Abstract
Charles Darwin first provided a lucid explanation of how gender differences evolve nearly 140 years ago. Yet, a disconnect remains between his theory of sexual selection and the mechanisms that underlie the development of males and females. In particular, comparisons between representatives of different phyla (i.e., flies and mice) reveal distinct genetic mechanisms for sexual differentiation. Such differences are hard to comprehend unless we study organisms that bridge the phylogenetic gap. Analysis of variation within monophyletic groups (i.e., amniotes) is just as important if we hope to elucidate the evolution of mechanisms underlying sexual differentiation. Here we review the molecular, cellular, morphological, and physiological changes associated with sex determination in reptiles. Most research on the molecular biology of sex determination in reptiles describes expression patterns for orthologs of mammalian sex-determining genes. Many of these genes have evolutionarily conserved expression profiles (i.e., DMRT1 and SOX9 are expressed at a higher level in developing testes vs. developing ovaries in all species), which suggests functional conservation. However, expression profiling alone does not test gene function and will not identify novel sex-determining genes or gene interactions. For that reason, we provide a prospectus on various techniques that promise to reveal new sex-determining genes and regulatory interactions among these genes. We offer specific examples of novel candidate genes and a new signaling pathway in support of these techniques.
Key Words: Genotypic sex determination, Gonad, Molecular genetics, Temperature-dependent sex determination
Introduction
Individuals come in one of two distinct forms, male or female, in most metazoans. Yet, the mechanisms underlying the development of morphological, physiological, and behavioral differences between the sexes have been elucidated in detail in just a few species. Comparison of mechanisms among the most intensively studied species (i.e., nematodes, fruit flies, and mice) reveals much less conservation than expected for a fundamental biological trait like sexual phenotype [Cline, 1993; Cline and Meyer, 1996; Marín and Baker, 1998; Swain and Lovell-Badge, 1999; Wilhelm et al., 2007]. For example, sex in the fruit fly, Drosophila melanogaster, is determined by the ratio of numerator and denominator genes found on the X chromosomes and autosomes, respectively [Cline, 1993]. Proteins produced by numerator genes (Sisterless-a, Sisterless-b, and Sisterless-c) function as transcriptional activators of a gene called sex lethal (Sxl) while denominator proteins inhibit numerator proteins [Harrison, 2007]. Hence, Sxl is transcribed and translated in flies with a high X chromosome to autosome ratio (XX:AA individuals become females), but is not expressed in flies with a low X to autosome ratio (XY:AA individuals become males). In contrast to this chromosome-counting mechanism, a single Y-linked gene called sex-determining region of the Y (Sry) determines sex in mice and most other mammals [Swain and Lovell-Badge, 1999; Wilhelm et al., 2007; Wallis et al., 2008]. This gene acts as a dominant male-determining factor and is not related to any sex-determining genes in flies. Moreover, transcription of Sry is regulated by a set of genes (Wt1, Gata/Fog2, and insulin receptors) distinct from fly numerator and denominator genes [Wilhelm et al., 2007].
Downstream targets of Sxl and Sry are also unique: SXL regulates female-specific splicing of transformer (tra) mRNA in flies [Lopez, 1998], while SRY induces male-specific transcription of the sex-determining region Y-box 9 (Sox9) gene in mice [Sekido and Lovell-Badge, 2009]. The next gene in the fly pathway, doublesex (dsx), is alternatively spliced to produce female-specific and male-specific isoforms of DSX proteins, which are master regulators of the female and male transcriptome, respectively [Arbeitman et al., 2004]. An entirely different set of genes takes on this key role in mice (i.e., genes involved in sex steroid signaling) [Ellegren and Parsch, 2007; Wilson and Davies, 2007]. A dsx homolog called doublesex and mab-related transcription factor 1 (Dmrt1) is involved in testis development in mice [Fahrioglu et al., 2006]. However, Dmrt1 does not play a pivotal role in sex determination in mice like dsx does in flies.
These observations lead to broader questions about sex-determining mechanisms. First, to what extent are sex-determining genes and gene networks evolutionarily conserved or unique? Second, can we reconstruct the specific molecular events responsible for the evolution of different sex-determining mechanisms? Given the extensive divergence between phyla (arthropods vs. chordates), the best way to address these questions is to study more closely related organisms that still display diverse modes of sex determination. Reptiles fit these criteria and represent one of the most interesting groups to study from a phylogenetic perspective [Bull, 1980, 1983; Korpelainen, 1990; Janzen and Paukstis, 1991; Valenzuela, 2004]. At the same time, however, researchers face significant challenges when working with reptiles. In this article, we outline what is known, and what is not known, about sexual differentiation in this group. We focus on the molecular and cellular mechanisms underlying sex determination. Most studies to date have been descriptive in nature and have examined homologs of sex-determining genes first identified in mammals. Such work provides important baseline data on conserved genes and should be encouraged, but it will not identify unique sex-determining genes or novel gene regulatory interactions. Therefore, we also provide a prospectus on alternative approaches that promise to reveal new candidate genes and to elucidate functional interactions among these genes.
