Abstract
Of the superfamily Muroidea (31 genera, 1578 species), the Sigmodontinae (74 genera, 377 species) is the second largest subfamily in number of species and represents a significant radiation of rodent biodiversity. Only 2 of the 74 genera are found in both North and South America (Sigmodon and Oryzomys) and the remainder are exclusively from South America. In recent molecular studies, the genus Sigmodon (Cricetidae, Sigmodontinae) has been considered sister to many other South American Sigmodontines [Steppan et al., 2004]. We examine the chromosomal evolution of 9 species of Sigmodon utilizing chromosomal paints isolated from S. hispidus, proposed to be similar to the ancestral karyotype [Elder, 1980]. Utilizing a phylogenetic hypothesis of a molecular phylogeny of Sigmodon [Henson and Bradley, 2009], we mapped shared chromosomal rearrangements of taxa on a molecular tree to estimate the evolutionary position of each rearrangement. For several species (S. hirsutus, S. leucotis, S. ochrognathus, S. peruanus, and S. toltecus), the karyotype accumulated few or no changes, but in three species (S. arizonae, S. fulviventer, and S. mascotensis) numerous karyotype rearrangements were observed. These rearrangements involved heterochromatic additions, centric fusions, tandem fusions, pericentric inversions, as well as the addition of interstitial DNA not identified by chromosome paints or C-banding. The hypothesis that the ancestral karyotype for this complex had a diploid number of 52, a fundamental number of 52, and a G-band pattern of which most, if not all are similar to that present in modern day S. hispidus fails to be rejected. This hypothesis remains viable as an explanation of chromosomal evolution in Sigmodontine rodents.
Key Words: Ancestral karyotype, Comparative cytogenetics, DAPI banding, Karyotype conservation, Muroid rodents, Phylogeny, Sigmodontinae, Tandem fusions
Introduction
The definition of the primitive karyotype is critical to establishing direction, nature, and phylogenetic position of chromosomal change, such definition has been established through chromosome painting for many New World mammals including canids [Graphodatsky et al., 2001; Nash et al., 2001], bats [Volleth et al., 1999; Pieczarka et al., 2005], bears [Nash et al., 1998], anteaters and sloths [Yang et al., 2006], marsupials [Rens et al., 2003], mustelids [Hameister et al., 1997; Nie et al., 2002], primates [Richard et al., 1996; Stanyon et al., 2001; Garcia et al., 2002], and New World rodents [Dawson et al., 1999; Stanyon et al., 2003; Li et al., 2004; Sitnikova et al., 2007; Hass et al., 2008]. Yet, New World muroid rodents have been poorly studied as Zoo-Fish is difficult to conduct on the extensively rearranged karyotypes and the few that have been conducted utilize paints isolated from closely related rodent species. As a paucity of comparative chromosome painting exists in New World muroid rodents (family Cricetidae), studies of their rearranged and conservative karyotypes would be useful in eutherian comparative cytogenetics and reconstruction of ancestral karyotypes of other groups. This study is designed to develop a hypothesis of the primitive chromosomal condition for one of the most specious muroid taxa, the Sigmodontinae.
Within muroid rodents, the second largest subfamily is the Sigmodontinae (New World rats and mice) composed of 377 species [Musser and Carleton, 2005]. This subfamily is a tremendously successful radiation of rodents that has been documented to be monophyletic, and has many examples of chromosomal evolution. Of the 74 genera in this subfamily, the morphological, molecular, and cytogenetic variation of the genus Sigmodon is well studied. Phylogenetically, the genus Sigmodon is the basal clade diverging from the common ancestor of this complex [Steppan et al., 2004]. Determining the characteristics of a primitive karyotype for Sigmodon will provide an hypothesis of the primitive karyotype condition for this subfamily and provide a starting point for other studies of chromosomal evolution within monophyletic radiations of Sigmodontine rodents.
Cotton rats of the genus Sigmodon [Rodentia: Cricetidae, Sigmodontinae – Musser and Carleton, 2005] were among the earliest rodent genera described in the Western Hemisphere [Say and Ord, 1825]. Easily recognizable by the ‘S-shaped’ cusp pattern on molar teeth, members of the genus are distributed throughout the southwestern to southeastern United States, Mexico, Central America, and northern South America, typically in grassy habitats [Bailey, 1902; Nelson and Goldman, 1933; Baker, 1969; Anderson, 1972; Voss, 1992; Carleton et al., 1999; Peppers et al., 2002; Carroll et al., 2005]. Throughout their distribution, cotton rats are often cyclical in population densities as a response to climatic change and food availability. Although one of the more commonly encountered rodents, morphological similarities among species hindered taxonomic identification of several species and establishment of phylogenetic relationships among taxa lagged until the advent of chromosomal and molecular sequence data [Zimmerman, 1970; Peppers and Bradley, 2000; Peppers et al., 2002; Carroll et al., 2005; Henson and Bradley, 2009].
In a recent review, 13 species of cotton rats were recognized by Musser and Carleton [2005]: S. alleni, S. alstoni, S. arizonae, S. fulviventer, S. hirsutus, S. hispidus, S. inopinatus, S. leucotis, S. mascotensis, S. ochrognathus, S. peruanus, S. planifrons, and S. toltecus. They also recognized S. zanjonensis which they concluded may or may not be specifically distinct [Carleton et al., 1999; Musser and Carleton, 2005]. Of these 13 species, karyotypes for 9 (S. alleni, S. arizonae, S. fulviventer, S. hirsutus, S. hispidus, S. leucotis, S. mascotensis, S. ochragnathus, and S. toltecus) [Hsu and Benirschke, 1968; Kiblisky, 1969; Lee and Zimmerman, 1969; Zimmerman, 1970; Carroll et al., 2005] have been described with non differentially stained chromosomes, G-bands, and C-bands [Zimmerman, 1970; Elder, 1980; Elder and Lee, 1985]. Diploid numbers ranged from 22 to 82 and fundamental number (total number of autosomal arms) ranged from 34 to 82; G-band data indicate that S. hispidus, S. leucotis, S. alleni, S. ochragnathus, S. arizonae, and S. mascotensis form a species group to the exclusion of S. fulviventer which Elder and Lee [1985] placed in its own species group. Incomplete chromosomal data exists for S. alstoni [Reig, 1986; Voss, 1992; only diploid number described] and no chromosomal data are available for S. inopinatus, S. peruanus, and S. planifrons.
The karyotype of Sigmodon hispidus has been recognized as very similar to the ancestral karyotype that gave rise to the karyotype diversity within Sigmodon. Elder [1980] using detectable G-band homologies proposed the G-band karyotype of S. hispidus as primitive, and using mostly fusions, reconstructed the karyotypes of S. arizonae (2n = 22/24)and S. mascotensis (2n = 28). The type of rearrangements detected was mostly tandem fusions in S. arizonae and S. mascotensis, though many more rearrangements including pericentric and paracentric inversions, and centric fusions had occurred in S. arizonae than S. masotensis. In addition, Elder [1980] concluded that S. arizonae or S. mascotensis could not be directly derived from the other species, as some of their chromosomal rearrangements involved different combinations of ancestral S. hispidus chromosomes.
