Abstract
In the developing CNS, neurons and glia are sequentially produced from the ventricular neural progenitor cells. One fundamental question in developmental neurobiology is what signals or factors control the developmental switch from neurogenesis to gliogenesis. Here we report that microRNAs (miRNAs) play an essential role in this important developmental process. Inhibition of miRNA formation in Olig1Cre-mediated Dicer conditional knock-out mice disrupted both oligodendrogenesis and astrogliogenesis in the ventral neuroepithelial cells. By contrast, the early patterning and development of motor neurons were not affected in the mutant spinal cord tissue.
Introduction
During early neural development, neurons and glial cells are sequentially generated from neural progenitor cells in the ventricular zone throughout the entire CNS. It has been well documented that different domains of neural progenitor cells produce distinct subtypes of neurons and macroglial cells (Miller, 2002; Rowitch, 2004; Richardson et al., 2006). In the ventral spinal cord, the ventricular zone is subdivided into five progenitor domains, with each domain expressing a unique combination of transcription factors and producing a distinct neuronal subtype (Briscoe et al., 2000). Motor neurons are first generated from the Olig1/2+ pMN domain in a Shh-dependent mechanism (Mizuguchi et al., 2001; Novitch et al., 2001). Beginning at around E12.5, the Olig1/2+ ventral neural progenitor cells in the pMN domain cease producing motor neurons; instead, they start to produce oligodendrocyte progenitor cells (OPCs or OLPs) (Lu et al., 2000; Zhou et al., 2000; Takebayashi et al., 2002). Early OPCs delaminate from the ventricular zone and subsequently undergo nondirectional migration to populate all regions of the spinal cord. By contrast, astrocyte precursor cells originate from other domains of neuroepithelial cells and reach their final destinations through radial migration (Lu et al., 2002; Zhou and Anderson, 2002; Hochstim et al., 2008).
Although the spatiotemporal events of neurogenesis and gliogenesis in the developing CNS have been well described, the molecular mechanisms underlying the binary switch from neurogenesis to gliogenesis have been under intensive investigation. Recent studies have implicated microRNAs (miRNAs) in the control of the timing of many developmental programs in both animals and plants (Banerjee and Slack, 2002; De Pietri Tonelli et al., 2008), raising the possibility that miRNAs may also play an important role in the timely generation of neurons and glia from the same pool of neural progenitor cells. miRNAs are a class of small noncoding RNA molecules (∼22 base pairs) that are derived from longer RNA precursors through cleavage by the ubiquitously expressed Dicer RNase (Carthew and Sontheimer, 2009). They function as the major regulators of posttranslational gene expression (Ouellet et al., 2006) by binding to the 3′-untranslated region of specific mRNA targets, directing their degradation and/or repressing their translation (He and Hannon, 2004). In the present study, we provided the first line of genetic evidence that miRNAs are essential for the initiation of gliogenesis process in the developing spinal cord. Conditional ablation of Dicer gene in the Olig1+ ventral spinal neuroepithelium inhibited both oligodendrogenesis and astrogliogenesis, but had little effect on early neural patterning and neurogenesis.
Materials and Methods
Mouse lines and genotyping.
Mice were housed under standard laboratory conditions at the animal facility of the University of Louisville. All experimental procedures conformed to National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee at the University of Louisville.
Olig1Cre/+ knock-in mice (Lu et al., 2002) were crossed with R26R mice (Soriano, 1999) to obtain Olig1Cre/+/R26R for LacZ histochemical analysis. Dicerflox/flox mice (Murchison et al., 2005) were mated to Olig1Cre/+ to obtain the Olig1Cre/+Dicerflox/+ double heterozygous mice, and conditional Dicer mutants was generated by interbreeding of double heterozygotes. For mouse genotyping, genomic DNA was extracted from embryonic tissues or mouse tails and subsequently used for genotyping by Southern analysis or by PCR. Genotyping protocols for Olig1Cre and Dicerflox mouse lines were described previously (Lu et al., 2002; Murchison et al., 2005).
In situ RNA hybridization and immunofluorescent staining.