Sex Determination
Although the specific molecular mechanism that determines sex has not been revealed in any reptilian species, general modes of sex determination can be described [Bull, 1980; Janzen and Paukstis, 1991]. An individual's genotype at one or more loci can control whether it develops testes or ovaries. Species that display this mechanism are said to have genotypic sex determination, or GSD. Species with GSD may or may not have distinct sex chromosomes (ZZ males and ZW females or XY males and XX females). A frequent alternative to GSD is environmentally triggered polyphenism (i.e., a single individual can develop testes or ovaries depending upon environmental conditions). Such species are said to have environmental sex determination, or ESD. Various environmental factors, including photoperiod, social environment, and temperature, influence sex determination across the animal kingdom [Bull, 1983; Korpelainen, 1990]. However, temperature is the only environmental variable that has been conclusively shown to determine sex in reptiles [Bull, 1980; Janzen and Paukstis, 1991; Valenzuela, 2004]. This form of ESD is called temperature-dependent sex determination, or TSD.
The overall pattern of evolutionary transitions between GSD and TSD in reptiles is not yet clear, but phylogenetic analyses suggest TSD is an ancestral trait in turtles and that GSD is derived in this group [Janzen and Krenz, 2004; Organ and Janes, 2008]. The ancestral trait in crocodilians is most likely TSD because all species have this mode of sex determination [Lang and Andrews, 1994; Deeming, 2004]. The two extant species of tuatara have TSD [Nelson et al., 2004]. In contrast, all snakes have GSD and distinct sex chromosomes (ZZ males and ZW females) [Matsubara et al., 2006]. Lizards deserve closer scrutiny because GSD and TSD can co-occur within the same family (e.g. Agamidae and Eublepharidae) [Viets et al., 1994; Harlow, 2004]. Phylogenetic and molecular analyses suggest sex-determining mechanisms can evolve quite rapidly in this group [Janzen and Krenz, 2004; Organ and Janes, 2008; Ezaz et al., 2009; Pokorná and Kratochvíl, 2009]. In fact, recent reports show that high temperatures can override chromosomal sex determination in 2 species of lizards: XX females become phenotypic males in one species while ZZ males become phenotypic females in the other [Shine et al., 2002; Quinn et al., 2007; Radder et al., 2008; Quinn et al., 2009]. Conversely, there is evidence for genetic variation in thermal sensitivity in TSD species, including several turtles, American alligators, and the leopard gecko [Bull et al., 1982; Janzen, 1992; Rhen and Lang, 1998; Janes and Wayne, 2006]. Taken together, these observations indicate that genes transduce temperature into a biological signal for sex determination.
Thus, the distinction between GSD and TSD in reptiles is not as dramatic as some have argued [for a more detailed discussion of this subject see Sarre et al., 2004]. Indeed, it is hard to imagine wholesale changes in gene regulatory networks (à la flies and mice) between closely related lizards with different sex-determining mechanisms or within monophyletic groups like turtles. It is more likely on theoretical and empirical grounds that the initial trigger (e.g., a single gene or small number of genes) is labile and that the underlying gene regulatory networks for producing a testis versus an ovary are largely conserved [Bull, 1983; Barske and Capel, 2008]. We explore this idea by examining the molecular, cellular, morphological, and physiological changes that occur during gonadogenesis.
Gonad Differentiation
The basic pattern of gonad development is conserved in reptiles [Raynaud and Pieau, 1985; Morrish and Sinclair, 2002]. The gonad initially develops as a bipotential primordium that is identical in all embryos regardless of genotype or temperature. This primordium, also called the genital ridge, forms as an outgrowth of cells from the mesonephric portion of the embryonic kidney. The genital ridge consists of coelomic epithelium, underlying mesenchymal cells, as well as primordial germ cells that migrate into the ridge from the embryonic yolk sac [Hubert, 1971, 1976; Fujimoto et al., 1979]. The genital ridge grows in size as somatic and germ cells proliferate. The next step occurs when the bipotential primordium commits to develop as a testis or an ovary, a process commonly referred to as sex determination. Depending upon species, the sex-determining period can end before any sign of gonad differentiation or may overlap with the early stages of gonad differentiation. The following generic description of testis and ovary morphogenesis is based on studies of turtle, crocodilian, and lizard embryos, and includes GSD and TSD species [Raynaud and Pieau, 1985; Wibbels et al., 1991; Smith and Joss, 1993; Pudney, 1995; Merchant-Larios et al., 1997; Greenbaum and Carr, 2001].
