in the single-stranded expansion product that protects against DNA repair by flap endonuclease 1 and mismatch repair. Melting studies of 100mer repeat tracts were performed to determine the abilities of the different repeat sequences to self-anneal. After boiling and slowly cooling the repeats tracts, UV absorbance at 260 nm wavelength was measured with increasing temperature (Fig. 4A). First derivative plots of the melting curves were used to determine the melting temperatures (Tms) for each repeat tract (Fig. 4B). The results are summarized in Table 1. The melting curves were found to be reproducible and relatively independent of concentration, as expected for mainly single-strand interactions involving self-annealing. The Tm for d(CCTG)25 was estimated because melting started at the lowest temperature and thus a good lower baseline could not be established. Thus, the true Tm for CCTG is that shown or somewhat lower. Thermodynamic values for melting the self-annealed structures were determined as described in Materials and Methods and summarized in Table 1. To independently measure hairpin formation, the 100mers were end-labeled and analyzed by native PAGE at room temperature (Fig. 5). The mobilities of the self-annealed structures were similar to that of double-strand molecular weight markers ∼50 bp in length.
Figure 4.
Determination of melting temperatures (Tms) for self-annealed 100mers of each repeat sequence. Oligonucleotides were annealed in buffer (50 mM Na-cacodylate pH 8, 10 mM MgCl2, 20 mM NaCl) and the absorbance was recorded as the temperature was increased from 2°C to either 75 or 98°C. (A) Plot of absorbance (λ = 260 nm) versus increasing temperature for d(TCTG)25 and d(CAGG)25. (B) First derivative plots of the melting curves in (A). Maxima in the first derivative plots indicate the melting temperatures.
Table 1. Melting temperatures and thermodynamic values for the loss of self-annealed DNA structure.
| Sequence | [µM] | Tm (°C) | ΔHo (kcal/mol) | ΔGo(37°C) (kcal/mol) | Annealed hairpina |
|---|---|---|---|---|---|
| d(GCTG)25 | 1.6 | 68 | 51 | 4.6 | 5′-GCTGGCTGGCTG→ |
| 3′-CGGTCGGTCG← | |||||
| d(CAGG)25 | 1.5 | 61 | 28 | 2.0 | 5′-CAGGCAGGCAGG→ |
| 3′-GGACGGACGGAC← | |||||
| d(CAGC)25 | 1.6 | 55 | 23 | 1.2 | 5′-CAGCCAGCCAGC→ |
| 4 | 52 | 19 | 0.9 | 3′-ACCGACCGACCG← | |
| d(ACTG)25 | 0.67 | 45 | 57 | 1.4 | 5′-ACTGACTGACTG→ |
| 3′-AGTCAGTCAGTC← | |||||
| d(CAGA)25 | 1.4 | 37 | 26 | 0 | 5′-CAGACAGACAGA→ |
| 3′-GACAGACAGAC← | |||||
| d(CCTG)25 | 3.1 | ≤35b | 15b | –0.1b | 5′-CCTGCCTGCCTG→ |
| 3′-GTCCGTCCGTCC← | |||||
| d(TCTG)25 | 1.6 | 31 | 28 | –0.6 | 5′-TCTGTCTGTCTG→ |
| 2.2 | 27 ± 1, n = 2 | 35 ± 1 | –1.2 ± 0.2 | 3′-TGTCTGTCTGTC← | |
| 3.2 | 31 | 30 | –0.6 | ||
| d(CAGT)25 | 1.6 | 24 | 39 | –1.7 | 5′-CAGTCAGTCAGT→ |
| 3′-GACTGACTGAC← |
aWatson–Crick base pairs are shown in bold.
bEstimated.
See Materials and Methods for buffer conditions.
Figure 5.
Native PAGE analysis of secondary structure formation by synthetic 100mers. Synthetic oligonucleotides composed of 25 repeats were annealed in buffer (50 mM Tris–HCl pH 8, 10 mM MgCl2, 20 mM NaCl), and loaded on an 8% native polyacrylamide gel in 1× TBE running buffer. Repeat sequences are shown above the gel. The far right lane contains a 10 bp ladder.