G-bands of rodent species with a diploid number of 48 to 52 from North and South America, Europe, Asia, and Australia were compared to S. hispidus for shared G-band patterns [Baker et al., 1983; Koop et. al., 1984]. The species with the karyotype of 2n = 48 to 52 shared similar G-band patterns in the largest 14 autosomes and the X. Many of these G-band patterns were shared with S. hispidus and were proposed as primitive for the common ancestor of this group.
The current study is designed to test the hypothesis that the karyotype of S. hispidus (2n = 52, FN = 52) is similar to the ancestral karyotype for Sigmodon. Evidence to support this hypothesis is that the karyotype of S. hispidus has a diploid number that is considered primitive for cricetid and murid rodents, has G-band patterns similar and often indistinguishable from those in other rodent species where the primitive karyotype for each respective genus has been proposed [Baker et al., 1983; Koop et al., 1984], and has been regarded as similar to the ancestral karyotype for Sigmodon through G-banding [Elder, 1980]. For this paper, we isolated chromosome paints from S. hispidus, used fluorescence in situ hybridization (FISH) for 9 species of Sigmodon, and mapped chromosomal rearrangements onto a statistically supported phylogenetic tree [Henson and Bradley, 2009] to determine the evolutionary position and direction of change for karyotypes of Sigmodon species.
Methods and Materials
Animals
Cell suspensions archived at Texas Tech University (TTU) were from the following specimens (table 1), and sequence data obtained from the cytochrome b, IRBP, and beta fibrinogen genes are available for some specimens (table 1).
Table 1.
The TK/TTU/CRD numbers, and locality information for the animals studied in this paper. Also included are the GenBank Accession numbers and accompanying citation of the animals that have been sequenced (1: Peppers et al., 2002; 2: Carroll and Bradley, 2005; 3: Henson and Bradley, 2009). TK numbers represent the data set and tissues collected from the animal. TTU (Genetic Resource Collection, Texas Tech University) and CRD (Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional Unidad Durango) numbers represent the skin and/or skull of the museum voucher.
| Species | TK # | TTU/CRD # | Sex | Locality | GenBank Accession # | Citation |
|---|---|---|---|---|---|---|
| S. arizonae | 70927 | CRD 2181 | M | Durango, Mexico | ||
| S. arizonae | 70928 | TTU 81699 | F | Durango, Mexico | AF155423/AY459383/EU635699 | 1, 2, 3 |
| S. fulviventer | 72394 | CRD 2153 | M | Durango, Mexico | ||
| S. hirsutus | 136882 | TTU 103971 | M | Colon, Honduras | ||
| S. hispidus | 93765 | M | Texas, USA | FJ232944 | ||
| S. hispidus | 93767 | M | Texas, USA | FJ232945 | ||
| S. hispidus | 121529 | TTU 108169 | M | Texas, USA | ||
| S. hispidus | 137315 | TTU 108155 | M | Tamaulipas, Mexico | EU073177/EU652895/EU635708 | 3 |
| S. leucotis | 72389 | TTU 81693 | F | Durango, Mexico | EU652909/EU652897/EU635712 | 3 |
| S. mascotensis | 93075 | TTU 82793 | F | Jalisco, Mexico | AF425215/AY459385/EU635715 | 2, 3 |
| S. mascotensis | 150633 | TTU 104775 | M | Michoacan, Mexico | ||
| S. ochrognathus | 48608 | TTU 75466 | F | Durango, Mexico | ||
| S. ochrognathus | 83130 | TTU 78696 | F | Texas, USA | ||
| S. peruanus | 134776 | TTU 103494 | F | Guayas, Ecuador | EU073179/EU652901/EU635719 | 3 |
| S. toltecus | 136365 | TTU 104421 | M | Cortes, Honduras | ||
| S. toltecus | 136997 | TTU 104086 | M | Colon, Honduras |
Chromosome Banding
Karyotypes were prepared according to Baker et al. [2003]. To establish a reference set of fluorescent G-banded chromosomes of S. hispidus, some chromosome preparations were only DAPI banded; whereas other chromosome preparations were DAPI stained following fluorescence in situ hybridization. To produce the reference set, blazed-dried chromosome preparations were treated according to the fluorescent G-band methods developed by M.J. Hamilton as described in Bowers et al. [1998] under the subheading ‘in situ hybridization and chromosome banding’. The chromosome preparations were treated with RNase, digested with pepsin, and dehydrated with a series of ethanol washes. This was not followed by any further chromosomal denaturation. After ethanol washes, slides were washed in 3 changes of 2× SSC and incubated in McIlvaine's buffer (0.2 M Na2HPO4, 0.1 M citric acid, pH 7.0) with 0.01 M MgCl2 for 15 min at room temperature. After draining the excess buffer from each slide we applied 100 μl of chromomycin A3 (100 μg/ml), covered slides with Parafilm® and incubated for 15 min at room temperature. The slides were then washed in 2 changes of McIlvaine's buffer for 2 min per wash, rinsed with DI H2O and allowed to dry. Approximately 35 μl of Vectashield® (Vector, Co) mounting medium with DAPI was added to each slide, and covered with a 24 × 60 coverslip to aid the spread of DAPI throughout the slide.
C-banding followed the procedures of Schmid (1978). The C-banded karyotypes of S. leucotis, S. hirsutus, S. peruanus, and S. toltecus were compared to the distribution of the S. hispidus paint in situ hybridization signal in each karyotype.
Cell Culture and Fluorescence in situ Hybridization
Fibroblast cultures were established from ear cartilage biopsies of a male S. hispidus (TTU 108168, TK 121545) following standard tissue culture techniques.
Chromosomes of a male S. hispidus (TTU 108168, TK 121545) were flow sorted, DOP-PCR amplified, and biotin labeled [Yang et al., 1995] into 22 individual probes at the Resource Center for Comparative Genomics, University of Cambridge. Each probe was individually in situ hybridized to S. hispidus following a modified version of Yang et al. [2003] with probes being denatured at 70°C for 5 min and slides denatured in 50% formamide, 2× SSC at 70°C for 2 min. Slides were counterstained with DAPI. Then the S. hispidus probes were individually hybridized in situ to S. arizonae, S. fulviventer, S. hirsutus, S. leucotis, S. mascotensis, S. ochrognathus, S. peruanus, and S. toltecus.
Analysis
Each DAPI image and painted chromosome image of the same metaphase spread as well as the reference DAPI images from different metaphase spreads were photographed with an Applied Imaging® camera. These images were captured using the Genus™ System 3.7 from Applied Imaging Systems (San Jose, CA).