Testes develop when the inner medullary region of the genital ridge grows and differentiates [Raynaud and Pieau, 1985; Wibbels et al., 1991; Greenbaum and Carr, 2001]. More specifically, this entails differentiation of a subpopulation of somatic cells into pre-Sertoli cells, which differentiate further into Sertoli cells [Smith and Joss, 1993]. These cells aggregate to form testis cords that surround primordial germ cells [Morrish and Sinclair, 2002]. Germ cells proliferate and then exit the cell cycle and arrest as spermatogonia. Meiosis and spermatogenesis do not commence until males reach sexual maturity [Pudney, 1995]. Another group of cells differentiates into peritubular myoid cells, which envelop the testis cords to form seminiferous tubules [Morrish and Sinclair, 2002]. A third cell type, the interstitial cells of Leydig, differentiates between the testis cords and seminiferous tubules [Doddamani, 2006]. The final distinguishing feature of testis morphogenesis is formation of the male-specific vasculature [Morrish and Sinclair, 2002].
In contrast, ovaries develop when the outer cortex (i.e., coelomic epithelium) grows and differentiates while the inner medulla regresses [Morrish and Sinclair, 2002]. The cortex thickens as the result of mitosis in epithelial and germ cells. Primordial germ cells in females proliferate longer than their counterparts in males and form nests of oogonia within the cortex [Raynaud and Pieau, 1985]. The next step in ovary development occurs when oogonia enter meiosis and arrest in prophase I to produce oocytes [Ditewig and Yao, 2005]. Primordial follicles form when a single layer of granulosa cells surrounds individual oocytes. The timing of meiosis initiation and follicle formation is species-specific, but usually occurs late in fetal life or shortly after birth [Moore et al., 2008]. Primordial follicles remain quiescent until females reach sexual maturity. At this time, a subset of follicles is recruited and initiates an extended period of growth, including the resumption of meiosis and ovulation of a fertilizable egg (or eggs) [Callebaut et al., 1997; Uribe and Guillette, 2000]. Thecal cells are a third key cell type in reptile ovaries, but it is not clear when these cells differentiate.
In addition, endocrine signals from the testes (i.e., androgens produced by Leydig cells and anti-Mullerian hormone produced by Sertoli cells) and ovaries (i.e., estrogens and progestins produced by thecal and granulosa cells) regulate sexual differentiation of all other somatic tissues, including the male and female reproductive tract and brain [Austin, 1994; Moore et al., 1998; Girling, 2002; Rhen and Crews, 2002; Lovern et al., 2004]. Thus, at the cellular, morphological, and physiological levels, reptile gonads develop and function in a manner homologous to bird and mammal gonads [reviewed in Morrish and Sinclair, 2002]. We next explore whether this homology extends to the molecular level.
Putative Sex-Determining Genes
Virtually all studies of the molecular biology of sex determination in reptiles have examined homologs of sex-determining genes first identified in mammals. The general approach is to clone partial or full-length cDNAs of these genes in TSD species. Sequences are then used to design primers for quantitative real-time PCR or as templates for synthesis of probes for Northern blots or in situ hybridization. Finally, mRNA expression is characterized in the gonads or adrenal-kidney-gonad complexes (AKGs) of embryos that have been incubated at male- and female-producing temperatures. Expression differences are taken as evidence that the gene of interest is involved in TSD. All told, 18 orthologs of mammalian sex-determining genes have been studied in this manner (table 1). Similar experiments have been conducted in a few GSD species [Valenzuela et al., 2006, this issue; Chakraborty et al., 2009]. Given the long list of genes that play a role in gonad development in mammals [Wilhelm et al., 2007], this will continue to be a valuable approach to the study of sex determination in reptiles. However, we will only briefly describe expression patterns for a few of the genes listed in table 1 because others have written recent and comprehensive reviews of this data [Place and Lance, 2004; Shoemaker and Crews, 2009; Merchant-Larios et al., this issue].
Table 1.
Reptilian orthologs of mammalian sex-determining genes
Estrogens are crucial for ovary development in all reptiles examined to date [reviewed in Crews, 1996; Pieau and Dorizzi, 2004; Lance, 2009; Ramsey and Crews, 2009]. Exogenous estrogens induce ovary development in embryos incubating at male-producing temperatures, while aromatase inhibition can induce testis development in embryos at female-producing temperatures. In accord with these pharmacological manipulations, expression of aromatase mRNA and protein is higher in gonads and AKGs from embryos incubated at female-producing temperatures than in embryos at male-producing temperatures (see references in table 1). The only variance in this pattern appears to be the timing of the temperature effect on aromatase, which occurs during the sex-determining period in some species but afterwards in others (i.e., during ovary differentiation) [see Valenzuela and Shikano, 2007 for discussion of these differences]. In some cases, the delay may be due to adrenal and kidney tissue obscuring expression differences in the gonads [Ramsey and Crews, 2007]. In any event, estrogen signaling is clearly necessary and sufficient for ovary development and is a key downstream component of the TSD pathway [Lance, 2009; Ramsey and Crews, 2009]. Like aromatase, FOXL2 is expressed at a higher level in gonads at female determining temperatures in TSD species (see references in table 1), which is in accord with its role in granulosa cell development in mammals [Crisponi et al., 2001; Schmidt et al., 2004].