Spinal cord tissues at the thoracic level were isolated from E11.5 to E18.5 mouse embryos and then fixed in 4% paraformaldehyde at 4°C overnight. Following fixation, tissues were transferred to 20% sucrose in PBS overnight, embedded in OCT media and then sectioned (16 μm thickness) on a cryostat. Adjacent sections from the control and mutant embryos were subjected to in situ hybridization (ISH) or immunofluorescent staining. Regular ISH was performed as described by Schaeren-Wiemers and Gerfin-Moser (1993) with minor modifications. 5′-Digoxigenin-labeled, locked nucleic acid (LNA)-modified anti-miR-9 (5′-TCATACAGCTAGATAACCAAAGA-3′) oligonucleotide probe was purchased from Exqion Inc. and used for in situ hybridization as described previously (Kloosterman et al., 2006). Double immunofluorescent procedures were described previously (Qi et al., 2001). The dilution ratio of antibodies is as follows: anti-Olig2 (1:6000), anti-MAG (Millipore Bioscience Research Reagents, 1:500), anti-GFAP (Millipore Bioscience Research Reagents, 1:50), anti-Nkx2.2 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA; 1:50) (Xu et al., 2000), anti-Sox10 (1:3000) (Stolt et al., 2002), anti-PDGFRα (Cell Signaling Technology, 1:400) and anti-S100β (Millipore Bioscience Research Reagents, 1:1000).
Results
Olig1Cre can induce reporter gene expression and Dicer deletion in the ventral spinal neuroepithelium
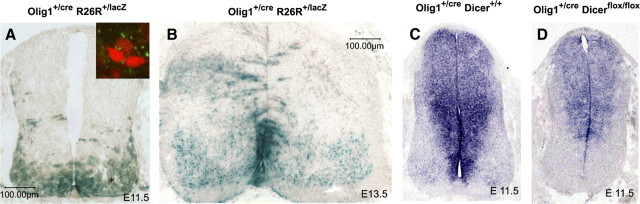
To determine the possible role of miRNAs in gliogenesis, we set out to disrupt miRNA formation in the ventral spinal cord using the Olig1Cre knock-in mouse line (Lu et al., 2002). Previous studies showed that Olig1 and Olig2 are initially expressed in a broad ventral region but later confined to the pMN domain (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2000). The initial broad expression of Olig1in the ventral ventricular zone was confirmed by the LacZ histochemical staining in Olig1cre/+/R26R (Rosa26-lox-lacZ) double transgenic reporter embryos (Soriano, 1999). At E11.5 and E13.5 stages, LacZ staining was predominantly detected in the ventral neuroepithelial cells including the pMN and p3 domains, although few LacZ+ cells could be found in E13.5 dorsal neuroepithelium as well (Fig. 1A,B). In addition, LacZ staining was also observed in the motor neurons derived from the pMN domain (Fig. 1A,B).
Figure 1.
Activation of β-galactosidase and Dicer activity in the ventral spinal cord by Olig1Cre driver mouse line. A, B, Spinal cord sections from E11.5 and E13.5 Olig1Cre/R26R:LacZ embryos were subjected to LacZ histochemical staining. Asterisks indicate the positions of motor neurons in the ventral spinal cord. The inset in A represents the LacZ+/HB9+ motor neurons in the ventral horn as detected by double immunofluorescence. C, D, Spinal cord sections from E11.5 Olig1Cre/+/Dicer-1flox/flox embryos were examined for miR-9 expression by in situ hybridization.
We next generated the Olig1Cre/Dicerflox/flox conditional knock-out animals by sequential cross-mating. For unknown reasons, the Olig1Cre/Dicerflox/flox mutants died immediately after birth. To confirm the selective elimination of Dicer function in the ventral neuroepithelium in the conditional mutants, we compared the expression of microRNA-9 (miR-9) in the ventral spinal cord between the control and Dicer mutants. miR-9 was originally identified to be expressed in oligodendrocyte progenitor cells (Lau et al., 2008). Our recent study revealed that miR-9 was initially expressed in the ventricular zone along the entire dorsal-ventral axis (Fig. 1C). As expected, expression of miR-9 expression was nearly completely absent in the ventral ventricular cells including the pMN and p3 domains in the mutants, whereas dorsal expression of miR-9 was largely spared (Fig. 1D). Therefore, Olig1Cre can effectively ablate Dicer function in the ventral neuroepithelium.