DAPI images were inverted with Image Pro Plus 4.5.1 22 (Media Cybernetics, Inc, MD) so that areas staining brightly with DAPI became the dark bands of classical G-bands. The G-bands were then enhanced with the HiGauss filter of Image Pro Plus. Banded chromosomes were arranged into a karyogram using the Genus™ System 3.7 software and numerically classified according to previous literature [Elder, 1980; Elder and Pathak, 1980; Elder and Lee, 1985]. Chromosomes that were stained with DAPI after FISH were compared to the reference set of fluorescent G-banded chromosomes. The DAPI stained chromosomes (which were identified from the same metaphase spread as the painted chromosomes) were used to identify each painted chromosome by comparison with the reference set of fluorescent G-banded chromosomes. Rearrangements were then mapped upon a molecular tree [Henson and Bradley, 2009].
Results
The S. hispidus paints (H1–H25 and HX/Y), the S. hispidus chromosomes that were contained in each paint [following the numerical order of Elder, 1980], and the chromosomes of S. arizonae, S. fulviventer, S. hirsutus, S. leucotis, S. mascotensis, S. ochrognathus, S. peruanus, and S. toltecus that were hybridized by each S. hispidus paint are given in table 2. Further explanation of the homology of S. hispidus paints to the other 8 species of Sigmodon is provided below.
Table 2.
The S. hispidus paints (H1-HX/Y) and the corresponding chromosome of S. arizonae, S. fulviventer, S. hirsutus, S. leucotis, S. mascotensis, S. ochrognathus, S. peruanus, and S. toltecus that hybridized to each paint. Question marks indicate unknowns.
| Paint | S. hispidus | S. ochrognathus | S. leucotis | S. hirsutus | S. peruanus | S. toltecus | S. fulviventer | S. mascotensis | S. arizonae |
|---|---|---|---|---|---|---|---|---|---|
| H1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 1 |
| H2 | 2 | 1 | 2 | 2 | 1 | 1 | 4 | 1 | 4 |
| H3 | 3 | 1 | 3 | 3 | 1 | 1 | 6 | 3 | 9 |
| H4 | 4 | ? | ? | 4 | ? | 4 | ? | 2 | 3 |
| H5 | 5 | 5 | 5 | 5 | 5 | 5 | 8 | 7 | 1 |
| H6 | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 5 | 3 |
| H7 | 7 | 7 | 7 | 7 | 7 | 7 | 10 | 6 | 6 |
| H8/9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 8, 9 | 3, 9 | 1, 9 | 5, 9 |
| H10 | 10 | 10 | 10 | 10 | 10 | 10 | 11 | 8 | 2 |
| H7/11 | 7, 11 | 7, 11 | 7, 11 | 7, 11 | 7, 11 | 7, 11 | 10, 13 | 6, 12 | 6, 2 |
| H12/14 | 12, 14 | 12, 14 | 12, 14 | 12, 14 | 12, 14 | 12, 14 | 10, 2 | 5, 3 | 7, 3 |
| H13/22 | 13, 22 | 13, 22 | 13, 22 | 13, 22 | 13, 22 | 13, 22 | 12, 8 | 8, 9 | 3, 8 |
| H15 | 15 | 15 | 15 | 15 | 15 | 15 | 1 | 2 | 4 |
| H16/20/22 | 16, 20, 22 | 16, 20, 22 | 16, 20, 22 | 16, 20, 22 | 16, 20, 22 | 16, 20, 22 | 7, 12, 8 | 10, 12, 9 | 10, 8 |
| H17 | 17 | 17 | 17 | 17 | 17 | 17 | 5 | 10 | 8 |
| H18 | 18 | 18 | 18 | 18 | 18 | 18 | 1 | 11 | 5 |
| H19/20/22 | 19, 20, 22 | 19, 20, 22 | 19, 20, 22 | 19, 20, 22 | ? | 19, 20, 22 | 1, 12, 8 | 11, 12, 9 | 8 |
| H21 | 21 | 21 | 21 | 21 | 21 | 21 | 13 | 7 | 3 |
| H23 | 23 | 23 | 23 | 23 | 23 | 23 | 1 | 9 | 4 |
| H24 | 24 | 24 | 24 | 24 | 24 | 24 | 5 | 13 | 8 |
| H25 | 25 | 25 | 24 | 25 | 25 | 25 | 14 | 2 | 1 |
| HX/Y | X, Y | X | X | X, Y | X | X, Y | X | X | X, 8 |
The most conserved chromosomal element among the species of Sigmodon was the X chromosome. The HX/Y paint consistently hybridized to a chromosome with a fluorescent G-banding pattern specific for the X chromosome in all 9 Sigmodon species (table 2). One rearrangement of the X chromosome was found in S. arizonae with the HX/Y chromosome paint hybridizing to both the X and to chromosome 8. Further, a small portion of the Y, possibly the pseudoautosomal region, hybridized with the HX/Y paint in S. hispidus, S. hirsutus, and S. toltecus. Hybridization of the HX/Y paint to the Y was not detected in S. fulviventer; and we did not have adequate chromosomal preps from male individuals of S. arizonae, S. leucotis, S. mascotensis, S. ochrognathus, and S. peruanus to test for cross hybridization to the Y.
Sigmodon hispidus (2n = 52; FN = 52)
Twenty-two whole chromosome paints of S. hispidus were produced for this study. Fifteen hybridized to a single chromosome in S. hispidus metaphases, whereas 7 hybridized to multiple chromosomes. Of the 7 paints, the combinations of multiple chromosomes are as follows: X/Y; 7/11; 8/9; 13/22; 12/14; 16/20/22; and 19/20/22.
The number of rearrangements in each species of Sigmodon was variable and ranged from zero to 16. In 5 species (S. hirsutus, S. leucotis, S. ochrognathus, S. peruanus, and S. toltecus) with a diploid number of 52, all paints hybridized to a single chromosome each; the order of the respective chromosomes were the same as found in S. hispidus. We concluded that no translocations have occurred between the karyotype of S. hispidus and these 5 species. In S. fulviventer, there were 11 rearrangements from S. hispidus, in S. mascotensis there were 12 rearrangements, and in S. arizonae there were 18 rearrangements. The following species descriptions detail the rearrangement combinations and hybridization patterns to S. hispidus starting with the least rearranged karyotypes to the most rearranged karyotypes. Paints were labeled by their corresponding S. hispidus chromosome following Elder [1980], for example, H1 is the S. hispidus paint corresponding to chromosome 1.
Sigmodon ochrognathus (2n = 52; FN = 66)
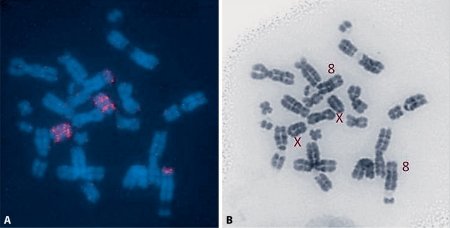
Twenty-one of the S. hispidus paints hybridized as homologous blocks to single chromosomes of S. ochrognathus (table 2). The lack of hybridization of H4 was probably due to the low concentration of the paint. Chromosomes 1, 5, 13–15, 19, 22, 24–25 completely hybridized to the S. hispidus paints similar to the pattern of the S. hispidus paints to S. hispidus. Otherwise, the centromeric heterochromatic regions of chromosomes 3, 6 (fig. 1), 7–12, 16–18, 20, 21, 23, and the X did not display any hybridization signal to the S. hispidus paints. Other than the heterochromatic short arms (which change the FN in this species), no other rearrangements were detected.