The picture is less consistent when we examine expression of estrogen receptor α. There is a temperature effect on ERα mRNA levels in some species but not others (see references in table 1). Moreover, the direction of the temperature effect can vary from higher expression at female-determining temperatures to higher expression at male-determining temperatures [Rhen et al., 2007; Ramsey and Crews, 2009]. There are also developmental changes in ERα expression at a given temperature. Temperature effects on estrogen receptor β have only been examined in one species to date: ERβ expression varies during development, generally increasing as the ovaries differentiate [Ramsey and Crews, 2009]. Although there is subtle variation in the pattern and timing of temperature effects, the bottom line is that estrogen receptors are expressed in gonads at both male and female temperatures, which is in harmony with the finding that embryos at male temperatures are responsive to exogenous estrogens.
A recent discovery is that temperature influences androgen receptor (AR) mRNA levels in at least 2 TSD turtles [Rhen et al., 2007; Ramsey and Crews, 2009]. Expression of AR is higher in gonads from embryos incubated at female-determining temperatures than in embryos at male-determining temperatures. At first glance, this seems like a strange finding because androgens and AR are well-known for regulating sexual differentiation and reproduction in male vertebrates. Yet, it is becoming clear that a fine balance of androgen signaling is vital for normal ovary development. Androgen excess is a cardinal feature of polycystic ovary syndrome in women [Ehrmann et al., 1995]. Female rodents, sheep, and primates treated prenatally or postnatally with dihydrotestosterone develop a phenotype that mimics polycystic ovary syndrome [Abbott et al., 2005; Xita and Tsatsoulis, 2006]. On the other hand, a minimal level of androgen signaling is required for full fertility in females: mice lacking AR suffer premature ovarian failure and are less fertile than wild type mice [Hu et al., 2004; Shiina et al., 2006; Walters et al., 2007]. Likewise, X chromosome abnormalities at or near the AR locus in humans are associated with premature ovarian failure [Kimura et al., 2007]. Other studies demonstrate that androgens promote early follicular growth in various species. Thus, elevated AR expression at female-producing temperatures may be important for ovary formation in TSD species. Indeed, dihydrotestosterone, a non-aromatizable androgen, can feminize snapping turtle embryos [Rhen and Lang, 1994; Rhen, unpublished data].
We now move from genes associated with ovary development to genes that may play a role in testis formation. Two genes, DMRT1 and SOX9, are expressed at higher levels in gonads at male-determining temperatures versus gonads at female-determining temperatures in several TSD species. This is consistent with their role in testis development in mammals [Kim et al., 2007; Sekido and Lovell-Badge, 2009]. A recent study examined the platelet-derived growth factor (PDGF) signaling system [Rhen et al., 2009], which is involved in embryonic and postnatal development of testes in mammals and birds [Gnessi et al., 2000; Brennan et al., 2003; Smith et al., 2005]. Expression of PDGF-B mRNA was higher in gonads at a male-determining temperature than in gonads at a female-determining temperature in the common snapping turtle, a species with TSD [Rhen et al., 2009]. In contrast, gonadal expression of PDGF-A, a paralog of PDGF-B, was not influenced by incubation temperature. It is interesting that PDGF-A expression, rather than PDGF-B expression, is sexually dimorphic in mouse and chicken embryos [Brennan et al., 2003; Smith et al., 2005]. Even though expression patterns of the 2 ligands have evolved, the PDGF signaling system still seems to play a conserved role in testis formation in vertebrates. Expression of the receptors for these ligands, PDGFR-α and PDGFR-β, was not affected by embryonic temperature. However, expression of the PDGF-B ligand alone may be sufficient for temperature-dependent activation of this signaling pathway, as described above for aromatase, estrogen, and estrogen receptors. These results support the idea that DMRT1, PDGF-B, and SOX9 are involved in testis development in reptiles.
Lance [2009] identified a shortcoming of such studies: a candidate gene approach that depends on homologs of mammalian sex-determining genes will not identify new or unique sex-determining genes in reptiles. Shoemaker and Crews [2009] pointed out another failing: gene expression profiling studies are purely descriptive and do not test gene function. Just because a gene is expressed in a tissue at the correct time does not mean it is necessary for development of that tissue. Conversely, just because a gene is not differentially expressed between incipient testes and ovaries does not mean it is not involved in sex determination. Gene function can be regulated by post-transcriptional mechanisms (i.e., alternative splicing) and post-translational modifications (i.e., protein cleavage, phosphorylation, acetylation, glycosylation, and ubiquitination to name a few). Finally, expression profiling alone cannot reveal functional interactions between genes because co-expressed genes may not be linked in the same gene regulatory network. We clearly need to use different methods to reveal novel candidate genes, to test the hypothesis that these genes are involved in sex determination, and to elucidate functional interactions among sex-determining genes. Next, we present some approaches that have the potential to accomplish these aims.
