Abstract
Through its interactions with proteins and proteoglycans, thrombospondin-1 (TSP-1) functions at the interface of the cell membrane and the extracellular matrix to regulate matrix structure and cellular phenotype. We have previously determined the structure of the high affinity heparin-binding domain of TSP-1, designated TSPN-1, in association with the synthetic heparin, Arixtra. To establish that the binding of TSPN-1 to Arixtra is representative of the association with naturally occurring heparins, we have determined the structures of TSPN-1 in complex with heparin oligosaccharides containing eight (dp8) and ten (dp10) subunits, by x-ray crystallography. We have found that dp8 and dp10 bind to TSPN-1 in a manner similar to Arixtra and that dp8 and dp10 induce the formation of trans and cis TSPN-1 dimers, respectively. In silico docking calculations partnered with our crystal structures support the importance of arginine residues in positions 29, 42, and 77 in binding sulfate groups of the dp8 and dp10 forms of heparin. The ability of several TSPN-1 domains to bind to glycosaminoglycans simultaneously probably increases the affinity of binding through multivalent interactions. The formation of cis and trans dimers of the TSPN-1 domain with relatively short segments of heparin further enhances the ability of TSP-1 to participate in high affinity binding to glycosaminoglycans. Dimer formation may also involve TSPN-1 domains from two separate TSP-1 molecules. This association would enable glycosaminoglycans to cluster TSP-1.
Thrombospondin-1 (TSP-1)4 is a secreted glycoprotein that functions during the tissue remodeling that is associated with development, wound healing, synaptogenesis, angiogenesis, and cancer. Through its interactions with proteins and proteoglycans, TSP-1 functions at the interface of the cell membrane and the extracellular matrix to regulate matrix structure and cellular behavior. The N-terminal domain of TSP-1 (TSPN-1) plays an integral role in the interaction of TSP-1 with other secreted and transmembrane proteins (1, 2). TSPN-1 contains binding sites for 1) glycosaminoglycans, 2) low density lipoprotein receptor-related protein-1, 3) various integrins, 4) calreticulin, and 5) fibrinogen (2). β1 integrins that contain the α3, α4, α6, and α9 subunits have all been reported to interact with TSPN-1 (3-6). The glycosaminoglycan-binding site is involved in the association of TSP-1 with syndecan-1, -3, and -4, perlecan, decorin, and other proteoglycans (7). In some cases, the TSPN-1 domain may also interact with the core protein (8). TSPN-1 has been shown to be required for the uptake of TSP-1 through a mechanism that involves proteoglycans and lipoprotein receptor-related protein-1 (9). Because proteins, such as matrix metalloproteinases and vascular endothelial cell growth factor, bind to TSP-1, it is possible that they are taken up along with this protein (1, 10). The regulation of matrix metalloproteinases and vascular endothelial cell growth factor levels by this mechanism can affect angiogenesis in the tumor microenvironment. The domains of TSP-1 also have direct effects on endothelial cell migration and apoptosis (11). Whereas the type 1 repeats of TSP-1 are potent inhibitors of angiogenesis, TSPN-1 stimulates angiogenesis (12, 13). Endothelial cell tube formation, an in vitro model of angiogenesis, is stimulated by TSPN-1. This effect is mediated by the proteoglycan syndecan-4 (12). TSPN-1 also stimulates angiogenesis through integrin engagement. Immobilized TSPN-1 promotes endothelial cell proliferation and survival through α4β1 (4). Synthetic peptides have been used to map the α4β1 binding site to a loop that follows the β12 strand of TSPN-1 (4). The α9β1 integrin on human dermal microvascular endothelial cells also binds TSPN-1 (14). This interaction reportedly stimulates cell proliferation and migration in vitro and angiogenesis in vivo.
We have previously reported the structures of TSPN-1 and TSPN-1 in complex with Arixtra, a synthetic pentameric heparin (15). From these structures, we have defined a major heparin-binding site on the base of the TSPN-1 globular β sandwich domain and observed that the sulfate groups of Arixtra bind to residues Arg-29, Arg-42, and Arg-77, but its polysaccharide chain was largely disordered. The fact that the Arixtra is partially disordered in the complex structure may suggest that the binding of the synthetic small heparin species to TSPN-1 is not well specified. The major TSPN-1·Arixtra associations seem to be from ionic interactions between heparin’s sulfate groups and TSPN-1’s positively charged residues in the heparin binding site, while the more specific hydrogen bonds between TSPN-1 and the backbone of the Arixtra molecule are not observed and/or are largely missing. The longest dimension of TSPN-1’s heparin-binding site is close to the length of a pentasaccharide (15). This is consistent with the minimum size (tetrasaccharide) of heparin that can bind to TSPN-1. The affinity of TSPN-1 for heparin increases with the length of the oligosaccharide up to a decasaccharide (16, 17). In our early studies, we also discovered a long flexible linker (~35 residues) between the TSPN-1 domain and the helical region that is involved in trimer formation (15). This structure raises the possibility that the three TSPN-1 domains within one TSP-1 molecule, leashed by their long flexible linkers, can bind ligands either independently or cooperatively. Compared with the TSPN-1 domain alone, an intact TSP-1 molecule reportedly binds to heparin with a 20-fold higher affinity, suggesting multivalent interaction of trimeric TSP-1 to heparin (9, 18).
In this study, we introduced a fractionated heparin species containing eight (dp8) or ten (dp10) monosaccharides into our co-crystallization experiments. Here we report two structures of TSPN-1 in complex with these fractionated heparins. These crystal complexes, which reveal two distinct dimeric binding modes for TSPN-1 bound to these heparin species, have been further supported by extensive in silico docking calculations that provide quantitative estimates of these binding interactions. We have also compared these complexes to another native TSPN-1 structure to demonstrate that it is the presence of the dp8 and dp10 heparin species that leads to the formation of these dimeric binding modes.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant TSPN-1
A recombinant version of the TSPN-1 (amino acids 1–240 of human TSP-1) was prepared as described previously (15). Limited digestion with α-chymotrypsin (1:200 w/w) was performed with intact TSPN-1 for 20 h at 0 °C and was stopped by adding 1 mm phenylmethylsulfonyl fluoride (15). The principal proteolytic fragment was purified by high-performance liquid chromatography or heparin-Sepharose affinity chromatography. Mass spectral analysis indicates that the proteolysis occurs between residues Cys-214 and Asn-230, resulting in the loss of the N-linked glycan that is attached to Asn-230, and the polyhistidine tail. This proteolysis appears to be necessary for crystallization on TSPN-1 (15). All proteins were further purified by size exclusion chromatography in protein buffer of 200 mm NaCl and 20 mm HEPES at pH 7.8.
Size Exclusion Chromatography
Size exclusion chromatography was performed on a Superdex 75 HR 10/30 column (Amersham Biosciences). The column was pre-equilibrated with buffer (20 mm HEPES, pH 7.5, 200 mm NaCl) and calibrated with pre-mixed protein standards, including myoglobulin (17,000 Da) and ovalbumin (44,000 Da). TSPN-1 (final volume of 200 μl) was applied to the column in the absence of dp10 or after mixing TSPN-1 (6 mg/ml, ~0.31 mm) and dp10 (2.9 mm) in a 1:1 molar ratio at 4°C. The chromatography was carried out at 4 °C at a flow rate of 0.8 ml/min. The calibration curve of Kav versus log molecular weight was prepared using the equation Kav = (Ve − Vo)/(Vt − Vo), where Ve = elution volume for the protein, Vo = column void volume, and Vt = total bed volume.