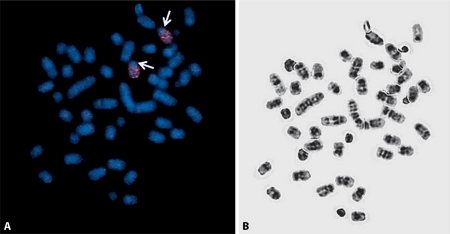
Fig. 1.
A Metaphase spreads of S. ochrognathus indicating the hybridization pattern of paint H6. The arrows indicate regions of nonhybridization on chromosome 6. B The respective DAPI spread.
Sigmodon leucotis (2n = 52; FN = 52)
Twenty-one of the S. hispidus paints hybridized as homologous blocks to single chromosomes of S. leucotis (table 2). The lack of hybridization of H4 was due to the low concentration of the paint. Chromosomes 1–3, 5–16, 18–23, 25 and the X completely hybridized to the S. hispidus paints similar to the pattern of the S. hispidus paints to S. hispidus. No rearrangements were detected and only a portion of chromosome 17 and 24 did not hybridize to the S. hispidus paints. The chromosomal preparations of S. leucotis were too poor to make an adequate assessment of C-banding.
Sigmodon hirsutus (2n = 52; FN = 52)
All 22 S. hispidus paints hybridized to the karyotype of S. hirsutus. Twenty-one S. hispidus paints hybridized as homologous blocks to single chromosomes of S. hirsutus (table 2). Chromosomes 1–5, 9–16, 19–21, 25 and the X completely hybridized to the S. hispidus paints similar to the pattern of the S. hispidus paints to S. hispidus. The distribution of constitutive heterochromatin in S. hirsutus is concentrated in the centromeric regions. Other regions of constitutive heterochromatin include one pair of small autosomes of S. hirsutus (fig. 2A). The regions of chromosome 6, 7, 17, 18, 23, 24 and the Y in S. hirsutus that did not hybridize to single copy paints of S. hispidus correspond well with regions of constitutive heterochromatin. Chromosome 18, which displayed a low level of hybridization to the S. hispidus paints, is most parsimoniously explained as corresponding to the small autosomal pair that is darkly C-banded. Other than heterochromatic additions, no rearrangements were detected.
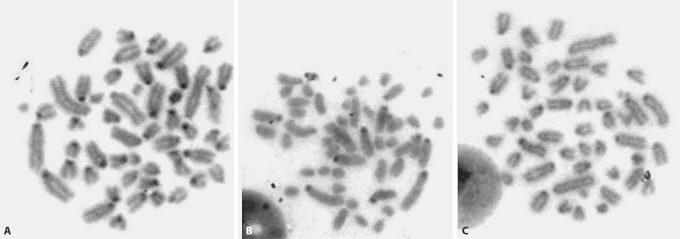
Fig. 2.
C-banded metaphase spreads of (A) S. hirsutus, (B) S. peruanus, and (C) S. toltecus. Centromeric regions stain darkly.
Sigmodon peruanus (2n = 52; FN = 50)
Twenty of the S. hispidus paints hybridized as homologous blocks to single chromosomes of S. peruanus (table 2). We could not detect hybridization to paint H4 and H19 probably due to the low concentration of these paints and the lack of adequate chromosomal preparations of S. peruanus. Chromosomes 5–7, 9, 11, 12, 14, 15, 18, 20, 21, and 24 completely hybridized to the S. hispidus paints similar to the pattern of the S. hispidus paints to S. hispidus. The distribution of constitutive heterochromatin in S. peruanus is concentrated in the centromeric regions(fig. 2B). These constitutive heterochromatin regions corresponded well to the areas of chromosomes 1–3, 10, 13, 16, and 17 that did not hybridize to any S. hispidus paints. Other than these heterochromatic additions, no rearrangements were detected.
Sigmodon toltecus (2n = 52; FN = 52)
All 22 S. hispidus paints hybridized to the karyotype of S. toltecus (table 2). All of these S. hispidus paints hybridized as homologous blocks to single chromosomes of S. toltecus. Chromosomes 1, 2, 4–6, 11–16, 18, 20, 22, 23, and 25 completely hybridized to the S. hispidus paints similar to the pattern of the S. hispidus paints to S. hispidus. No euchromatic rearrangements were detected. The distribution of constitutive heterochromatin in S. toltecus is concentrated in the centromeric regions and the smallest pair of autosomes (fig. 2C). The mid-sized and smaller autosomes of S. toltecus stained darkly in the centromeric regions after C-banding, corresponding well with the centromeric regions of 3, 7, 10, 17, 19, and 24 that did not hybridize to paints from S. hispidus. The smallest pair of autosomes that stained the darkest may be chromosome pair 21 in S. toltecus. There was a low level of hybridization of the S. hispidus paints to this chromosome as it may be mainly constitutive heterochromatin such as repetitive elements. Rearrangements in S. toltecus were restricted to heterochromatic additions as noted above.
Sigmodon fulviventer (2n = 28 or 30; FN = 34)
Twenty-one of the S. hispidus paints hybridized to the karyotype of S. fulviventer (table 2). All of these S. hispidus paints hybridized as homologous blocks to single chromosomes of S. fulviventer. The lack of hybridization of H4 to S. fulviventer was probably due to the low concentration of the paint as its hybridization signal was weak in all the Sigmodon species. Only by reducing the stringency of the washes, was signal detected in some species. Paints that completely hybridized to a single chromosome in S. fulviventer were paint H2 to S. fulviventer chromosome 4, paint H3 to S. fulviventer chromosome 6, paint H7 to S. fulviventer chromosome 10, paint H10 to S. fulviventer chromosome 11, paint H25 to S. fulviventer chromosome 14, and HX/Y paint to the S. fulviventer X. Ten fusions and 1 inversion were documented in the S. fulviventer karyotype from the S. hispidus paints (fig. 3). The S. hispidus chromosomes 23, 15, 19, and 18 fused together to form chromosome 1 in S. fulviventer; H1 and H14 fused to form chromosome 2; H8 and H6 fused to form chromosome 3; H24 and H17 fused to form chromosome 5; H16 and H12 to form chromosome 7; H5 and H22 to form chromosome 8; H13 and H20 to form chromosome 12; and H11 and H21 to form chromosome 13.