DISCUSSION
To determine the effects of DNA sequence on repeat expansion, four different tetranucleotide-repeat sequence tracts and their complements were synthesized to give a total of eight different repeat tracts as substrates for DNA pol β. Results of replication showed expansion rapidly increasing with time for the first 10–20 min and then a slower increase until the reactions were stopped at 60 min (Fig. 2). The results showed that, based on their relative potential for expansion, the repeats could be roughly divided into two groups. The members of the first group all contained the triplet CAG. This group demonstrated the highest expansion levels clustered between 30 and 40% expansion products after 60 min of replication (Fig. 2C). Members of the second group contained the complementary triplet CTG and demonstrated lower expansion levels spread over a relatively wide (∼6-fold) range of between 4% (for TCTG and GCTG) and 23% (for CCTG) expansion products. Variation within each group depended on the identity of the fourth base. C as the fourth base produced the largest amounts of expansion products in each group. T as the fourth base, on the other hand, produced the least (or close to the least) amount of expansion within each group. Overall, the greatest difference in amounts of expansion products was the 10-fold greater amount for CAGC compared with TCTG/GCTG (the pair showed similar values). Expansion products were defined as all replication products greater than the largest predicted fill-in product. In these reactions, products ≥56 nt were considered expansion products. The 56mer products could be the result of replication from a partially annealed primer in which the two 5′ repeats in the primer bound to the 3′ end of the template next to the unique sequence, leaving a single repeat overhang. The 56mer products are considered to be produced by expansion since primer and template were annealed under equilibrium conditions, and primer misannealing cannot be ruled out as a significant contributer to DNA slippage. Quantitation of expansion without the 56mer, of course, lowered all expansion levels. CAGG remained highest (18%), while in the middle (spanning 5–10% expansion) were CAGA, CCTG, CAGC, CAGT and ACTG in descending order. TCTG and GCTG gave close to undetectable levels of expansion. The difference between expansion of CCTG and TCTG was at least 10-fold, which is larger than the 6-fold difference seen when the 56mer band was included, indicating a strong preference for the expansion of the disease-associated repeat regardless of whether or not initial primer misalignment is included in the quantitation of expansion products.
Expansion (including the 56 nt product) was also evaluated at a single time point (60 min) so that relative expansion for all of the repeats could be compared on a single gel (Fig. 3). Compared with Figure 2, the results are similar: CAGC exhibits the highest level of expansion by both methods and is closely followed by CAGA and CAGG. ACTG, TCTG and GCTG, in the same relative order as in Figure 2, showed the smallest amounts of expansion. The one difference between the two different sets of results is that the neighboring CCTG and CAGT sequences switched places with respect to which expands more. We believe the difference is caused by the increased experimental error in comparing results obtained using two different methods. Using either method, however, we conclude that there is a large difference, 6- to >10-fold (depending on whether or not the 56 nt product is included, respectively) in expansion potential between equivalent sized CCTG and TCTG repeats.
Hairpin formations within the template strand of the repeat tract and the single-strand repeat expansion intermediate have been predicted to play an important role in the DNA expansion process. Hairpin formation in the template strand is predicted to block replication and increase DNA slippage (6). Hairpin formation in the expanded DNA is predicted to protect the expanded DNA from cellular DNA repair activities such as flap endonuclease 1 (26,27) and mismatch repair (28). Therefore, we determined the ability of each of the repeat tracts to form hairpins. The results of stability measurements and gel analysis showed a wide range of hairpin-forming abilities among the different repeats. GCTG and CAGG hairpins had the highest hairpin stabilities, as measured by ΔGo and Tm (Table 1), whereas CAGT and TCTG hairpins had the least. It was possible that hairpin formation in the nascent strand helped expansion by balancing the loss of hydrogen bonding and stacking interactions that occurred during DNA slippage. Alternatively, hairpin formation in the template strand has been proposed to pause replication, allowing more opportunity for DNA slippage. Comparisons of the relative stabilities of the structures formed by the different repeat sequences with the relative amounts of expansions occurring during DNA replication demonstrated a lack of correlation. For example, GCTG and CAGC are complementary and both formed stable secondary structures by themselves (Table 1), but GCTG did not expand well compared with CAGC (Figs 2 and 3), whether or not the 56 nt product was considered an expansion product. Thus, the ability to expand during DNA replication did not correlate with the ability of the expanded DNA product to form hairpins. The lack of correlation implies that hairpin formation neither in the nascent strand nor in the template strand is required for DNA slippage under our conditions.