Inhibition of miRNA formation in the ventral spinal cord did not affect neural patterning and motor neuron development
To assess the role of miRNAs in ventral neural patterning and neurogenesis, we first examined the expression of three progenitor identity genes Pax6, Olig2 and Nkx2.2 in the ventral spinal cord of Dicer conditional mutants. During neurogenesis stages, Olig2 and Nkx2.2 specifically mark the pMN domain and p3 domain, respectively; whereas Pax6 is expressed in domains dorsal to Nkx2.2 (Briscoe et al., 1999, 2000). Immunostaining results revealed a nearly identical pattern of Olig2, Nkx2.2 and Pax6 expression in E11.5 ventral neuroepithelium between the control and Dicer mutants (Fig. 2A–D). Consistently, a similar number of HB9+ and Islet-1+ motor neurons were generated in the ventral horn in both genotypes (Fig. 2E–I). Similarly, the expression of several ventral interneuron markers such as Chox10, Evx1 and Pax2 was not significantly altered in the conditional mutants (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Together, these results demonstrated that miRNAs did not play a significant role in the initial neural patterning and subsequent neurogenesis in the spinal cord.
Figure 2.
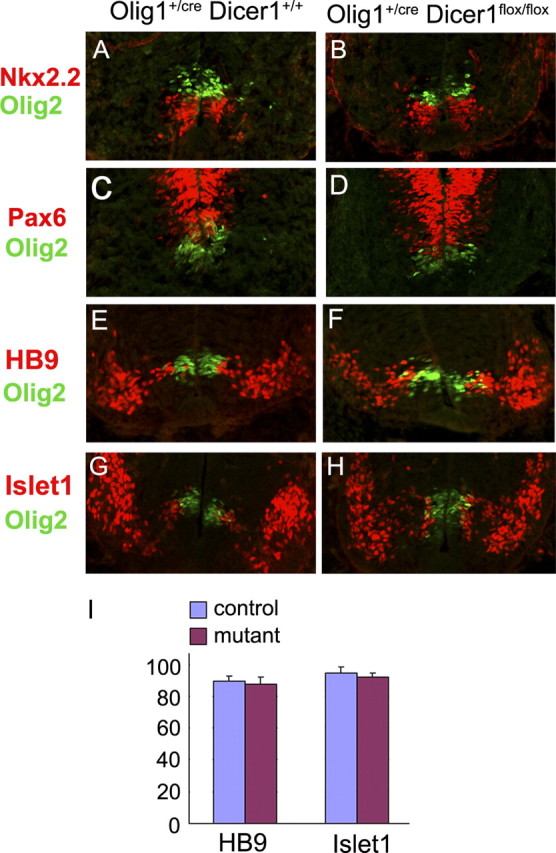
Normal neural patterning and motor neuron development in Dicer mutant spinal cord. Spinal cord sections from E11.5 Olig1Cre/+/Dicer+/+ and Olig1Cre/+/Dicerflox/flox embryos were immunostained simultaneously with anti-Olig2 and anti-Pax6 (A, B), anti-Nkx2.2 (C, D), anti-HB9 (E, F) or anti-Isl-1(G, H). I, Quantification of HB9+ and Islet1+ motor neurons in the control and Dicer mutant tissues (n = 3).