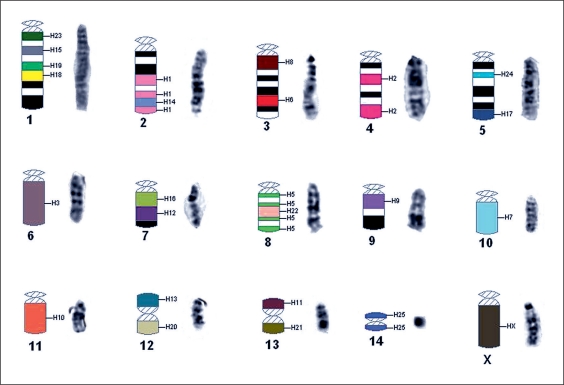
Fig. 3.
An ideogram showing where the paints from S. hispidus, labeled H1–H25, hybridized to the karyotype of S. fulviventer. Regions of chromosomes 1–5, 7 and 9 that did not hybridize to the paints are indicated by black and white bands that correspond to G-positive and G-negative bands. A DAPI banded chromosome follows its respective in situ chromosome.
Regions of chromosomes 1, 2, 3–5, 7, and 9 demonstrated no hybridization to S. hispidus paints(fig. 3). In all of the 8 species of Sigmodon examined, this is the greatest amount of nonhybridization detected. The smallest chromosome of S. hispidus (chromosome 25) corresponds to the smallest chromosome in S. fulviventer (chromosome 14), the centromeric position in this chromosome is now central, producing a metacentric resulting from an inversion.
Sigmodon mascotensis (2n = 28; FN = 28)
All 22 of the S. hispidus paints hybridized to blocks within single chromosomes of S. mascotensis. Only 3 chromosomes of S. mascotensis completely hybridized to a single paint: chromosome 4 by paint H1, chromosome 13 by paint H24, and X of S. mascotensis by the HX/Y paint. A portion of chromosome 1 and chromosome 6 in S. mascotensis did not hybridize to the S. hispidus paints(fig. 4). Twelve fusions resulting from the combination of 2 acrocentric chromosomes to produce a larger acrocentric were documented in the S. mascotensis karyotype from the S. hispidus paints (fig. 4). The S. hispidus chromosomes 2 and 8 fused to form chromosome 1 in S. mascotensis; H15, H25, and H4 fused to form chromosome 2; H14 and H3 fused to form chromosome 3; H12 and H6 fused to form chromosome 5; H5 and H21 fused to form chromosome 7; H13 and H10 fused to form chromosome 8; H23, H9, and H22 fused to form chromosome 9; H16 and H17 fused to form chromosome 10; H19 and H18 fused to form chromosome 11; and H20 and H11 fused to form chromosome 12.
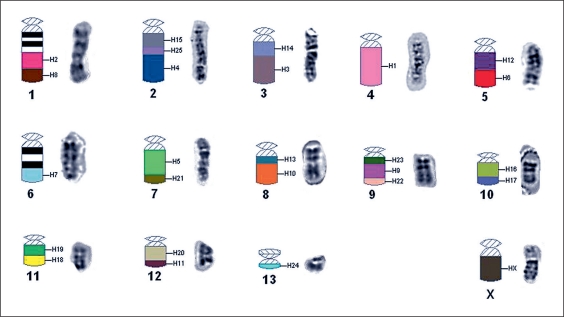
Fig. 4.
An ideogram showing where the paints from S. hispidus, labeled H1–H25, hybridized to the karyotype of S. mascotensis. Regions of chromosomes 1 and 6 that did not hybridize to the paints are indicated by black and white bands that correspond to G-positive and G-negative bands. A DAPI banded chromosome follows its respective in situ chromosome.
Sigmodon arizonae (2n = 22 or 24; FN = 38)
This species had the largest number of chromosomal rearrangements as well as the greatest diversity of types of rearrangements. All 22 of the S. hispidus paints hybridized to the karyotype of S. arizonae. Twenty-one of the paints hybridized as homologous blocks to single chromosomes of S. arizonae. The only paint that hybridized to multiple chromosomes was the HX/Y paint. It hybridized to the X chromosome of S. arizonae as well as the most distal region of chromosome 8 (fig. 5). Paints that completely hybridized to a single chromosome in S. arizonae were H7 to S. arizonae chromosome 6, H12 to S. arizonae chromosome 7, H16 to S. arizonae chromosome 10, and the HX/Y paint to the X of S. arizonae.
Fig. 5.
A Metaphase spreads of S. arizonae. The HX/Y paint hybridized to the X and to the distal end of chromosome 8 of S. arizonae. B The respective DAPI spread.
Fifteen fusions, one partial arm translocation, and 2 inversions were documented in the S. arizonae karyotype from the S. hispidus paints (fig. 6). The S. hispidus chromosomes 1, 5, and 25 fused to form chromosome 1 in S. arizonae; H10 and H11 formed a portion of chromosome 2; H4, H6, H13, H21, and H14 fused to form chromosome 3; H23, H15, and H2 fused to form chromosome 4; H18 and H8 fused to form chromosome 5; inversions were detected in chromosomes 6 and 7; the fusions of H24, H19, H20, H22, and H17 plus the whole arm translocation of HX/Y formed chromosome 8; and H9 and H3 fused to form chromosome 9. Regions that did not hybridize to the single-copy S. hispidus paints were found on portions of chromosome 2, chromosome 4 and chromosome 5 in S. arizonae (fig. 6).
Fig. 6.
An ideogram showing where the paints from S. hispidus, labeled H1–H25, hybridized to the karyotype of S. arizonae. Regions of chromosomes 2, 4, and 5 that did not hybridize to the paints are indicated by black and white bands that correspond to G-positive and G-negative bands. A DAPI banded chromosome follows its respective in situ chromosome.
Discussion
There is a high degree of karyotype conservation within the 8 species of Sigmodon to the karyotype of S. hispidus, the proposed ancestral karyotype of Sigmodon. Other than heterochromatic additions, no rearrangements were detected in 5 of the species (S. hirsutus, S. leucotis, S. ochrognathus, S. peruanus, and S. toltecus); and in 3 species with chromosomal rearrangements (S. arizonae, S. fulviventer, and S. mascotensis), most of the chromosomes were composites of whole arms present in the ancestral chromosomes. There are few studies to document such chromosomal conservation in the radiation of a new world rodent genus.
Within muroid rodents, many studies exist regarding chromosomal evolution in old world muroids (Muridae) due to the availability and utility of Mus musculus paints, but only 3 chromosome painting studies have been conducted to examine the chromosomal evolution of new world muroid species. One study utilized both unique rodent paints and Mus paints [Sitnikova et al., 2007 Microtus]; and 2 other studies utilized just Mus paints [Dawson et al., 1999 Peromyscus; Hass et al., 2008 Akodon and Oligoryzomys]. As human paints fail to hybridize to the extensively rearranged karyotypes of muroid rodents, Mus paints are likely a resource for comparative cytogenetics. Though, as murids and cricetids probably shared a common ancestor 31.1 mya [Adkins et al., 2001], the hybridization of Mus paints to cricetids may not be as successful, although more experiments are warranted.