Crystallization
The purified protein was concentrated to ~10–15 mg/ml for crystallization with the vapor diffusion hanging drop method. Native protein crystals grew from buffer containing 30% polyethylene glycol 1500 and 0.08 m sodium acetate at pH 4.6. The same crystallization buffer was also used for the co-crystallization with heparins as discussed below. The majority of native crystals from the α-chymotryptic degradation product of the TSPN-1 construct were thin plates with the space group of P1 (15). The preparation of heparin oligosaccharides containing eight (dp8) and ten (dp10) subunits by heparinase treatment of low molecular weight heparin is described elsewhere (19). For TSPN-1 and fractionated heparin complexes, the digested TSPN-1 was used for co-crystallization with dp8 and dp10 with a molar ration of 1:2. The TSPN-1·dp 8 co-crystal is chunky and has a trapezoid-like shape while TSPN-1·dp10 crystal has a well defined long tetragon-like shape with pointed tips at both ends. In this report, we describe the stable TSPN-1·dp8 and TSPN-1·dp10 complex structures and the TSPN-1 native P1 form structure.
Data Collection
Diffraction data sets were collected from pre-frozen crystals at 100 K at the 19ID beamline of the Structure Biology Center at the Advanced Photon Source, Argonne National Laboratory. All diffraction data sets were processed and reduced using the HKL2000 suite (Table 1) (20).
TABLE 1.
Crystallographic statistics
| TSPN-1·dp10 | TSPN-1·dp8 | TSPN-1 (P1 form) | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | C2 | P1 |
| Unit cell | |||
| a (Å) | 40.32 | 95.50 | 40.02 |
| b (Å) | 41.07 | 42.06 | 41.71 |
| c (Å) | 241.8 | 52.97 | 59.99 |
| α (°) | 90 | 90 | 73.63 |
| β (°) | 90 | 91.88 | 89.59 |
| γ (°) | 90 | 90 | 75.73 |
| Wavelength (Å) | 0.99187 | 1.03320 | 1.07812 |
| Resolution (Å) | 50-2.4 | 50-1.9 | 50-1.85 |
| Number of unique reflections | 14,771 | 14,441 | 29,499 |
| Redundancy | 4.4 | 5.4 | 2.4 |
| Completeness (%) | 88.8 (65.9)a | 86.1 (53.5)a | 94.0 (75.4)a |
| Rmerge (%) | 4.6 (26.8)a | 6.0 (40.6)a | 5.1 (24.0)a |
| I/σ(I) | 29.87 (3.64)a | 34.40 (2.01)a | 21.11 (2.90)a |
| Refinement | |||
| Resolution | 25-2.4 | 40-1.9 | 30-1.85 |
| Reflections (work/test) | 13,083 (1,140) | 12,136 (1,379) | 27,640 (1,741) |
| Rcrystal/Rfree | 25.62 (29.62) | 25.01 (27.12) | 18.67 (24.98) |
| Bond length (Å)/angle (°) r.m.s.d. from ideal geometry | 0.0065/1.62b | 0.0067/1.73b | 0.018/1.81c |
| Protein atoms average B value (Å2), main chain/side chain | 47.23/48.59 | 57.85/60.00 | 21.231/24.575 |
Last resolution bin.
Refined with CNS.
Refined with Refmac.
Structure Determination and Refinement
Both TSPN-1·dp10 and TSPN-1·dp 8 co-crystal structures, as well as the native structure in P1 form, were solved using the refined TSPN-1 structure (PDB code: 1Z78) as the search model with the program Molrep in the CCP4 suite (21). The model rebuilding and final refinement of the structure were done using the programs O (22) and CNS (23), respectively (Table 1). In the TSPN-1·dp10 complex structure, several sulfate groups were built into bulky densities associated with the major heparin-binding site as discussed under “Results.” Because O-sulfate and N-sulfate groups of these fractionated heparins are indistinguishable in the structure for the partially disordered molecule, all of them were modeled as O-sulfate groups and designated SO groups.
In the final native TSPN-1 model, we observed broken or poorly resolved densities that had been described previously for the first native TSPN-1 structure (15). The coordinates of the complexes TSPN-1·dp10 and TSPN-1·dp8 and the native TSPN-1 in P1 have been deposited in the Protein Data Bank (PDB) under the access codes 2OUH, 2OUJ, and 2ES3, respectively.
Docking
To further evaluate the binding mode and interaction interface between TSPN-1 and the two fractionated heparin oligomers dp8 and dp10, we carried out in silico docking calculations using AutoDock4 (24). TSPN-1 structures from their co-crystallized complexes with heparin ligands dp8 and dp10 were used as the starting coordinates for our docking calculations. The heparins dp8 and dp10 were constructed from known heparin structures that are deposited in the PDB; prior to their use in our docking studies, they were corrected for bond order and orientation, and the refined models were then protonated. Dp8 and dp10 were subsequently energy minimized using a standard Tripos force field that employs Powell minimization and simplex optimization with a distance-dependent dielectric function and an energy gradient of 0.05 kcal/molÅ. AutoDock-Tool 1.4.5 was then used to compute the Gasteiger atomic charges followed by a merge of all non-polar hydrogens such that the charge of these hydrogens were assigned to the atom to which they are directly bonded (25, 26).
To evaluate the structure of the TSPN-1 heparin complexes, we have used grid dimensions of 60 Å × 72 Å × 60 Å for both dp8 and dp10 in complex with TSPN-1. Docking grid maps were calculated using Autogrid4. The grid spacing for the dp8 TSPN-1 complex structure was 0.708 Å, and for the dp10 TSPN-1 complex structure, it was 0.802 Å. The interface of each TSPN-1 dimer complex was selected as the grid center for both the dp8 and dp10 TSPN-1 complexes. AutoDock4 uses a standard Lamarckian genetic algorithm that couples a typical Darwinian genetic algorithm for global searching and the Solis and Wets algorithm for local searching for this docking protocol. The Lamarckian genetic algorithm parameters used for the present docking study of both dp8 and dp10 heparin oligomers docked to the TSPN-1 dimer structure were as follows: the initial population of 50 randomly placed individuals, a maximum number of 25 × 105 energy evaluations or a maximum number of 27,000 generations, a mutation rate of 0.02, a crossover ratio of 0.80; an elitism value of 1, probability of performing local search on an individual was set to a frequency of 0.06, a maximum number of consecutive success or failures before doubling or halving the local search step size was 4, and a maximum of 800 iterations per local search. For each of the TSPN-1 complexes we calculated 20 independent docking runs, and only the lowest energy confirmation is presented.