Identification of New Candidate Sex-Determining Genes
While it certainly makes sense to study homologs of genes first identified in mammals, researchers investigating sex determination in reptiles should not limit themselves to this approach. First, there is no guarantee that the primary sex-determining genes in reptiles will be orthologous to sex-determining genes in mammals. Such genes could be entirely unique to reptiles. This scenario would be analogous to SRY evolution in therian mammals [Wallis et al., 2008]. Alternatively, the primary sex-determining genes in reptiles could have mammalian orthologs, but those genes may play no part in sex determination in mammals. Thus, it is possible that we would never discover a primary sex-determining gene in reptiles. Second, by waiting for the discovery of bona fide sex-determining genes in mammals, we limit the rate of discovery of candidate sex-determining genes in reptiles. Wilhelm et al. [2007] list ∼30 genes that produce some sort of gonadal phenotype when mutated in mammals. These genes represent a tiny fraction of the ∼24,000 genes in the mammalian genome. Functional characterization of all these genes via reverse genetics in mice will be a very slow process. As a result, identification of new sex-determining genes in mammals (and reptiles) will be sporadic.
Fortunately, there are a large number of techniques for unbiased screening of the transcriptome and proteome. Older methods for identifying differentially expressed genes are relatively inexpensive and simple to use (i.e., differential display PCR; suppressive subtractive hybridization), while newer methods tend to be more expensive and require specialized equipment and expertise (i.e., construction and sequencing of a cDNA library to produce cDNA or oligonucleotide microarrays; transcriptome characterization using next generation sequencing; 2D protein electrophoresis combined with matrix-assisted laser desorption/ionization). There are advantages and disadvantages to each method beyond expense and technical difficulty, but we leave it to the reader to explore these by reading the literature and approaching colleagues that use (or have used) these methods. Here we provide examples of novel candidates that were discovered via unbiased screens.
We have used differential display PCR to identify genes that are differentially expressed in gonads early in the temperature-sensitive period in the common snapping turtle. Eggs were collected and incubated as previously described [Rhen et al., 2007, 2009]. In brief, we dissected gonads from embryos incubated at 26.5°C (a male-producing treatment) and from siblings that were first incubated at 26.5°C and then shifted to 31°C for 6 days beginning at stage 16 of development (a female-producing treatment). We sampled embryos from both thermal regimes on days 2, 3, 4, and 5 of the shift and stored gonads in RNAlater solution (Ambion, Austin, Tex., USA). We extracted and analyzed total RNA from gonad pairs isolated from individual embryos as previously described [Rhen et al., 2007, 2009]. Total RNA (100 ng) from each pair of gonads was reverse transcribed using a one-base anchored oligo-dT primer (T11M, where M can be G, A, or C) as per the manufacturer's instructions (RNAimage Kit; GenHunter, Nashville, Tenn., USA). This cDNA was then used as template for PCR with the same oligo-dT primer and an arbitrary 13mer as the forward primer (RNAimage Kit). We ran PCR products on 6% polyacrylamide gels in TBE buffer for approximately 2 h at 60 V (Invitrogen, Carlsbad, Calif., USA). After electrophoresis, SYBR Gold was used to stain gels and resolve differentially expressed PCR bands (rather than radiolabeling during PCR). We ran 3 biological replicates for each of the 8 groups (2 temperatures × 4 days) on the same gel and measured optical density of PCR products using LabWorks software on the AutoChemi gel visualization system (UVP, Upland, Calif., USA). We used univariate and multivariate ANOVA to identify bands that were differentially expressed between temperatures. Intra-assay coefficients of variation for band intensity ranged from 6 to 16% with 3 biological replicates in each group. Differentially expressed bands were cut out of the gel, extracted, and re-amplified as per the manufacturer's instructions (RNAimage Kit). Bands were cloned and sequenced as previously described [Rhen et al., 2007, 2009]. This approach is significantly more rigorous than most differential display PCR studies because prior studies did not use biological replicates (i.e., most studies only compare one treatment sample vs. one control sample, which leads to a very high rate of false positives).
So far, we have used our method to clone cDNA fragments for nearly 50 candidate TSD genes. We have confirmed that temperature influences expression of these genes using independent biological samples and quantitative PCR using the methods described in Rhen et al. [2007, 2009]. Interestingly, 18% of the genes display some homology to sex-linked genes in mammals or birds, even though there are no sex chromosomes in snapping turtles. Such genes may play a conserved role in gonad development and may have evolved sex linkage in mammals or birds as a result of this role. A few other genes also deserve mention. Calreticulin expression was significantly higher at a male-versus a female-determining temperature, which is consistent with a putative role in spermatogenesis in rats [Nakamura et al., 1993]. It is even more significant that calreticulin is expressed in sperm in C. elegans and that calreticulin null-mutants display temperature-dependent reproductive defects [Park et al., 2001]. Temperature influences expression of the snapping turtle proteasome 26S subunit, ATPase, 3 (PSMC3). Altered Psmc3 expression in mouse testes is associated with male infertility [Rockett et al., 2004]. Expression of the turtle ribosomal protein L13a also differed between temperatures. This gene is involved in the regulation of translation [Mazumder et al., 2003], which may be noteworthy because the first gene we isolated with differential display PCR also regulates translation.