In other mammalian taxa, especially canids, karyotype conservation has been reported within and between Carnivore families [Wurster-Hill and Gray, 1975], and similarly to the chromosome evolution in Sigmodon, the ancestral arms were conserved with few inner-arm rearrangements [Fronicke et al., 1997]. Karyotype conservation can also be found within the fellow cricetid rodent, Microtus. Similar to the karyotype repatterning in Sigmodon species, all 21 autosomal paint probes and the X probe of the higher diploid number species (Microtus agrestis) hybridized to the lower diploid numbered species (M. oeconomus), and 18 paints hybridized to single chromosomal segments [Sitnikova et al., 2007]. Chromosome painting between distantly related cricetids Mesocricetus auratus (golden hamster) to M. oeconomus documented less karyotype conservation as only 8 paints of M. auratus hybridized to single chromosomes of M. oeconomus, whereas the other 13 paints hybridized to between 2 to 4 different chromosomes.
The chromosomal painting data provide no evidence to reject the hypothesis that the 8 species of Sigmodon evolved from a common ancestor with a 2n = 52 and the ancestral chromosomal segments have been conserved in each. Paracentric or pericentric inversions may have caused the differences in the fundamental numbers in these species, but the nature of any intrachromosomal rearrangements is difficult to establish by whole chromosome paints. We propose that technical problems were the reason that the H4 paint did not hybridize to chromosomes of S. leucotis, S. ochrognathus, and H4 and H19 to chromosomes of S. peruanus. Chromosomal change for some of the species with a 2n = 52 karyotype was indicated by the absence of signal in some interstitial regions that did not hybridize to the paints. These are described below.
Heterochromatin Additions in S. hirsutus, S. ochrognathus, S. peruanus, and S. toltecus
Only the centromeric regions of S. ochrognathus did not hybridize to the S. hispidus paints. These results support the conclusions of Elder and Lee [1985] based on G-band data that the karyotype of S. ochrognathus and S. hispidus are essentially indistinguishable in euchromatin, the only difference being the greater amount of heterochromatin in S. ochrognathus located near the centromeric regions. This pattern also was present in the karyotypes of S. hirsutus, S. peruanus, and S. toltecus. When these species were C-banded, the regions of nonhybridization corresponded well to heterochromatin rich regions. The lack of hybridization to heterochromatin rich regions in S. hirsutus, S. leucotis, S. peruanus, and S. toltecus suggests that these regions have unique tandem repeats in the heterochromatin that are not present in the genome of S. hispidus.
Chromosome Painting to the Chromosomes of S. arizonae, S. fulviventer, and S. mascotensis
Though the karyotype of S. hispidus is likely conserved in each of these 3 species, regions of nonhybridization to the S. hispidus paints were documented, especially in S. fulviventer. Not as many rearrangements were detected in S. fulviventer, compared to S. arizonae and S. mascotensis. More autosomes of S. fulviventer hybridized to a single S. hispidus paint than S. arizonae and S. mascotensis (6 chromosomes in S. fulviventer, 3 in S. arizonae, 3 in S. mascotensis) and more chromosomal areas of S. fulviventer did not hybridize to the S. hispidus paints (chromosomes 1–5, 7 and 9) compared to S. arizonae (chromosomes 2, 4 and 5) and S. mascotensis (chromosomes 1 and 6). The centromeric regions of chromosomes 3, 4, 5, and 6 of S. arizonae did not hybridize to the paints of S. hispidus but these regions correspond well to areas rich in heterochromatin [see fig. 3b in Elder, 1980]. The distribution of constitutive heterochromatin in S. fulviventer corresponds to the centromeric regions and a few interstitial bands in chromosomes 5 and 7 [Elder and Lee, 1985]. But regions of nonhybridization to the paints of S. hispidus only match the heterochromatin distribution in chromosome 4 and 5 of S. fulviventer. All other regions of nonhybridization are located interstitially in the chromosomes of S. fulviventer and may indicate the presence of other unique types of repetitive elements.
The regions that did hybridize to the S. hispidus paints in S. arizonae, S. fulviventer, and S. mascotensis were indicative of fusions, either centric or tandem, which had the effect of reducing the ancestral diploid number of 52 to 22, 30, and 28. In S. mascotensis, the most parsimonious explanation for most of the rearrangements is by tandem fusions as whole ancestral chromosomes fused together to form these species specific chromosomes (fig. 4). With chromosome painting data, this is the only type of rearrangement that was detected in S. mascotensis using the S. hispidus paints which confirms the conclusion of Elder [1980] that mainly tandem fusions had occurred in S. mascotensis. In S. fulviventer, the most parsimonious explanation for the rearrangements is by centric fusions, tandem fusions, and an inversion; Elder and Lee [1985] also confirmed fusions and an inversion. In S. arizonae, 15 of the rearrangements are fusions (centric and/or tandem), 2 are inversions, and one is a partial arm translocation as a portion of the HX/Y paint was found to hybridize to chromosome 8 in S. arizonae. Most of these types of rearrangements, with the exception of the partial arm translocation also were documented by Elder [1980].
Phylogenetic Associations to Ancestral Karyotype
Other than centromeric and interstitial additions of heterochromatin, the only rearrangements detected in the 8 species of Sigmodon examined in this study were in S. arizonae, S. fulviventer, and S. mascotensis. Further, each rearrangement only became established in the common ancestor of a single species after they diverged from all other members of the genus thus far studied (fig. 7). Other studies have documented a similar location to chromosomal rearrangements in other species. Baker et al. [1983] studied the systematic relationships among 11 species of Oryzomys. Of the 55 rearrangements that were documented during the chromosomal evolution of this complex, 43 were present in a single species in the phylogenetic tree (see their fig. 4, p. 412), although study of more species of Oryzomys may find rearrangements shared by 2 or more species. In the outgroup species (Nectomys squamipes and Sigmodon hispidus), the majority of the rearrangements also occurred recently in their evolutionary history.
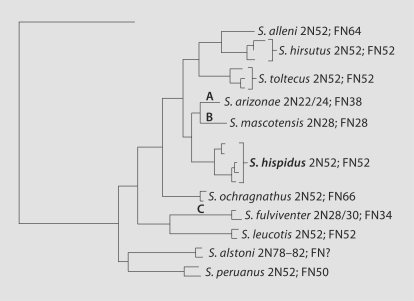
Fig. 7.
A neighbor-joining gene tree of cytochrome b, beta fibrinogen, and IRBP sequences from Henson and Bradley [2009]. Letters A–C represent the branches where the rearrangements occurred in the common ancestor of respective species. Rearrangements for A (S. arizonae) are: H5/H25/H1; H10/H11; H4/H6/H13/ H21/H14; H23/H15/H2; H18/H8; H24/H19/H20/H22/H17/HXY; H3/H9. Rearrangements for B (S. mascotensis) are: H2/H8; H15/H25/H4; H14/H3; H12/H6; H5/H21; H13/H10; H23/H9/H22; H16/H17; H19/H18; H20/H11. Rearrangements for C (S. fulviventer) are: H23/H15/H19/H18; H1/H14; H8/H6; H24/H17; H16/H12; H5/H22; H13/H20; H11/H21. Rearrangement H23/H15 shared by S. arizonae and S. fulviventer and H19/H18 shared by S. mascotensis and S. fulviventer are homoplastic in this interpretation.