miRNAs are essential for oligodendrogenesis in the spinal cord
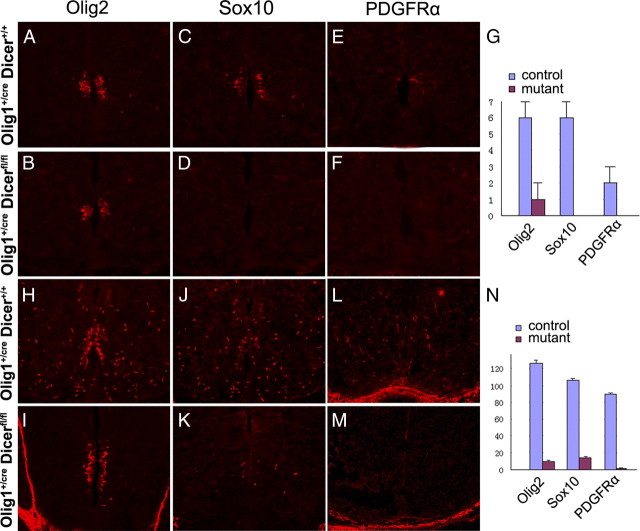
At around E12.5, neuroepithelial cells in the pMN domain cease producing motor neurons and start to give rise to migratory OPC cells. In the control embryos, Olig2+ cells started to migrate away from the pMN domain into the surrounding region (Fig. 3A). Expression of two other OPC markers Sox10 (Stolt et al., 2002) and PDGFRα, was also detected next to the pMN domain (Fig. 3B,C). Surprisingly, in Olig1Cre/+/Dicerflox/flox mutants, expression of Olig2 was only observed in the ventricular zone, and expression of Sox10 and PDGFRα was not detected at all (Fig. 3D–F,M). At E14.5, a large number of Olig2+, Sox10+ and PDGFRα+ OPCs were dispersed throughout the entire spinal cord in the control embryos (Fig. 3G–I). By contrast, in the mutant embryos, expression of Olig2 was predominantly confined to the ventricular zone and ventrally expanded, and few Olig2+ cells detached from the ventral ventricular zone (Fig. 3J). The blockade of OPC generation in Dicer mutants was further confirmed by the drastic reduction of Sox10+ cells and the complete absence of PDGFRα+ cells in the spinal cord parenchyma (Fig. 3K,L,N). Together, these results indicated that oligodendrogenesis in the ventral spinal cord was suppressed by the absence of miRNA formation.
Figure 3.
Inhibition of oligodendrogenesis in Olig1cre/Dicer flox/flox mutant spinal cord. Transverse spinal cord sections from E12.5 (A–F) and E14.5 (G–L) Olig1Cre/+/Dicer+/+ and Olig1Cre/+/Dicer flox/flox embryos were immunostained with anti-Olig2 (A, D, G, J), anti-Sox10 (B, E, H, K) and anti-PDGFRα (C, F, I, L). M, N, Statistical analysis of Olig2+, PDGFRα+, and Sox10+ OPCs in E12.5 (M) and E14.5 (N) control and Dicer mutant tissues (n = 3).
miRNAs are required for astrogliogenesis in the ventral spinal cord
To address the role of miRNA function in astrocyte development, we examined the expression of the well defined mature astrocyte marker GFAP in the Dicer mutant spinal cord. In E18.5 control pups, GFAP immunofluorescent staining was observed in the entire white matter region of the spinal cord. Strikingly, GFAP immunostaining in Olig1Cre/+/Dicerflox/flox animals was completely absent in a triangular region immediately flanking the floor plate (Fig. 4A,B). Similarly, expression of another two astrocyte markers ID3 and S100β (Langley et al., 1984) was also absent in the same region (Fig. 4C–F). These results indicated that the conditional ablation of Dicer disrupted the development of a subset of astrocytes derived from the ventral Olig1+ neuroepithelium, probably from the ventralmost p3 progenitor domain. In support of this idea, the expression of Slit 1, a specific marker for the ventralmost white matter astrocytes derived from the p3 domain (Hochstim et al., 2008), was completely inhibited in the Dicer mutants (supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
Figure 4.