RESULTS
TSPN1·dp10 Co-crystal Structure
The fractionated heparin dp10 was co-crystallized with TSPN-1 under the condition of 30% polyethylene glycol 1500, 0.08 m sodium acetate at pH 4.6. The TSPN-1·dp10 co-crystal is in the space group of P212121 (Table 1), the same as the previously reported co-crystal of TSPN-1·Arixtra (15). The unit cell dimension along the c axis of the TSPN-1·dp10 co-crystal is more than double that of TSPN-1·Arixtra co-crystal (Table 1). As a consequence, there are two TSPN-1 domains related by a non-crystallographic 2-fold axis almost perpendicular to the c-axis in each asymmetric unit (Fig. 1). There are no major structural variations between the two TSPN-1 domains or between them and the structure we reported earlier (15). The two TSPN-1 domains can be superimposed on each other with an r.m.s.d. as low as 0.3 Å. Structural variation were only observed in a few loop regions, primarily the α1_β2 loop (G20AARKGSG) that is generally very flexible. The two TSPN-1 domains have a small hydrophobic interface that is mainly composed of residues from the β2_β3 loop of TSPN-1, including residues Leu-30, Pro-36, Ser-37, and Pro-39 (Fig. 1). With the exception of a few water molecule bridges, there are no specific hydrogen bonds that are well defined by the observed electron densities between the two domains. The buried surface area resulting from the contact is ~632 Å2, much less than the value of 1600 ± 400 Å2 that is generally believed to be of physiological significance (27). Thus, the small hydrophobic interface between these two TSPN-1 domains seems to be insufficient to lead to dimer formation. The two TSPN-1 domains are orientated in such a way that their major heparin binding sites are aligned to form an extended positively charged patch of ~20 × 60 Å2 (Fig. 1B). The electric dipole of the two TSPN-1 domains is located between two molecules, along the pseudo-2-fold axis and perpendicular to the positively charged patch. The dipole moment is calculated to be 235.6 Angstrom electron charge units (28). The potential distribution and dipole moment orientation could help guide the approaching and eventual binding of negatively charged heparin molecules onto the extended heparin binding site (29, 30). From TSPN-1 packing in the crystal, the space next to its positively charged patch has enough room to accommodate a heparin molecule, such as dp10. Extra electron densities, mostly in the form of large globular shapes, are observed around the positively charged patch, especially associated with the residue Arg-42, but also with Arg-29 and Arg-77 from both TSPN-1 domains (Fig. 1C). A sole continuing density is found around the Arg-42 of one of the domains, designated as TSPN-1A in Fig. 1A. These densities can’t be interpreted as solvent molecules. Some bulky electron densities are located in the positions marked with yellow crosses in Fig. 1B. These uncharacterized densities are very similar to those observed in the TSPN-1·Arixtra complex crystal structure where they are interpreted to be sulfate groups from the partially disordered pentameric oligosaccharide, Arixtra (15). Both Arg-42 and Arg-29 seem to play an important role in the interaction of TSPN-1 with dp10 or Arixtra. In the TSPN-1·Arixtra complex, there are three bulky electron densities associated with Arg-42 that were interpreted to be from the sulfate groups of Arixtra (15). In the TSPN-1·dp10 complex, there are only two of these bulky electron densities associated with Arg-42.
FIGURE 1. The dimeric TSPN-1 structure in its co-crystal with heparin dp10.
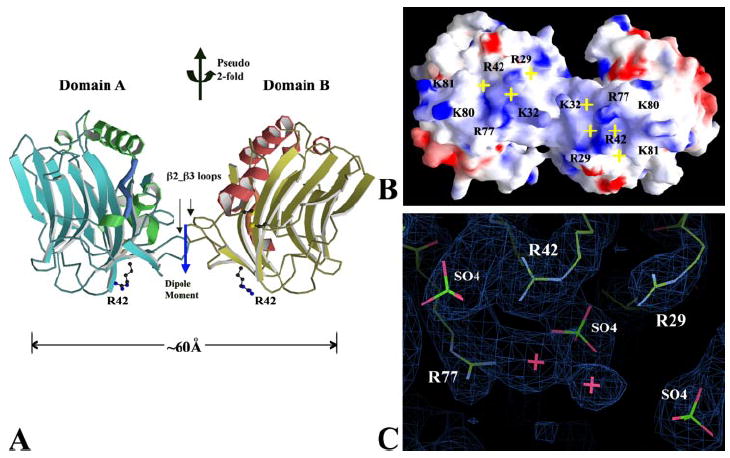
A, a ribbon drawing of a dimeric TSPN-1, in which two monomers are related by a pseudo 2-fold rotation axis. Residue Arg-42 from the major heparin-binding site at the bottom of each domain is drawn in ball-and-stick form. The two TSPN-1 domains contact each other to form a small hydrophobic interface contributed mainly by residues on the β2_β3 loop, including Leu-30, Pro-36, Ser-37, and Pro-39. The electric dipole moment of the dimeric TSPN-1 is shown by the blue arrow and centered at the beginning of the arrow. B, an electrostatic potential surface representation of the dimeric TSPN-1. The dimeric TSPN-1 forms an extended positively charged patch (~20×60Å2) by bridging the major heparin-binding sites of two TSPN-1 domains. The orientation of the figure is related to the position of the dimeric TSPN-1 in panel A by a rotation of ~90° around the horizontal axis. Some extra bulky electron densities associated with the extended positively charged patch are located in the positions marked by yellow crosses. They were assigned to SO4 groups from dp10 and presumed to provide gluing points between TSPN-1 and dp10. C, the major heparin-binding site of TSPN-1 domain A with a 2Fo – Fc map contoured at the 1.3σ. The map is colored in blue. The two SO4 groups associated with residue Arg-42 are also seen in the TSPN-1·Arixtra complex structure (15). For refinement purposes, several water molecules, as shown in pink crosses, were positioned into the uncharacterized continuous densities from these SO4 groups. Figs. 1A, 2A, 3A, 4A, and 5A were prepared using the program MolScript (50), Figs. 1B, 3B, and 5B were prepared using the program GRASP (28). Figs. 1C and 3C were prepared using the program COOT (51).
The association of these bulky densities with the arginines within the heparin-binding sites of both of the TSPN-1 domains suggests that each domain is interacting with dp10. Because two TSPN-1s bind the heparin on the same side, we call this binding mode the cis-dimer binding mode. The lack of well defined electron densities for dp10, especially the heparin backbone of dp10, could be due to the presence of multiple heparin binding modes, partial disordering, and/or chemical heterogeneity of the heparin species being studied. Like the TSPN-1·Arixtra complex structure, the flexibility of the carbohydrate backbone causes partial disorder that results in the presence of only “gluing atoms” (i.e. sulfate groups) within the densities. Moreover, because dp10 is almost twice as long as the positively charged patch of TSPN-1, it is likely that two TSPN-1 domains within a cis-dimer bind one heparin (a molar ratio of 2:1) with the heparin having two possible orientations that are anti-parallel to one another. These two orientations of the heparin may further explain why there is no continuous electron density for the heparin molecule’s backbone. The proposal that a single dp10 binds two TSPN-1 molecules is further supported by the fact that the hydrophobic interface between the two TSPN-1 molecules is too small to support dimer formation, as discussed above. In this proposed 2-to-1 binding model, it is the protein (two TSPN-1 domains) binding to heparin (one dp10) that provides the major “gluing” force of the TSPN-1 dimerization. Two TSPN-1 domains properly align themselves so that they (1) create a small hydrophobic interface to provide additional binding energy, (2) maximize the interface for their binding to dp10, and (3) create a combined dipole perpendicular to the extended positively charged patch to facilitate heparin binding.
We have used size exclusion chromatography to determine whether or not dimerization of TSPN-1 occurs in solution. In the absence of dp10, TSPN-1 is eluted as a single sharp peak with an apparent molecular weight of 17,690 (in comparison to 23,630 predicted from the sequence). Preincubation of TSPN-1 with dp10 results in a shift in the apparent molecular weight of the peak to 26,600 (data not shown). Because the molecular weight of dp10 (excluding sodium ions) is ~2,870, the shift of the elution volume of the TSPN-1 in the presence of dp10 can’t be simply attributed to the addition of the heparin to monomeric TSPN-1. The mixture TSPN-1 with the pentameric heparin Arixtra did not cause any significant shift of elution peak.5 The protein peak eluted in the presence of dp10 is significantly broader than the one that is observed in its absence, suggesting a state of monomer-dimer equilibrium for TSPN-1 in the presence of dp10. The elution volume shift caused by the formation of a weak dimer has been observed in other protein preparations (31, 32).