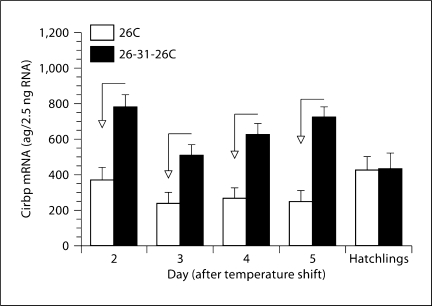
We cloned and sequenced the full-length cDNA for this gene using the 5′ RACE System for Rapid Amplification of cDNA Ends (Invitrogen, Carlsbad, Calif., USA) and definitively identify it as cold-inducible RNA-binding protein (CIRBP). This is significant because CIRBP expression is influenced by temperature in other species and because it regulates expression of other genes via translational repression [Nishiyama et al., 1997; De Leeuw et al., 2007]. Moreover, CIRBP physically interacts with an X-linked RNA binding protein (RBMX) [Rual et al., 2005], which has a Y-linked allele (RBMY) crucial for sperm development in mammals [Elliott, 2004]. Hence, CIRBP may regulate expression or activity of other sex-determining genes in the snapping turtle. We measured CIRBP mRNA expression using the methods described in Rhen et al. [2007, 2009]. CIRBP mRNA was differentially expressed early in the sex-determining period (fig. 1), but in opposition to DMRT1 [Rhen et al., 2007]. Differential expression of CIRBP preceded temperature effects on aromatase, ERα, AR, and SOX9 expression. We are conducting additional studies of this gene to explore its potential role in TSD in the snapping turtle.
Fig. 1.
Cold-inducible RNA binding protein (CIRBP) expression in gonads from snapping turtle embryos and hatchlings. One group of eggs was incubated at 26.5°C throughout development, which produces only males (26C). Another group of eggs was incubated at 26.5°C until stage 16, shifted to a female-producing temperature of 31°C for 6 days, and returned to 26.5°C for the remainder of development (26-31-26C). This brief exposure to 31°C is enough to produce exclusively females. Levels of CIRBP mRNA are least squares means (± 1 SE) for each temperature regime and time point. Arrows indicate a difference in CIRBP expression between incubation temperatures.
Our finding that 2 translational regulators, ribosomal protein L13a and CIRBP were differentially expressed at male-versus female-determining temperatures is particularly interesting in light of an early report by Harry et al. [1990]. These authors used antibodies to study temperature-dependent expression of heat shock protein 70 and heat shock protein 90 in the loggerhead sea turtle, a TSD species. Expression of these proteins was similar in gonads from embryos incubated at male- and female-determining temperatures. However, antibodies cross-reacted with other proteins that were differentially expressed between incubation temperatures. The authors went on to identify these proteins as heterogeneous nuclear ribonucleoprotein particles (hnRNPs). These proteins are generally involved in mRNA processing, including alternative splicing of transcripts [Smith and Valcárcel, 2000; Cáceres et al., 2002]. Although Harry et al. [1990] were motivated by the specific hypothesis that heat shock proteins are involved in TSD, their results highlight the potential for proteomics analyses to reveal novel genes that are differentially expressed.
Functional Analysis of Sex-Determining Genes
The list of candidate sex-determining genes in reptiles is growing and will lengthen with the application of genome-wide screens. Genes that are differentially expressed at male-versus female-producing temperatures are likely to be involved in TSD, either as the thermosensitive gene itself, as direct targets of such a gene, or as downstream components of a larger gene regulatory network. However, some candidate genes may not be involved in TSD per se. We must therefore distinguish genes that are causally related to TSD from genes whose expression is only correlated with TSD. Several approaches could be used to distinguish these genes, including linkage studies, gene perturbation studies (i.e., knockouts, transient knockdowns, and ectopic gene expression), and pharmacological manipulations. Unfortunately, maps of molecular markers or visible phenotypes have not been constructed in any reptile. Long generation times hamper such studies in turtles, snakes, and crocodilians, although some lizards mature in their first year. A careful screening of such species for other key traits like high fecundity, small body size, and ease of captive breeding may eventually lead to the first model species for reptile genetics.
In the short term, however, the most promising route for functional studies of specific genes is to manipulate their expression via RNA interference and transfection of expression vectors in cultured embryonic gonads (i.e., knock down and over-express genes in gonads at male- and female-producing temperatures to determine if we can alter their developmental fate). Methods are available for studying fetal and neonatal mouse gonads in culture [Eppig and O'Brien, 1996; Livera et al., 2006]. These methods have been extended to the study of TSD in gonads from sea turtle embryos [Moreno-Mendoza et al., 2001]. Yao et al. [2004] also used gonad explants and gonad-mesonephros chimeras to reveal that cell migration from the mesonephros into the gonads is not required for testis cord formation in turtles. Using procedures similar to Moreno-Mendoza et al. [2001], we have cultured gonads from snapping turtle embryos for up to 6 days and introduced fluorescent siRNA in a dose-dependent manner into gonads [Rhen and Schroeder, unpublished data]. Such studies suggest that we may be able to carry out functional studies of candidate TSD genes. At this time, however, we are not able to induce gonad differentiation in culture. Development of such an in vitro system will require extensive optimization. Nevertheless, these studies highlight the potential power of studying gonad development in organ culture.