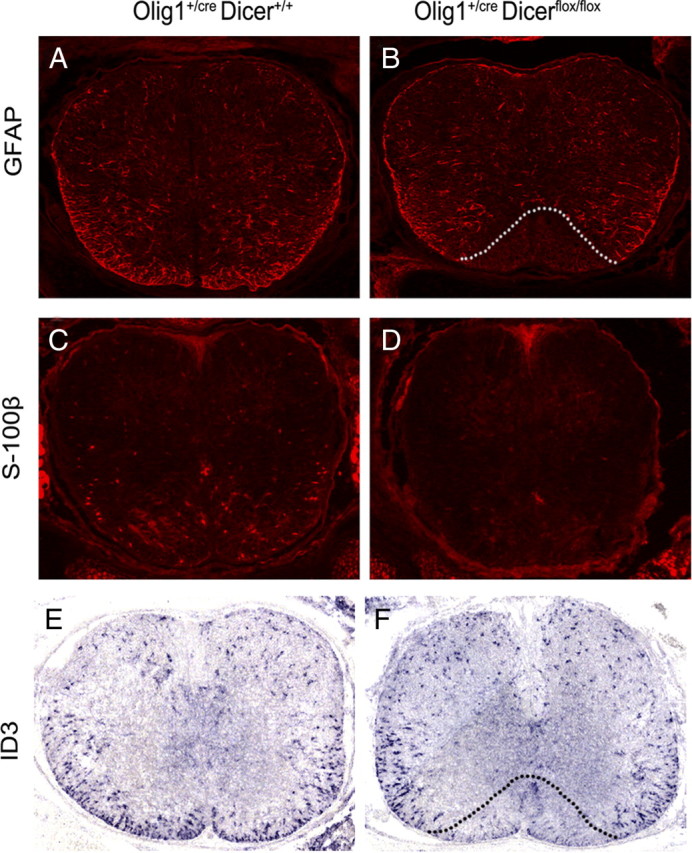
Abnormal development of astrocytes in Olig1cre/Dicer flox/flox mutant spinal cord. Transverse spinal cord sections at the thoracic level from E18.5 wild-type and mutant embryos were immunostained with anti-GFAP (A, B) and anti-S100β (C, D) antibodies, or subjected to ID3 in situ RNA hybridization. (E, F). The region devoid of GFAP or ID3 signal in the mutants is outlined by dashed lines.
Discussion
In the developing CNS, different progenitor domains as defined by their unique combinatorial expression of progenitor transcription factors give rise to distinct subtypes of neurons and glia at different time windows. The molecular events underlying the sequential generation of neurons and glia remain one of the most important questions in developmental neurobiology. In the present study, we provided the genetic evidence that miRNAs play an essential role in the transition from neurogenesis to gliogenesis. In the Olig1Cre/ Dicerflox/flox conditional knock-out mice, miRNA biosynthesis is largely inhibited in the ventral neuroepithelium due to the Cre-mediated ablation of Dicer gene, as confirmed by the absence of miR-9 expression in the ventral ventricular zone of the Dicer conditional mutants (Fig. 1). In association with the absence of miRNA biogenesis, the production of oligodendrocyte progenitor cells was nearly completely suppressed. In Dicer mutants, very few Olig2+ neural progenitor cells were able to delaminate from the ventral ventricular zone during gliogenesis stages to form migratory OPC cells. As a result, expression of oligodendrocyte progenitor markers Sox10 and PDGFRα was almost completely absent in the conditional mutants (Fig. 3). Interestingly, at E14.5, Olig2 expression in the ventricular zone was ventrally expanded in the mutant embryos (Fig. 3H,K). The increased number of Olig2+ neural progenitor cells was probably due to the lack of OPC emigration from this zone, as we failed to detect significant differences in the proliferation or cell death of Olig2+ neural progenitor cells between the control and conditional mutants (our unpublished observations).
Similarly, development of astrocytes was also disrupted in the ventral spinal cord of the Dicer mutants, as evidenced by the complete lack of expression of two astrocyte markers, GFAP and S100β, in the ventral region surrounding the floor plate (Fig. 4A–D). In addition, we found that ID3 transcription factor has specific expression in spinal cord astrocyte cells, and its expression was also absent in the ventral triangular region as observed in GFAP expression (Fig. 4E,F). In the conditional mutants, Cre activity was observed in both the pMN domain and the p3 domain (Fig. 1A,B). Since pMN domain only produces only oligodendrocytes (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2002), the most likely explanation for the lack of expression of astrocyte markers in the ventralmost white matter area is that the ventral neuroepithelial cells from the p3 domain failed to produce astrocyte cells in the Dicer conditional mutants. Consistent with this idea, expression of VA3 astrocyte marker Slit1 (Hochstim et al., 2008) was inhibited in the mutants (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Thus, in the absence of miRNA formation, the pMN domains failed to generate OPC cells, and the p3 domain failed to produce astrocytes.