TSPN-1·dp10 Docking
Although challenging, rigid body docking has been previously used to evaluate the interactions between heparin sulfate moieties and cognate heparin binding sites localized on known proteins, including antithrombin, basic fibroblast growth factor, fibroblast growth factor 1, and interleukin-8 (25, 33, 34). In support of the crystal complex ofTSPN-1with dp10 we determined the docked complex of TSPN-1·dp10. Of the 20 calculated structures, 18 were clustered around the heparin binding site illustrated in Fig. 2, identifying a common mode and/or orientation of binding between TSPN-1 and dp10. The outlying structures were determined to be in a reversed orientation resulting in a loss of critical interactions; however, the majority of the residues involved in mediating the interaction are conserved as determined by similar ΔG values for all complexes. The minimum energy structure from our rigid body docking calculations (Fig. 2, A and B) identified that the dp10 heparin moiety forms critical contacts with residues in both TSPN-1 domain A and B. Although the conformational flux and/or presence of multiple interactions precluded atomic resolution of the complex interface in the crystal structure, a phenomenon that is well established for heparin-binding proteins, our docked conformation supports the presence of a localized hydrogen bonding network that is in agreement with the previously noted “bulky electron density” patch. These interactions involve several positively charged amino acids within both domains. The low energy structure for the interaction between TSPN-1 and dp10, as illustrated in Fig. 2, supports the presence of a hydrogen bonding network involving residues Arg-29, Lys-32, Arg-42, and Lys-80 of domain A as well as Lys-32 of domain B within the TSPN-1 dimer. Importantly, the low energy conformer of these non-biased rigid body docking calculations, which has a ΔG of −8.04 kcal/mol and an estimated Ki of 1.28 nm, supports the crystallographic structure of this TSPN-1 complex, which identified a cis-dimer conformation in the presence of dp10 that is mediated by several of these residues. Our docked conformation supports the presence of both backbone amide and side-chain amide interactions with the proposed gluing atoms (sulfate groups) within the dp10 heparin species. Important contacts are observed for the 1st, 4th, 5th, and 6th sugar sulfates and/or carbonyl functional groups with residues in domain A and the 7th sugar moiety with Lys-32 in domain B (Table 2). Importantly, Arg-42 is a hydrogen bond acceptor that is involved in seven unique atomic interactions with the dp10 ligand. Hydrogen bonding networks were defined using auto-monitor in Sybyl7.3 (Tripos) with interaction scores and probabilities defined using MOE (Molecular Operating Environment, Chemical Computing Group, Montreal, Canada).
FIGURE 2. Docked structure of dp10 heparin in complex with TSPN-1.
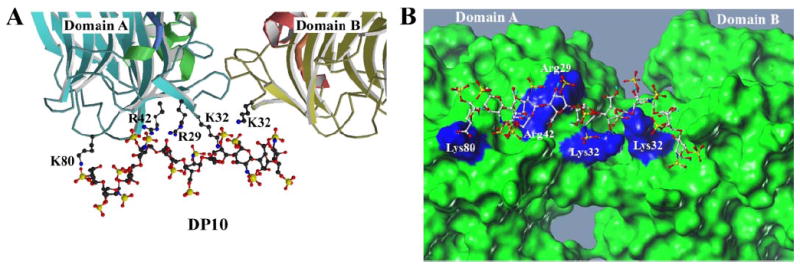
A, TSPN-1·dp10 low energy complex identified using AutoDock4. TSPN-1 residues involved in the hydrogen bonding network are rendered in ball-and-stick. Each residue involved in the hydrogen bonding network is annotated. For clarity hydrogen bonds are not displayed. Each domain is labeled and rendered as a ribbon drawing, with α-helices colored green (domain A) or magenta (domain B), and β-strands and random coil are colored cyan (domain A) or yellow (domain B). The heparin moiety dp10 is rendered in ball and stick representation. B, Molcad surface of the TSPN-1·dp10 complex with residues involved in the hydrogen bonding network colored in blue. This view of the TSPN-1·dp10 complex is the result of rotating the complex −90° around the x-axis (out of the plane of the page or screen) to provide a view of this complex from the heparin binding site, which is located at the base of complex in panel A. Figs. 2B and 4B were prepared using the SYBYL suite of programs (Tripos).
TABLE 2. TSPN-1 heparin interactions for dp8 and dp10 within their proposed heparin binding sites.
The table shows residues involved in the hydrogen bonding network.
| Complex | |||
|---|---|---|---|
| TSPN-1·dp8 |
TSPN-1·dp10 |
||
| Domain A | Domain B | Domain A | Domain B |
| Lys-24 | Lys-24 | Arg-29 | Lys-32 |
| of 2nd sugar | O- in 6th sugar ring | OH of 5th sugar | of 7th sugar |
| 6–7 O-linkage | of 4th sugar | ||
| Arg-42 | COO− of 7th sugar | ||
| COO− of 5th sugar | Lys-32 | ||
| O- of 5th sugar ring | Arg-29 | OH of 7th sugar | |
| of 6th sugar | of 5th sugar | of 6th sugar | |
| Arg-77 | Arg-42 | ||
| of 6th sugar | of 4th sugar | ||
| COO− of 5th sugar | |||
| Lys-80 | |||
| COO− of 1st sugar | |||
TSPN-1·dp8 Co-crystal Structure
The fractionated heparin dp8 was also co-crystallized with TSPN-1 under the same crystallization condition as that of TSPN-1·dp10. The TSPN-1·dp8 co-crystal is chunky and has a trapezoid-like morphology with a space group of C2. There is only one TSPN-1 molecule in one asymmetric unit. The interesting feature of this structure is that the heparin-binding sites of the TSPN-1 domain and its 2-fold symmetry related partner are oriented in such a way that they face each other, and there is a shallow groove between the two domains, where the extra electron densities are observed (Fig. 3). This symmetry-related dimer also has a small hydrophobic interface composed of residues on the β2_β3 loop of TSPN-1, including Leu-30 and Val-31. The buried surface resulting from the contact of the two TSPN-1 molecules is ~657 Å2 and is also unlikely to be sufficient for the formation of TSPN-1 dimers in the absence of heparin. We want to emphasize here that these small hydrophobic interfaces are unique to both the TSPN-1·dp10 and TSPN-1·dp8 complex structures. By contrast, in the P21 and P1 forms of the native TSPN-1 structures (see below), and, in the TSPN-1·Arixtra structure, the molecular interactions between neighboring symmetry-related molecules are largely hydrophilic involving multiple hydrogen bonds. Similarly, the two domains, with their heparin-binding sites facing each other, also create an electric dipole moment equal to 276.84 Angstrom electron charge units along the 2-fold symmetric axis that is perpendicular to the groove.
FIGURE 3. The dimeric TSPN-1 structure in its co-crystal with heparin dp8.