The number of rearrangements that were found in each species of Sigmodon were mapped on a phylogenetic tree based on nuclear and mitochondrial genes [Henson and Bradley, 2009] (fig. 7). Only one rearrangement was shared between S. arizonae and S. fulviventer (23/15) and only one between S. mascotensis and S. fulviventer (19/18), but if these are truly shared from a single ancestral chromosomal rearrangement, a loss of the rearrangement would be required in some species if the Henson and Bradley [2009] tree is the phylogeny. A more parsimonious explanation is that in each species these rearrangements were acquired independently by convergent evolution. Convergence would require fewer events than the reversal in the other species in the Henson and Bradley [2009] tree. This would explain the lack of these rearrangements in the recent common ancestor to S. arizonae, S. fulviventer, and/or S. mascotensis.
The phylogenetic arrangement of Henson and Bradley [2009] based on the sequence data from nuclear and mitochondrial genes takes into account previous studies conducted for this genus [Peppers and Bradley, 2000; Peppers et al., 2002; Carroll and Bradley, 2005; Carroll et al., 2005]. Results from these DNA sequence-based studies were used to elevate S. toltecus and S. hirsutus to species level, although they are almost indistinguishable karyotypically and morphologically from S. hispidus. In this study, the apparent conservation of the S. hispidus karyotype is retained in S. toltecus, further illustrating the need for multiple datasets to determine phylogenetic associations.
Acknowledgements
We would like to thank H. Garner and K. MacDonald of the NSRL, Museum of Texas Tech for the quick accession and storage of voucher specimens and tissue loans; and Amber Matthews of the Animal Care Services, Texas Tech University for assistance during the consecution of the animal care and use protocol. Thanks to M. Hamilton and J. Salazar-Bravo for providing editorial comments on earlier drafts of the manuscript.
Field work in Ecuador, Honduras, Mexico, and Texas was supported by grants from James Sowell and NIH grant, A141435-07, from Robert D. Bradley. Many thanks to the Texas Tech University field parties of Mexico 1997, 2000, 2005, and 2006; Ecuador 2004; and Honduras 2004. Thanks to R.K. Baker and F. Anwarali for assistance in collection of S. hispidus in Texas.
References
- Adkins RM, Gelke EL, Rowe D, Honeycutt RL. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol Biol Evol. 2001;18:777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- Anderson S. Mammals of Chihuahua: Taxonomy and distribution. Bull Am Mus Nat History. 1972;148:149–410. [Google Scholar]
- Bailey VE. Synopsis of the North American species of Sigmodon. Proc Biol Soc Washington. 1902;15:101–116. [Google Scholar]
- Baker RH. Cotton rats of the Sigmodon fulviventer group. Miscell Pub, Museum Nat History, University of Kansas. 1969;51:177–232. [Google Scholar]
- Baker RJ, Koop BF, Haiduk MW. Resolving systematic relationships with G-bands: a study of five genera of South American Cricetine rodents. Syst Zool. 1983;32:403–416. [Google Scholar]
- Baker RJ, Hamilton M, Parish DA. Preparations of mammalian karyotypes under field conditions. Occasional papers, Museum of Texas Tech University. 2003;228:1–8. [Google Scholar]
- Bowers KL, Hamilton MJ, White SM, Baker RJ. Origins of heterochromatic repatterning in white-footed mice, Peromyscus leucopus. J Mammal. 1998;79:725–735. [Google Scholar]
- Carleton MD, Fisher RD, Gardner AL. Identification and distribution of cotton rats, genus Sigmodon (Muridae: Sigmodontinae), of Nayarit, Mexico. Proc Biol Soc Washington. 1999;112:813–856. [Google Scholar]
- Carroll DS, Bradley RD. Systematics of the genus Sigmodon: DNA sequences from beta-fibrinogen and cytochrome b. Southwest Naturalist. 2005;50:342–349. [Google Scholar]
- Carroll DS, Peppers LL, Bradley RD. Molecular systematics and phylogeography of the Sigmodon hispidus species group. In: Sanchez-Cordero V, Medellin RA, editors. Contribuciones mastozoologicas en homenajea a Bernardo Villa-R. Mexico: Instituto de Biologia e Instituto de Ecologia, UNAM CONABIO; 2004. pp. 87–100. [Google Scholar]
- Dawson WD, Young SR, Wang Z, Liu LW, Greenbaum IF, et al. Mus and Peromyscus chromosome homology established by FISH with three mouse paint probes. Mamm Genome. 1999;10:730–733. doi: 10.1007/s003359901080. [DOI] [PubMed] [Google Scholar]
- Elder FFB. Tandem fusion, centric fusion, and chromosomal evolution in the cotton rats, genus Sigmodon. Cytogenet Cell Genet. 1980;26:199–210. doi: 10.1159/000131441. [DOI] [PubMed] [Google Scholar]
- Elder FFB, Lee MR. The chromosomes of Sigmodon ochrognathus and S fulviventer suggest a realignment of Sigmodon species groups. J Mammal. 1985;66:511–518. [Google Scholar]
- Elder FFB, Pathak S. Light microscope observations on the behavior of silver-stained trivalents in pachytene cells of Sigmodon fulviventer (Rodentia, Muridae) heterozygous for centric fusion. Cytogenet Cell Genet. 1980;27:31–38. doi: 10.1159/000131461. [DOI] [PubMed] [Google Scholar]
- Fronicke L, Muller-Navia J, Romanakis K, Scherthan H. Chromosomal homeologies between human, harbor seal (Phoca vitulina) and the putative ancestral carnivore karyotype revealed by Zoo-Fish. Chromosoma. 1997;106:108–113. doi: 10.1007/s004120050230. [DOI] [PubMed] [Google Scholar]
- Garcia F, Ruiz-Herrera A, Egozcue J, Ponza M, Garcia M. Chromosomal homologies between Cebus and Ateles (Primates) based on ZOO-FISH and G-banding comparisons. Am J Primatol. 2002;57:177–188. doi: 10.1002/ajp.10047. [DOI] [PubMed] [Google Scholar]
- Graphodatsky AS, Yang F, O'Brien PCM, Perelman P, Milne BS, et al. Phylogenetic implications of the 38 putative ancestral chromosome segments for four canid species. Cytogenet Cell Genet. 2001;92:243–247. doi: 10.1159/000056911. [DOI] [PubMed] [Google Scholar]
- Hameister H, Klett C, Bruch J, Dixkens C, Vogel W, Christensen K. Zoo-FISH analysis: the American mink (Mustela vison) closely resembles the cat karyotype. Chromosome Res. 1997;5 doi: 10.1023/a:1018433200553. [DOI] [PubMed] [Google Scholar]