An older but powerful technique is pharmacological manipulation of signaling pathways in ovo. One can treat individual embryos with almost any small molecule by dissolving it in the appropriate solvent and placing a drop of the solvent (i.e., 5 μl) on the eggshell. The compound is presumably carried through the shell, absorbed by the chorioallantoic vasculature, and transported to the embryo. Crews and colleagues [1991] pioneered this procedure to administer estrogens to the developing embryo. Many researchers subsequently used the technique to study the impact of other steroids, steroid receptor antagonists, and steroidogenic enzyme inhibitors on sex determination. This approach has recently been used to deliver busulfan to ablate germ cells within the developing gonad [Dinapoli and Capel, 2007]. A general caveat for any study using pharmacological agonists and inhibitors is that these compounds may activate or inhibit related signaling pathways. Extra care should be taken in the design of experimental controls when compounds developed for mammals are used in reptiles. Finally, one should validate the assumption that compounds reach the embryo at concentrations that reflect the amount applied to the eggshell [Muller et al., 2007].
Modeling the Gene Regulatory Network for Sex Determination
Functional characterization of individual genes is clearly one of the biggest challenges facing researchers that study reptiles. An exciting new approach that deserves special consideration is modeling of gene regulatory networks. The most complete models integrate a vast array of data, including a list of all genes expressed in the cell or tissue of interest, the molecular function of those genes, information on cis regulatory elements and trans regulatory factors, and gene expression profiles after numerous gene perturbation experiments [Bolouri and Davidson, 2002; Friedman, 2004]. However, it is also possible to develop models from time series data for a smaller number of genes with fewer experimental perturbations [Bansal et al., 2006, 2007]. A variety of statistical techniques, including Bayesian analyses and structural equation modeling, are used to develop explicit models of gene regulatory networks [Rockman, 2008]. Here, we use temperature-dependent expression profiles for 3 genes and a simple statistical method to illustrate one method for modeling gene regulatory networks.
We previously observed that aromatase and FOXL2 expression increased in snapping turtle embryos shifted from a male- to a female-determining temperature [Rhen et al., 2007]. Androgen receptor (AR) expression also increased in embryos shifted to the female-determining temperature, but the increase was delayed one day relative to the increase in aromatase and FOXL2 expression [Rhen et al., 2007]. We reanalyzed the data from that study and found simple correlations among these genes were very high at the female-producing temperature (aromatase-AR ρ = 0.75, p < 0.0001, n = 42; aromatase-FOXL2 ρ = 0.64, p < 0.0001, n = 42; AR-FOXL2 ρ = 0.68, p < 0.0001, n = 44), but were not significant at the male-producing temperature (aromatase-AR ρ = 0.22, p = 0.18, n = 41; aromatase-FOXL2 ρ = 0.31, p = 0.052, n = 41; AR-FOXL2 ρ = 0.25, p = 0.12, n = 41). These patterns suggest that aromatase and FOXL2 might regulate each other and that one or both genes might be upstream of AR in the ovary-determining pathway. Yet, a simple form of structural equation modeling suggested just the opposite: the partial correlation between aromatase and FOXL2 expression at the female-determining temperature was not significant after controlling for AR expression (ρ = 0.26, p = 0.10, n = 42). Moreover, the partial correlations between AR and aromatase (ρ = 0.56, p = 0.0001, n = 42) and between AR and FOXL2 were still significant (ρ = 0.40, p = 0.0007, n = 44). This suggests that AR independently regulates aromatase and FOXL2.
We tested this hypothesis by incubating snapping turtle eggs at a temperature (28.2°C) that produces a mixed sex ratio and treating embryos with dihydrotestosterone (an androgen that cannot be aromatized) and/or flutamide (an AR antagonist). We used the same procedures for collecting and incubating eggs and administering hormones as described in Rhen and Lang [1994]. In brief, equal or approximately equal numbers of viable eggs from each clutch were assigned to one of 4 hormone treatments to control for clutch differences. Eggs were placed in containers filled with moist vermiculite and then randomly positioned within foam box incubators set at 28.2°C. A few eggs were sampled periodically to monitor development and make sure treatments were applied at the correct stage. Upon reaching stage 15, eggs received one of 4 treatments. The vehicle treated controls received 5 μl of 100% ethanol dropped onto the surface of the eggshell. Dihydrotestosterone (50 μg) and/or flutamide (100 μg) were dissolved in 5 μl of 100% ethanol and dropped onto the surface of the eggshell. As described above, this is a well-established method of delivering hormones to embryos that develop inside eggs. Gonads from equal numbers of embryos were collected at stage 15, 16, 17, and 18 and placed in RNAlater solution (Ambion, Austin, Tex., USA). Total RNA was extracted from gonads and aromatase and FOXL2 mRNA levels measured using reverse transcription and quantitative PCR exactly as described in Rhen et al. [2007, 2009]. In addition, a subset of eggs was allowed to hatch to determine the effect of hormone treatments on sex ratios.