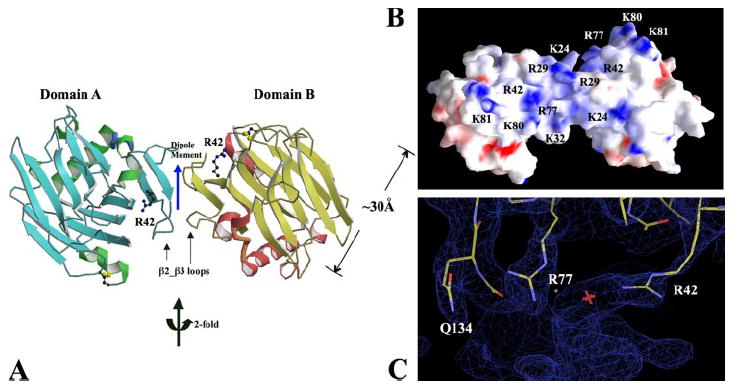
A, a ribbon drawing of a dimeric TSPN-1, in which two monomers are related by a symmetric 2-fold rotation axis. Residue Arg-42 from the major heparin-binding site of each domain is drawn in ball-and-stick form. The two TSPN-1 domains contact each other to form a small hydrophobic interface contributed mainly by residues on the β2_β3 loop, including Leu-30 and Val-31. The electric dipole moment of the dimeric TSPN-1 is shown by the blue arrow and centered at the beginning of the arrow. B, an electrostatic potential surface representation of the dimeric TSPN-1. The dimeric TSPN-1 forms an extended positively charged shallow groove (~30 Å long) by bridging the juxtaposing major heparin-binding sites of two TSPN-1 domains. The orientation of the figure is similar to the position of the dimeric TSPN-1 in panel A. C, the major heparin-binding site of TSPN-1 with a 2Fo – Fc map contoured at the 0.9σ. The map is colored in blue. For refinement purpose, a water molecule, as shown with a pink cross, has been positioned into the uncharacterized continuous densities around the positively charged residues.
The extra electron densities observed along the shallow groove formed by the heparin binding sites of two TSPN-1 domains implies that the dp8 runs along this shallow groove. From the connection of the bulky densities between two domains in TSPN-1·dp8 structure (Fig. 3), residues Arg-42 and Arg-77 appear to play a major role as they do in the structure of TSPN-1·dp10 (Fig. 1). The densities start from the tips of their side chains to large bulky densities (Fig. 3C). Residues Arg-42 and Arg-77 always have well defined densities themselves, and they seem to provide essential heparin-anchoring sites. Interestingly, in the TSPN-1·dp8 structure, a nearby residue of these arginines, Gln-134, also seems to be involved in the interaction with heparin (Fig. 3C). This observation is not surprising, because protein-heparin interactions also involve some specific hydrogen bonds in addition to ionic interactions. Gln-134 seems to be selectively involved in heparin binding in TSPN-1·dp8 structure. Additionally, residue Arg-29, which is involved in Arixtra binding, is in the middle of the combined heparin-binding site in this structure (Fig. 3B). The tip of its side chain does not have density, and it is not clear where it points. The residues, Lys-24 and Lys-32, also appear to be involved in heparin binding. They also have poor densities for their side chains. Unlike Arg-42 and Arg-77, residues Arg-29, Lys-24, and Lys-32 seem to be less specified in heparin binding, and they may have several binding modes.
Based on the above observation that the heparin lies between two TSPN-1 domains, we designate this binding mode as the trans-dimer binding mode. For reasons previously provided and discussed for the TSPN-1·dp10 structure, the observed complex is not compatible with a 1:1 molar ratio of TSPN-1: dp8. Thus, we propose that in the TSPN-·dp8 structure involves a single dp8 heparin moiety bound to two TSPN-1 domains in a trans-dimer mode of interaction. Again, similar to what is discussed in the structure of TSPN-1·dp10, the asymmetric heparin molecule has two alternative anti-parallel binding modes and the crystal symmetry smears out the electron densities of the ligand, especially its backbone in the TSPN-1·dp8 complex.
TSPN-1·dp8 Docking
The low energy conformation of the 20 calculated TSPN-1·dp8 complexes, which has an estimated binding energy, ΔG of −7.86 kcal/mol and a Ki of 1.73 nm, provides significant additional data that further supports the importance of residues Lys-24, Arg-42, and Arg-77 in domain A and residues Lys-24 and Arg-29 in domain B in mediating the hydrogen bonding network with dp8 (Fig. 4). Of the 20 calculated complexes we had complete convergence and overlap of the critical TSPN-1 residues that mediate this interaction. These TSPN-1 residues are along the narrow groove that, as previously discussed, is formed in this trans-dimer. These TSPN-1 residues make hydrogen bonding interactions with the 5th, 6th, and 7th sugar moieties of dp8. Although the side-chain electron density for Lys-24 is not observed in our crystal complex, this docked conformation suggests that the Nζ side-chain amide of this residue is a hydrogen bond acceptor for the second sugar moiety of dp8. It is possible that the conformational flux exhibited at the N and C termini for known heparins contributes to the poorly resolved electron density for Lys-24. Our docked complex (Fig. 4) supports a critical role for Arg-42 of domain A, which makes a series of hydrogen bonding interactions with the 5th and 6th sugars interacting with both the sulfate atoms of the 6th sugar and the carbonyl of the 5th. Taken together these data support a unique mechanism of interaction for both dp8 and dp10 with the interactions of dp8 clustered along the narrow groove (Fig. 4B), whereas the interactions of dp10 extend along the continuous interface provided by the cis-dimer (Fig. 2B). Figs. 2 and 4 highlight these unique complexes and provide the orientation of these heparin moieties within these defined interfaces. Although the interactions between TSPN-1 and dp8 (Fig. 4) and dp10 (Fig. 2) appear to involve similar amino acids there is a unique orientation and alignment for each of these ligands within the well defined, unique interfacial binding sites identified in our crystallography studies. The docked complexes were rendered in Sybyl7.3.
FIGURE 4. Docked structure of dp8 heparin in complex with TSPN-1.
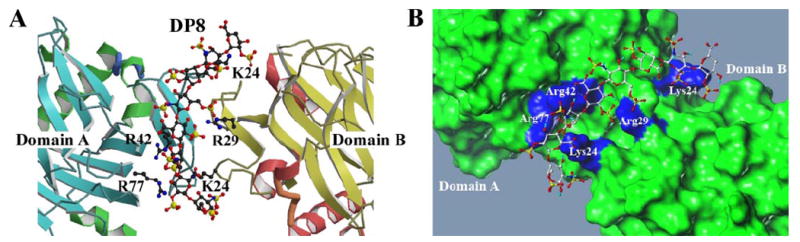
A, TSPN-1·dp8 low energy complex identified using AutoDock4. TSPN-1 residues involved in the hydrogen bonding network are rendered in ball-and-stick and annotated to identify those residues involved in the hydrogen bonding network. For clarity hydrogen bonds are not displayed. Each domain is labeled and rendered as a ribbon drawing, with α-helices colored green (domain A) or magenta (domain B), and β-strands and random coil colored cyan (domain A) or yellow (domain B). The heparin moiety dp8 is rendered in ball and stick. B, Molcad surface of the TSPN-1·dp8 complex with residues involved in the hydrogen bonding network colored blue. The TSPN-1·dp8 complex in panel B has been rotated 90° around the x-axis and 45° around the y-axis (out of the plane of the page or screen) to provide a view of this complex from the heparin binding site at the top of the complex as illustrated in A.