- Hass I, Sbalqueiro IJ, Muller S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetida) from southern Brazil established by Zoo-Fish using Mus musculus (Muridae) painting probes. Chromosome Res. 2008;16:75–88. doi: 10.1007/s10577-007-1211-5. [DOI] [PubMed] [Google Scholar]
- Henson DD, Bradley RD. Molecular systematics of the genus Sigmodon: results from mitochondrial and nuclear gene sequences. Canad J Zool. 2009;87:211–220. doi: 10.1139/z09-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TC, Benirschke K. An Atlas of Mammalian Chromosomes. vol 2. Berlin: Springer-Verlag; 1968. [Google Scholar]
- Kiblisky P. The chromosomes of the hispid cotton rat, Sigmodon hispidus, from two localities in Venezuela. J Mammal. 1969;50:810–811. [PubMed] [Google Scholar]
- Koop BF, Baker RJ, Haiduk MW, Engstrom MD. Cladistical analysis of primitive G-band sequences for the karyotype of the ancestor of the Cricetidae complex of rodents. Genetica. 1984;64:199–208. [Google Scholar]
- Lee MR, Zimmerman EG. Robertsonian polymorphism in the cotton rat, Sigmodon fulviventer. J Mammal. 1969;50:333–339. [PubMed] [Google Scholar]
- Li T, O'Brien PCM, Biltueva L, Fu B, Wang J, et al. Evolution of genome organization of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res. 2004;12:317–335. doi: 10.1023/B:CHRO.0000034131.73620.48. [DOI] [PubMed] [Google Scholar]
- Musser GG, Carleton MD. Superfamily Muroidea. In: Wilson DE, Reeder DM, editors. Mammal Species of the World. Baltimore: Johns Hopkins University Press; 2005. pp. 894–1531. [Google Scholar]
- Nash WG, Wienberg J, Ferguson-Smith MA, Menninger JC, O'Brien SJ. Comparative genomics: tracking chromosome evolution in the family Ursidae using reciprocal chromosome painting. Cytogenet Cell Genet. 1998;83:182–192. doi: 10.1159/000015176. [DOI] [PubMed] [Google Scholar]
- Nash WG, Menninger JC, Wienberg J, Padilla-Nash HM, O'Brien SJ. The pattern of phylogenomic evolution of the Canidae. Cytogenet Cell Genet. 2001;95:210–224. doi: 10.1159/000059348. [DOI] [PubMed] [Google Scholar]
- Nelson EW, Goldman EA. Three new rodents from southern Mexico. Proc Biol Soc of Washington. 1933;46:195–198. [Google Scholar]
- Nie W, Wang J, O'Brien PCM, Fu B, Ying T, et al. The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding. Chromosome Res. 2002;10:209–222. doi: 10.1023/a:1015292005631. [DOI] [PubMed] [Google Scholar]
- Peppers LL, Bradley RD. Cryptic speciation in Sigmodon hispidus: evidence from DNA sequences. J Mammal. 2000;81:332–343. [Google Scholar]
- Peppers LL, Carroll DS, Bradley RD. Molecular systematics of the genus Sigmodon (Rodentia: Muridae): evidence from the mitochondrial cytochrome-b gene. J Mammal. 2002;83:396–407. [Google Scholar]
- Pieczarka JC, Nagamachi CY, O'Brien PCM, Yang F, Rens W, et al. Reciprocal chromosome painting between two South American bats: Carollia brevicauda and Phyllostomus hastatus (Phyllostomidae, Chiroptera) Chromosome Res. 2005;13:339–347. doi: 10.1007/s10577-005-2886-0. [DOI] [PubMed] [Google Scholar]
- Reig OA. Diversity patterns and differentiation of high Andean rodents. London: Oxford University Press; 1986. [Google Scholar]
- Rens W, O'Brien PCM, Fairclough H, Harman L, Graves JAM, Ferugson-Smith MA. Reversal and convergence in marsupial chromosome evolution. Cytogenet Genome Res. 2003;102:282–290. doi: 10.1159/000075764. [DOI] [PubMed] [Google Scholar]
- Richard F, Lombard M, Dutrillaux B. Zoo-Fish suggests a complete homology between human and Capucin monkey (Platyrrhini) euchromatin. Genomics. 1996;36:417–423. doi: 10.1006/geno.1996.0486. [DOI] [PubMed] [Google Scholar]
- Say T, Ord G. Description of a new species of Mammalia, whereon a new genus is proposed to be founded. J Acad Nat Sci Philadelphia. 1825;4:352–356. [Google Scholar]
- Schmid M. Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma. 1978;66:361–388. [Google Scholar]
- Sitnikova NA, Romanenko SA, O'Brien PCM, Perelman PL, Fu B, et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res. 2007;15:447–456. doi: 10.1007/s10577-007-1137-y. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Consigliere S, Bigoni F, Ferguson-Smith M, O'Brien PCM, Wienberg J. Reciprocal chromosome painting between a New World primate, the woolly monkey, and humans. Chromosome Res. 2001;9:97–106. doi: 10.1023/a:1009274802086. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Stone G, Garcia M, Froenicke L. Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics. 2003;82:245–249. doi: 10.1016/s0888-7543(03)00109-5. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Adkins RM, Anderson J. Phylogeny and divergence-data estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Volleth M, Klett C, Kollak A, Dixkens C, Winter Y, et al. Zoo-Fish analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae) Chromosome Res. 1999;7:57–64. doi: 10.1023/a:1009227428727. [DOI] [PubMed] [Google Scholar]
- Voss RS. A revision of the South American species of Sigmodon (Mammalia: Muridae) with notes on their natural history and biogeography. Am Mus Nov. 1992;3050:1–56. [Google Scholar]
- Wurster-Hill DH, Gray CW. The interrelationships of chromosome banding patterns in procyonids, viverrids, and felids. Cytogenet Cell Genet. 1975;15:306. doi: 10.1159/000130528. [DOI] [PubMed] [Google Scholar]
- Yang F, Carter NP, Shi L, Ferguson-Smith MA. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma. 1995;103:642–652. doi: 10.1007/BF00357691. [DOI] [PubMed] [Google Scholar]
- Yang F, Alkalaeva EZ, Perelman PL, Pardini AT, Harrison WR, et al. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc Natl Acad Sci USA. 2003;100:1062–1066. doi: 10.1073/pnas.0335540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Graphodatsky AS, Li T, Fu B, Dobigny G, et al. Comparative genome maps of the pangolin, hedgehog, sloth, anteater and human revealed by cross-species chromosome painting: further insight into the ancestral karyotype and genome evolution of eutherian mammals. Chromosome Res. 2006;14:283–296. doi: 10.1007/s10577-006-1045-6. [DOI] [PubMed] [Google Scholar]
- Zimmerman EG. Karyology, systematics and chromosomal evolution in the rodent genus, Sigmodon. Vol. 4. Pub Museum-Michigan State University; 1970. pp. 385–454. [Google Scholar]