The stability of hairpin formation (Table 1) was generally strongest for those sequences that could produce two neighboring base pairs, a G:C followed by a C:G (Table 1). This included GCTG, CAGG (which, because of the nature of repeats, can also be written GCAG) and CAGC. The two base mismatches for each repeat were, respectively, neighboring G/Ts, A/Gs and A/Cs, in decreasing order of stability. The one exception was CCTG (GCCT), which is predicted to produce two neighboring G:Cs followed by C/T and T/C mismatches. This repeat had a significantly lower stability than the other repeats in this class. The other class of repeats could also make G:C and C:G base pairs, but interrupted in each case by a mismatch. This class included, in the order of decreasing Tm and ΔGo, ACTG, CAGA, TCTG and CAGT. The mismatches in each case included T/T and A/A for ACTG, two A/As for CAGA, two T/Ts for TCTG, and an A/A and a T/T for CAGT. Thus, greatest stability was gained by uninterrupted nearest-neighbor, or stacking, interactions between the GC and CG base pairs. Native gel electrophoresis confirmed the high stability of the self-annealed secondary structures formed by GCTG and CAGG (Fig. 5), and the gel mobilities of the structures indicated sizes half that of the full-length oligomer. The results are consistent with hairpin formation. In general, the results are in agreement with thermodynamic studies of the effects of base–base mismatches on double helix formation (29,30). The thermodynamic data indicate that, based on nearest-neighbor analysis, mismatches are destabilizing. The most stable mismatches are those containing guanine (GT, GG and GA); the least stable contain cytosine and pyrimidine–pyrimidine mismatches (29,30).
The sequence dependence of expansion, during replication in vitro, while not correlated with hairpin formation, may be a factor in determining which tandem repeat tracts expand in vivo. For example, sequence effects may play a role in the bias for CCTG repeat expansion versus its neighboring TCTG repeat tract at the DM2 locus. Normal alleles have CCTG tracts at the DM2 locus with single repeat interruptions that are absent in the expanded alleles (3). Loss of interruptions within the CCTG tract are predicted to play a role in expansion of that tract (20), based on similar observations in SCA1 (31,32) and fragile X syndrome (32–35). The bias of sequence effects on expansion determined in this study enables us to speculate that another factor in the observed bias for CCTG expansion at the DM2 locus may be the 6- to >10-fold greater potential for expansion of the CCTG repeat tract over a TCTG repeat tract of the same size. The strand complementary to that containing the CCTG and TCTG repeats contains CAGG and CAGA repeats, both of which were found to expand well. Therefore, for the bias observed in our lagging strand model to be manifested in vivo, the CCTG/TCTG tracts would need to be on the nascent strand being synthesized by lagging-strand replication. This placement is consistent with studies that associate repeat expansion with the free 5′ ends and single-strand regions associated with lagging-strand replication (5–12) and with models of expansion associated with Okazaki fragments (6,33,36).
DNA sequence repeats occur throughout the human genome (1); the factors that play a role in which repeats suffer expansion and the biological consequences are just starting to be recognized. Assuming that the evidence indicating that expansion occurs during DNA synthesis is correct, sequence bias takes its place in line along with lagging strand replication, repeat tract interruptions and repeat tract length as factors that must be considered as influencing repeat expansion potential.
REFERENCES
- 1.Tóth G., Gáspári,Z. and Jurka,J. (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res., 10, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margolis R.L. and Ross,C.A. (2001) Expansion explosion: new clues to the pathogenesis of repeat expansion neurodegenerative diseases. Trends Mol. Med., 7, 479–482. [DOI] [PubMed] [Google Scholar]
- 3.Liquori C.L., Ricker,K., Moseley,M.L., Jacobsen,J.F., Kress,W., Naylor,S.L., Day,J.W. and Ranum,L.P.W. (2001) Myotonic dystrophy type 2 caused by CCTG expansion in intron 1 of ZNF9. Science, 293, 864–867. [DOI] [PubMed] [Google Scholar]
- 4.Matsuura T., Yamagata,T., Burgess,D.L., Ramussen,A., Grewal,R.P., Watase,K., Khajavi,M., McCall,A.E., Davis,C.F., Zu,L., Achari,M., Pulst,S.M., Alonso,E., Noebels,J.L., Nelson,D.L., Zoghbi,H.Y. and Ashizawa,T. (2000) Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nature Genet., 26, 191–194. [DOI] [PubMed] [Google Scholar]
- 5.Freudenreich C.H., Stavenhagen,J.B. and Zakian,V.A. (1997) Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol., 17, 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S., Jaworski,A., Ohshima,K. and Wells,R.D. (1995) Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E.coli. Nature Genet., 10, 213–218. [DOI] [PubMed] [Google Scholar]