The Second Native TSPN-1 Structure
The two dimeric forms of TSPN-1 in complex with fractionated heparins discussed above are unique crystal forms. No symmetric dimeric crystal forms are produced in the absence of longer heparins (n > 5). Here we report a second native TSPN-1 structure, from the most common crystal form of TSPN-1 grown from exactly the same crystallization condition. These crystals always form as thin plates that have an unstable lattice in the preparation of heavy atom derivatives. This structure provides us with another opportunity to compare TSPN-1 molecular packing in crystals.
The second native TSPN-1 structure with no ligand is in P1 space group, and there are two TSPN-1 molecules in a unit cell. The two TSPN-1 molecules are oriented upside-down relative to each other so that the heparin-binding sites of the two TSPN-1 molecules are on the opposite sides of the dimeric conformation. There is no pseudo-dyad between them, and there is no extra electron density associated with their two heparinbinding sites. The two molecules interact with each other through residues mainly from their edge strands, β8 and β11 (Fig. 5). The total buried surface from the interaction is ~1018Å2. Across this interface, there are 14 well defined hydrogen bonds and salt bridges and some water-bridged hydrogen bonds, as well as a few hydrophobic contacts. The crystals with this hydrophilic interaction-dominated interface (or packing) are unstable and susceptible to changes in the lattice to double the unit cell when heavy atom compounds were soaked into the crystals. There are no significant differences in the conformation of the P1 form as compared with the previously published structure (15). The two structures can be superimposed on each other with r.m.s.d. values of <0.75 Å. The atomic coordinates and structure factors of the second form of native TSPN-1 have been deposited in the Protein Data Bank with accession code 2ES3 (15).
FIGURE 5. The native TSPN-1 structure in P1 space group.
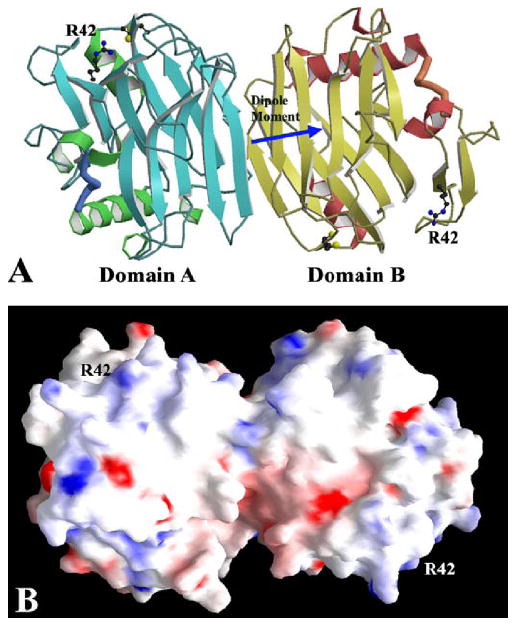
A, a ribbon drawing of the two TSPN-1 domains in one asymmetric unit. They are not related by any pseudo-symmetric operation. Residue Arg-42 from the major heparin-binding site of each domain is drawn in ball-and-stick form. The two TSPN-1 domains interact with each other extensively through their edge strands (see “Results”). The electric dipole moment of the two TSPN-1 domains is shown by the blue arrow and centered at the beginning of the arrow. B, an electrostatic potential surface representation of thetwoTSPN-1domainsin the nativeTSPN-1structure. Residue Arg-42 from the major heparin-binding site of each TSPN-1 domain is labeled to show the separation of the two major heparin-binding sites in this native structure. The orientation of the figure is similar to the one shown in panel A.
DISCUSSION
We have previously reported the structure of TSPN-1 in association with the synthetic heparin, Arixtra (15). Wechose Arixtra, because it is a homogeneous chemical species that we anticipated would be amenable to crystallization. However, Arixtra has two constituents that are rare in heparin, a 3-O-sulfate group that is essential for the anticoagulant activity and one glucuronic acid (35). To establish that the binding of TSPN-1 to Arixtra is representative of the association with naturally occurring heparins, we have determined the structures of TSPN-1 in complex with two different sized heparin fragments, dp8 and dp10. We have found that (1) dp8 and dp10 bind to TSPN-1 in a manner similar to Arixtra, and (2) dp8 and dp10 induce the formation of a trans and/or cis TSPN-1 dimer, respectively. Arginine residues in position 29, 42, and 77 appear to be involved in the binding of sulfate groups for all three forms of heparin as identified in our crystallographic and docking studies. In addition, Gln-134, Lys-24, and Lys-32 are positioned to interact with the sulfate groups of dp8. All of these residues fall within a positively charged patch on the bottom of the TSPN-1 domain. Lysine residues in position 80, 81, and 106 also contribute their positively charged side chains to the bottom of this domain. Whereas we have not observed an association of heparin with these residues in the crystal structures, the amide-bearing side chain of Lys-80 is identified as a hydrogen bond acceptor involved in the formation of a hydrogen bond with the carbonyl moiety of the first sugar in the dp10 complex in our docking studies. The amino acid Lys-80 has been reported to be involved in heparin binding through the use of site-directed mutagenesis (36).
The determination of the structure of heparin/protein complexes is impeded by the heterogeneity of the heparin and by the fact that, in some cases, multiple modes of interaction seem to exist. Natural heparin and heparan sulfate are large polydisperse molecules of extended, unbranched oligosaccharides. They are composed of disaccharide units that include a hexuronic acid subunit bound to a d-glucosamine subunit by an α1 – 4 glycosidic linkage. Heparin and heparans vary in the degree of sulfation and acetylation. The uronic acid subunit may either be underivatized or 2-O-sulfated. The α-d-glucosamine moiety may be either N-sulfated or N-acetylated. The N-sulfated glucosamines may also be O-sulfated at C3, C6, or both. TheN-acetylated glucosamines may or may not be O-sulfated at C6. Thus, each disaccharide monomer has one of six possible structures in the glucosamine position and four possible structures in the hexuronic acid position (37). In heparin, the hexuronic acid is mainly present as 2-O-sulfated iduronic acid. The multiple chemical and structural forms of natural heparan sulfates leads to complexity in the study of heparinprotein interactions by x-ray crystallography as we and others have previously reported. Whereas many heparin-binding proteins have been identified and studied using biochemical approaches, the number of available heparin·protein complex structures solved by x-ray crystallography and/or NMR spectroscopy remains small (38). The use of well defined synthetic heparins in protein·heparin complex crystallization studies has helped to reveal the interactions in detail (39-42). However, the specificity and varied binding modes of heparin species provide difficult obstacles for structural investigations of these complexes. It is possible that the partially disordered structure of Arixtra was the result of multiple binding modes, because the interactions between sulfate groups of the heparin and clustered positively charged residues of TSPN-1 provided anchoring sites for complex formation; however, the backbone of the heparin was not in a single well defined conformation. Our data here suggests that dp8 and dp10 are similarly partially disordered and/or prone to conformational fluctuations. These observations suggest that TSPN-1 has multiple binding modes to a variety of different heparins and probably also to heparan sulfates in which sulfated domains of dp8 and dp10 occur in all species examined to date. This versatility may enable TSP-1 to act as a ligand for proteoglycans in a wide range of tissues and biological contexts. TSP-1 functions in the tissue remodeling that is associated with development, wound healing, synaptogenesis, angiogenesis, and cancer. Thus, TSP-1 may have evolved to have high affinity glycosaminoglycan-binding sites with broad specificity and conformational plasticity permitting these varied binding conformations to be recognized. This conclusion is consistent with the observation that heparins that are fractionated into populations with a wide range of affinities for fibronectin, laminin, and type I collagen all display high affinity for TSP-1 (43). Another example of multiple protein-heparin binding modes is the heparin·aFGF complex, in which the interacting details of the decasaccharide with each aFGF domain in the heparin-linked aFGF dimers are very different (44). It has therefore been proposed that the variation in heparin-binding modes of proteins might reflect the biological diversity of the interactions, because the heparan sulfates in vivo are heterogeneously sulfated.