Two most recent papers (Dugas et al., 2010; Zhao et al., 2010) reported that miRNAs are essential for oligodendrocyte maturation, but not for OPC generation and astrocyte development in the brain. The apparent differences between our data and their findings could be attributed to the regional differences in Olig-Cre gene expression and the different developmental stages examined. Since Olig1-Cre and Olig2-Cre are not expressed in the ventricular zone of the cerebral cortex, it may not be surprising to see normal OPC generation and astrogliogenesis in this region, given that OPC cells and astrocytes can arise from the local subventricular zone of the embryonic cortex (Kessaris et al., 2006). In addition, any defect in early OPC generation in the cortex could be compensated at later postnatal stages by increased cell proliferation (Zhao et al., 2010).
Intriguingly, miRNA function did not appear to act in early neural patterning and the development of motor neurons, as suggested by the more or less normal expression of three ventral patterning genes (Pax6, Olig2 and Nkx2.2) and motor neuron-specific markers (HB9 and Islet1) in the conditional mutant spinal cord (Fig. 2). Despite the fact that gliogenesis is greatly suppressed in Dicer mutants, there was no prolonged or ectopic motor neuron formation, suggesting that neurogenesis and gliogenesis can be mechanistically separated. In addition, the lack of ectopic expression of Olig2 in postmitotic motor neurons (Fig. 2B) also indicated that miRNA-mediated silencing of gene expression did not account for the rapid downregulation of Olig2 mRNA or protein expression in these neurons. It is likely that gene regulation at the transcriptional level rather than the posttranscriptional level is responsible for the suppression of many progenitor genes in postmitotic neurons.
Although the present study clearly demonstrated that miRNAs are required for gliogenesis in the spinal cord, it has also raised several outstanding questions regarding the regulation of gliogenesis by miRNAs. First, which miRNA species are involved in this gliogenesis process? It would be interesting and important to know whether the same miRNA molecules control both oligodendrogenesis and astrogliogenesis. Second, what are the miRNA downstream target genes that are involved in gliogenesis? Considering that miRNAs are negative regulators of gene expression, it is plausible that miRNAs function to repress the expression of some inhibitory factors that normally suppress gliogenesis at early neurogenesis stages. The derepression mechanism appears to be a common phenomenon for transcriptional regulation of cell fate specification and differentiation (Muhr et al., 2001). Third, given the conserved mechanism for gliogenesis throughout the CNS, are miRNAs also essential for gliogenesis in the rostral brain regions. Answers to these questions in the future will provide important new insights into the mechanistic control of glial development and regeneration in the CNS.
Footnotes
This work is supported by the National Institutes of Health (Grant NS37717), the National Multiple Sclerosis Society (Grant RG 3275), and the Kentucky Spinal Cord and Head Injury Trust (Grant #8-12). We are very grateful to Drs. Charles Stiles, David Rowitch, and Richard Lu for generously providing the Olig1Cre mutant mouse lines. We also thank Drs. Charles Stiles and Michael Wegner for the anti-Olig2 and anti-Sox10 antibodies.
References
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx-2.2 and specification of neuronal identity by graded sonic hedgehog signaling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNA and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers J, Haffner C, Murchison E, Hannon G, Huttner W. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. J Neurosci. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Langley OK, Ghandour MS, Gombos G. Immunohistochemistry of cell markers in the central nervous system. In: Lajtha A, editor. Handbook of neurochemistry. New York: Plenum; 1984. [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic Hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–467. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labeled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Xu X, Cai J, Fu H, Wu R, Qi Y, Modderman G, Liu R, Qiu M. Selective expression of Nkx2.2 transcription factor in the migratory chicken oligodendrocyte progenitor cells and implications for the embryonic origin of oligodendrocytes. Mol Cell Neurosci. 2000;16:740–753. doi: 10.1006/mcne.2000.0916. [DOI] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]