- 7.Maurer D.J., O’Callaghan,B.L. and Livingston,D.M. (1996) Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R.E., Kovvali,G.K., Prakash,L. and Prakash,S. (1995) Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science, 269, 238–240. [DOI] [PubMed] [Google Scholar]
- 9.Freudenreich C.H., Kantrow,S.M. and Zakian,V.A. (1998) Expansion of length-dependent fragility of CTG repeats of yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- 10.Miret J.J., Pessoa-Brandao,L. and Lahue,R.S. (1998) Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 95, 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary J.D., Nichol,K., Wang,Y.H. and Pearson,C.E. (2002) Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nature Genet., 31, 37–46. [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer J.K. and Livingston,D.M. (1998) Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet., 7, 69–74. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy L. and Shelbourne,P.F. (2000) Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum. Mol. Genet., 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 14.Fortune M.T., Vassilopoulos,C., Coolbaugh,M.I., Siciliano,M.J. and Monckton,D.G. (2000) Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet., 9, 439–445. [DOI] [PubMed] [Google Scholar]
- 15.Kovtun I.V. and McMurray,C.T. (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet., 27, 407–411. [DOI] [PubMed] [Google Scholar]
- 16.Manley K., Shirley,T.L., Flaherty,L. and Messer,A. (1999) Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet., 23, 471–473. [DOI] [PubMed] [Google Scholar]
- 17.Jankowski C., Nasar,F. and Nag,D.K. (2000) Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA, 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakupciak J.P. and Wells,R.D. (2000) Gene conversion (recombination) mediates expansions of CTG·CAG repeats. J. Biol. Chem., 275, 40003–40013. [DOI] [PubMed] [Google Scholar]
- 19.van den Broek W.J.A.A., Nelen,M.R., Wansink,D.G., Coerwinkel,M.M., te Riele,H., Groenen,P.J.T.A. and Wieringa,B. (2002) Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 11, 191–198. [DOI] [PubMed] [Google Scholar]
- 20.Ranum L.P.W. and Day,J.W. (2002) Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev., 12, 266–271. [DOI] [PubMed] [Google Scholar]
- 21.Beard W.A. and Wilson,S.H. (1995) Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol., 262, 98–107. [DOI] [PubMed] [Google Scholar]
- 22.Marky L.A. and Breslauer,K.J. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- 23.Petruska J., Arnheim,N. and Goodman,M.F. (1996) Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res., 24, 1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons-Darden T. and Topal,M.D. (1999) Abasic sites induce triplet-repeat expansion during DNA replication in vitro. J. Biol. Chem., 274, 25975–25978. [DOI] [PubMed] [Google Scholar]
- 25.Heidenfelder B.L., Makhov,A.M. and Topal,M.D. (2003) Hairpin formation in Friedreich’s ataxia triplet repeat expansion. J. Biol. Chem., 278, 2425–2431. [DOI] [PubMed] [Google Scholar]
- 26.Spiro C., Pelletier,R., Rolfsmeier,M.L., Dixon,M.J., Lahue,R.S., Gupta,G., Park,M.S., Chen,X., Mariappan,S.V.S. and McMurray,C.T. (1999) Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell, 4, 1079–1085. [DOI] [PubMed] [Google Scholar]
- 27.Henricksen L.A., Tom,S., Liu,Y. and Bambara,R.A. (2000) Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem., 275, 16420–16427. [DOI] [PubMed] [Google Scholar]
- 28.Moore H., Greenwell,P.W., Liu,C.-P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form sedentary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aboul-ela F., Koh,D., Tinoco,I.,Jr and Martin,F.H. (1985) Base–base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res., 13, 4811–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung M.-Y., Ranum,L.P.W., Duvick,L.A., Servadio,A., Zoghbi,H.Y. and Orr,H.T. (1993) Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type 1. Nature Genet., 5, 254–258. [DOI] [PubMed] [Google Scholar]
- 32.Pearson C.E., Eichler,E.E., Lorenzetti,D., Kramer,S.F., Zoghbi,H.Y., Nelson,D.L. and Sinden,R.R. (1998) Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry, 37, 2701–2708. [DOI] [PubMed] [Google Scholar]
- 33.Eichler E.E., Holden,J.J.A., Popovich,B.W., Reiss,A.L., Snow,K., Thibodeau,S.N., Richards,C.S., Ward,P.A. and Nelson,D.L. (1994) Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nature Genet., 8, 88–94. [DOI] [PubMed] [Google Scholar]
- 34.Kunst C.B. and Warren,S.T. (1994) Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell, 77, 853–861. [DOI] [PubMed] [Google Scholar]
- 35.Snow K., Tester,D.J., Kruckeberg,KE., Schaid,D.J. and Thibodeau,S.N. (1994) Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum. Mol. Genet., 3, 1543–1551. [DOI] [PubMed] [Google Scholar]
- 36.Richards R.I. and Sutherland,G.R. (1994) Simple repeat DNA is not replicated simply. Nature Genet., 6, 114–116. [DOI] [PubMed] [Google Scholar]