In this study, we have observed that a single heparin molecule can bind two TSPN-1 domains in either a cis or trans orientation. The two TSPN-1 domains in the cis orientation are bound such that their positively charged residues are localized at the bottom of the domain and thus aligned to engage heparin in this conformation. In this dimer, the two TSPN-1 domains contact each other with the residues in each of their β2_β3 loops participating in hydrophobic interactions. This loop is very flexible and seems to be ideal to compromise any strain raised from the heparin-induced TSPN-1 dimers. However, the TSPN-1 domains are not arranged in a simple fashion along heparin’s helical orientation in the cis-binding mode. Instead, they are arranged in a dimeric form of a pseudo 2-fold symmetry. In this way, they form an extended heparin-binding site, with each monomer positioned to interact with one side of the heparin (cis-mode) moiety. Because the extended heparin-binding site of the dimeric TSPN-1 is ~60 Å, it can interact with at least three turns of heparin helix. The cis binding models are based on the close correspondence between the length of dp10 (two turns of the heparin helix) and the length of a TSPN-1 molecule (~30 Å). The binding of each TSPN-1 to dp10 is similar to the binding of a single TSPN-1 to the pentameric Arixtra, a finding that is supported by the contiguous hydrogen bonding network identified in our low energy docked conformation (Figs. 1 and 2). In this orientation, the β12 strands, the α4 helix, and the intervening loop face away from the dimer interface. Because this region has been identified as the integrin α4β1 and fibrinogen binding sites, the dimerization of TSPN-1 by heparin may increase the affinity of TSP-1 for these ligands by promoting multivalent interactions (4, 45).
The x-ray crystallography data presented here indicates that heparin can induce a second type of TSPN-1 dimer. In the trans dimer, dp8 lies between two TSPN-1 domains in a shallow groove. Heparin induces a similar trans dimer of the aFGF molecule (44). The combined heparin-binding sites in the TSPN-1·dp8 complex form this shallow groove, which permits the two TSPN-1 monomers to bind heparin from two sides (Figs. 3 and 4). The length of the combined binding site is shorter, corresponding to the shorter heparin species. The trans-binding models utilize the potential availability of two binding surfaces on the heparin helix to complex two TSPN-1 molecules per approximately five monosaccharide residues (or about one turn of the heparin helix). Our low energy TSPN-1·dp8 docked complex supports the presence of unique interfacial contacts for each monomer (Fig. 4). The heparin moiety dp8 is aligned from the top of the electrostatic surface (Fig. 4B) with Lys-24 of domain A hydrogen bonded to the second sugar of dp8 and Lys-24 of domain B interacting with the sixth and seventh sugars of dp8. This orientation runs along this shallow groove while the dp10 heparin traverses the electrostatic map commencing with a hydrogen bond involving Lys-80 in domain A to and through an interaction involving Lys-32 of domain B (Fig. 2B).
The interfaces of both types of dimer are formed from the predominantly hydrophobic contact of the β2_β3 loop regions. Two classes of heparin/protein binding modes have been proposed by others: those in which protein molecules bind to the same side of a heparin (cis-binding model) and those in which protein molecules bind to opposite sides of the heparin (trans-binding model) (46, 47). TSPN-1 represents the first example of a protein that can form both cis and trans dimers in the presence of heparin.
One of the features of both cis- and trans-binding modes is that the formation of each dimer produces a dipole moment that is perpendicular to the combined heparin-binding site. There are no studies on the relationship of the protein electric dipole moments and heparin-binding ability. However, a comparison between DNA-protein and heparin-protein interactions may inspire some thoughts on their common features. First, both DNA and heparin are negatively charged linear molecules. Second, both molecules are composed of a given number of repeated subunits. Protein-DNA recognition has been extensively studied. Some studies conclude that DNA-binding proteins have higher net positive charge and electric dipole moment than other proteins, suggesting a potential role of dipole moment of a protein in DNA binding (48). Theoretical approaches for identifying and characterizing heparin-binding proteins are very limited. From primary sequences, it is generally dependent on the search for hallmark sequences, which are not always present. From three-dimensional structures, a cluster of positively charged residues on the surface is usually assumed to be a potential heparin-binding site (38). These residues can be well separated in the protein’s primary sequence. The notion that a localized concentration of positively charged residues within a patch on the molecular surface could generate any impact on protein’s electric properties, such as dipole moment, remains open to discussion. Another seemingly comparable feature is that many proteins bind to DNA as either homodimers or heterodimers (49). Heterodimers can achieve the diversity of target-site recognition and functions. Homodimers, often bind to target DNA asymmetrically, with one monomer often contributing a greater amount of the binding energy, which leads to quasi-symmetric structures in which identical subunits adopt similar but not identical conformations as we observed in our docked conformations (48). The homodimers observed in this study seem to be comparable to this DNA-binding feature with both domains important for binding the dp8 and/or dp10 heparins, but with one of the TSPN-1 monomers involved in a disproportionate number of critical hydrogen bonding interactions.
The data presented here indicate that TSP-1 engages heparin in multiple ways. This flexibility probably enables TSP-1 to function as a ligand for a broad spectrum of glycosaminoglycans from a variety of tissue sources. The ability of several TSPN-1 domains to bind to glycosaminoglycans simultaneously probably increases the affinity of binding through multivalent interactions. The formation of cis and trans dimers of the TSPN-1 domain with relatively short regions of heparin probably further enhances the ability of TSP-1 to participate in high affinity binding to glycosaminoglycans. Dimer formation may also involve TSPN-1 domains from different TSP-1 molecules. This association would enable glycosaminoglycans to cluster TSP-1 molecules. Whereas our structural data raise the possibility of dimer formation, the existence of cis and trans dimers in the extracellular matrix or at the cell surface remains to be shown.
Acknowledgments
We thank Lydia Gregg and Sami Lawler for help in preparing the manuscript and Nijole Gasiunas for preparing the heparin oligonucleotides.
Footnotes
This work was support by Grants HL49081, HL48675, and HL68003 from NHLBI, National Institutes of Health.
The atomic coordinates and structure factors (code 2OUH and 2OUJ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University,NewBrunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: TSP-1, thrombospondin-1; TSPN-1, N-terminal domain of TSP-1; dp8, heparin oligosaccharide containing eight subunits; dp10, heparin oligosaccharide containing ten subunits; r.m.s.d., root mean square deviation; aFGF, acidic fibroblast growth factor.
K. Tan, J.-H. Wang, and J. Lawler, unpublished data.
References
- 1.Bornstein P, Agah A, Kyriakides TR. Int J Biochem Cell Biol. 2004;36:1115–1125. doi: 10.1016/j.biocel.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Herndon ME, Lawler J. Matrix Biol. 2000;19:597–614. doi: 10.1016/s0945-053x(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 3.Calzada MJ, Sipes JM, Krutzsch HC, Yurchenco PD, Annis DS, Mosher DF, Roberts DD. J Biol Chem. 2003;278:40679–40687. doi: 10.1074/jbc.M302014200. [DOI] [PubMed] [Google Scholar]
- 4.Calzada MJ, Zhou L, Sipes JM, Zhang J, Krutzsch HC, Iruela-Arispe ML, Annis DS, Mosher DF, Roberts DD. Circ Res. 2004;94:462–470. doi: 10.1161/01.RES.0000115555.05668.93. [DOI] [PubMed] [Google Scholar]
- 5.Krutzsch HC, Choe BJ, Sipes JM, Guo N, Roberts DD. J Biol Chem. 1999;274:24080–24086. doi: 10.1074/jbc.274.34.24080. [DOI] [PubMed] [Google Scholar]
- 6.Sargiannidou I, Qiu C, Tuszynski GP. Semin Thromb Hemost. 2004;30:127–136. doi: 10.1055/s-2004-822977. [DOI] [PubMed] [Google Scholar]
- 7.Elzie CA, Murphy-Ullrich JE. Int J Biochem Cell Biol. 2004;36:1090–1101. doi: 10.1016/j.biocel.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Winnemoller M, Schon P, Vischer P, Kresse H. Eur J Cell Biol. 1992;59:47–55. [PubMed] [Google Scholar]
- 9.Wang S, Herndon ME, Ranganathan S, Godyna S, Lawler J, Argraves WS, Liau G. J Cell Biochem. 2004;91:766–776. doi: 10.1002/jcb.10781. [DOI] [PubMed] [Google Scholar]
- 10.Greenaway J, Lawler J, Moorehead R, Bornstein P, Lamarre J, Petrik J. J Cell Physiol. 2007;210:807–818. doi: 10.1002/jcp.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawler J, Detmar M. Int J Biochem Cell Biol. 2004;36:1038–1045. doi: 10.1016/j.biocel.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari do Outeiro-Bernstein MA, Nunes SS, Andrade AC, Alves TR, Legrand C, Morandi V. Matrix Biol. 2002;21:311–324. doi: 10.1016/s0945-053x(02)00010-0. [DOI] [PubMed] [Google Scholar]
- 13.Taraboletti G, Morbidelli L, Donnini S, Parenti A, Granger HJ, Giavazzi R, Ziche M. FASEB J. 2000;14:1674–1676. doi: 10.1096/fj.99-0931fje. [DOI] [PubMed] [Google Scholar]
- 14.Staniszewska I, Zaveri S, Del Valle L, Oliva I, Rothman VL, Croul SE, Roberts DD, Mosher DF, Tuszynski GP, Marcinkiewicz C. Circ Res. 2007;100:1308–1316. doi: 10.1161/01.RES.0000266662.98355.66. [DOI] [PubMed] [Google Scholar]
- 15.Tan K, Duquette M, Liu JH, Zhang R, Joachimiak A, Wang JH, Lawler J. Structure. 2006;14:33–42. doi: 10.1016/j.str.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulatero C, Lyon M, Lawler J, Jayson G, Gallagher J. Proc Am Assoc Can Res. 2003;44:1430. [Google Scholar]
- 17.Yu H, Tyrrell D, Cashel J, Guo N, Vogel T, Sipes JM, Lam L, Fillit HM, Hartman J, Mendelovitz S, Panel A, Roberts DD. Arch Biochem Biophys. 2000;374:13–23. doi: 10.1006/abbi.1999.1597. [DOI] [PubMed] [Google Scholar]
- 18.Herndon ME, Stipp CS, Lander AD. Glycobiology. 1999;9:143–155. doi: 10.1093/glycob/9.2.143. [DOI] [PubMed] [Google Scholar]
- 19.Goger B, Halden Y, Rek A, Mosl R, Pye D, Gallagher J, Kungl AJ. Biochemistry. 2002;41:1640–1646. doi: 10.1021/bi011944j. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski ZMW, Minor W. In: Methods in Enzymology. Carter CW Jr, Sweet RM, editors. Academic Press; New York: 1997. pp. 307–326. [Google Scholar]
- 21.CCP4. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Morris GM, Goodsell DS, Huey R, Olson AJ. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 25.Bitomsky W, Wade RC. J Am Chem Soc. 1999;121:3004–3013. [Google Scholar]
- 26.Huey R, Morris GM, Olson AJ, Goodsell DS. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 27.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls A, Sharp KA, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 29.De Pascalis AR, Jelesarov I, Ackermann F, Koppenol WH, Hirasawa M, Knaff DB, Bosshard HR. Protein Sci. 1993;2:1126–1135. doi: 10.1002/pro.5560020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koppenol WH. Biophys J. 1980;29:493–507. doi: 10.1016/S0006-3495(80)85148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sergeev YV, Wingfield PT, Hejtmancik JF. Biochemistry. 2000;39:15799–15806. doi: 10.1021/bi001882h. [DOI] [PubMed] [Google Scholar]
- 32.Sinha D, Marcinkiewicz M, Lear JD, Walsh PN. Biochemistry. 2005;44:10416–10422. doi: 10.1021/bi050361x. [DOI] [PubMed] [Google Scholar]
- 33.Krieger E, Geretti E, Brandner B, Goger B, Wells TN, Kungl AJ. Proteins. 2004;54:768–775. doi: 10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- 34.Robinson CJ, Harmer NJ, Goodger SJ, Blundell TL, Gallagher JT. J Biol Chem. 2005;280:42274–42282. doi: 10.1074/jbc.M505720200. [DOI] [PubMed] [Google Scholar]
- 35.Bauer KA. Chest. 2003;124:364S–370S. doi: 10.1378/chest.124.6_suppl.364s. [DOI] [PubMed] [Google Scholar]
- 36.Lawler J, Ferro P, Duquette M. Biochemistry. 1992;31:1173–1180. doi: 10.1021/bi00119a029. [DOI] [PubMed] [Google Scholar]
- 37.Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, Harrenga A, Wells TN, Proudfoot AE. Structure. 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Capila I, Linhardt RJ. Angewandte Chemie. (Int) 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 39.Dementiev A, Petitou M, Herbert JM, Gettins PG. Nat Struct Mol Biol. 2004;11:863–867. doi: 10.1038/nsmb810. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Johnson DJ, Esmon CT, Huntington JA. Nat Struct Mol Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 42.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 43.San Antonio JD, Slover J, Lawler J, Karnovsky MJ, Lander AD. Biochemistry. 1993;32:4746–4755. doi: 10.1021/bi00069a008. [DOI] [PubMed] [Google Scholar]
- 44.DiGabriele AD, Lax I, Chen DI, Svahn CM, Jaye M, Schlessinger J, Hendrickson WA. Nature. 1998;393:812–817. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- 45.Voland C, Serre CM, Delmas P, Clezardin P. J Bone Miner Res. 2000;15:361–368. doi: 10.1359/jbmr.2000.15.2.361. [DOI] [PubMed] [Google Scholar]
- 46.Faham S, Linhardt RJ, Rees DC. Curr Opin Struct Biol. 1998;8:578–586. doi: 10.1016/s0959-440x(98)80147-4. [DOI] [PubMed] [Google Scholar]
- 47.Vives RR, Sadir R, Imberty A, Rencurosi A, Lortat-Jacob H. Biochemistry. 2002;41:14779–14789. doi: 10.1021/bi026459i. [DOI] [PubMed] [Google Scholar]
- 48.Sarai A, Kono H. Annu Rev Biophys Biomol Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 49.Goodsell DS, Olson AJ. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 50.Kraulis PJ. J Appl Crystallogr. 1991;24:946. [Google Scholar]
- 51.Emsley P, Cowtan K. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]