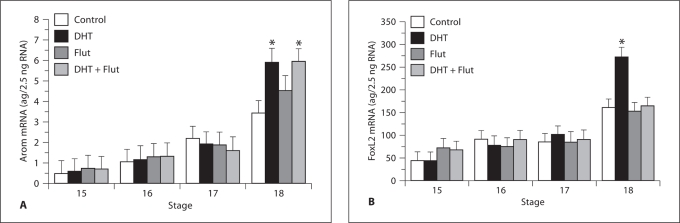
Administration of dihydrotestosterone to snapping turtle eggs significantly increased the proportion of hatchlings that were female (98%, n = 45) compared to controls (66% females, n = 44), replicating results from Rhen and Lang [1994]. Consistent with its feminizing effect on sex ratio, dihydrotestosterone increased aromatase mRNA and FOXL2 mRNA expression in embryonic gonads (fig. 2). Flutamide feminized sex ratio in some families but masculinized sex ratio in others, suggesting this compound can act as an AR agonist or antagonist in the snapping turtle. In fact, flutamide acted as a partial agonist to weakly induce aromatase mRNA (fig. 2A), but completely antagonized dihydrotestosterone induction of FOXL2 mRNA (fig. 2B). It is noteworthy that dihydrotestosterone and flutamide did not influence gene expression until stage 18, which corresponds to the stage when AR levels increased in embryos shifted to a female-producing temperature [Rhen et al., 2007]. These findings demonstrate that androgens and AR regulate aromatase and FOXL2 expression in embryonic gonads and that correlated expression of aromatase and FOXL2 is due, in part, to the pleiotropic effects of AR (a transcription factor). Androgens have been shown to regulate aromatase in other species, but the androgen-AR-FOXL2 pathway is a novel regulatory interaction that has never been described in any species. Yet, observations in mice suggest that this may be a conserved regulatory module: inactivation of AR leads to premature ovarian failure in female mice, as does inactivation of FOXL2 (i.e., mutations in different genes in the same biochemical pathway produce the same phenotype).
Fig. 2.
A Aromatase and B FOXL2 mRNA expression in gonads from snapping turtle embryos incubated at 28.2°C. Embryos were allocated to 1 of 4 groups at stage 15, including non-treated and vehicle-treated controls, a dihydrotestosterone- (DHT) treated group, a flutamide- (Flut) treated group, and a group treated with both DHT and Flut. Vehicle controls received 5 μl of 100% ethanol spotted on the surface of the eggshell. Dihydrotestosterone (50 μg) and/or flutamide (100 μg) were dissolved in 5 μl of 100% ethanol and spotted on the surface of the eggshell. Embryos were sampled at developmental stages 15–18 after treatment. Levels of aromatase and FOXL2 mRNA are least square means (± 1 SE) for each treatment group and developmental stage. Asterisks indicate a significant difference between the treated versus control groups.
Although this was a very simple example of how to model a gene regulatory network, it highlights the potential for techniques like path analysis and structural equation modeling to reveal novel regulatory relationships among sex-determining genes. However, it is important to note that these gene regulatory models are hypothetical and that they require experimental testing before firm conclusions are drawn.
Conclusions
Fundamental similarities in gonad development are evident at the genetic, molecular, cellular, developmental, and physiological levels in all vertebrates. Many, but not all, genes appear to play a conserved role in sex determination and morphogenesis of testes and ovaries. It is therefore reasonable to hypothesize that additional features of sex determination are conserved and that further study of orthologs of mammalian sex-determining genes are in order. However, researchers studying sex determination in reptiles should not be limited to this approach. We present a variety of methods that can be used to discover novel candidate genes at a genome-wide scale. We present specific examples of novel genes that we discovered using an unbiased screen for genes that are differentially expressed between male- and female-producing temperatures. Researchers should also endeavor to test the hypothesis that these genes are involved in sex determination. We present several experimental techniques that can be used to test gene function and to develop explicit models of the gene regulatory networks underlying testis and ovary formation. We must become more adventurous if we truly hope to identify primary sex-determining genes in reptiles with TSD or GSD.
Acknowledgements
This work was supported by Grant 5R21HD049486 from the National Institute of Child Health and Human Development to T. Rhen. We also wish to thank the Minnesota Department of Natural Resources for providing special permits for collection of snapping turtle eggs.
References
- Abbott DH, Barnett DK, Bruns CM, Dumesic DA. Androgen excess fetal programming of female reproduction: A developmental aetiology for polycystic ovary syndrome? Hum Reprod Update. 2005;11:357–374. doi: 10.1093/humupd/dmi013. [DOI] [PubMed] [Google Scholar]
- Anand A, Patel M, Lalremruata A, Singh AP, Agrawal R, et al. Multiple alternative splicing of Dmrt1 during gonadogenesis in Indian mugger, a species exhibiting temperature-dependent sex determination. Gene. 2008;425:56–63. doi: 10.1016/j.gene.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- Austin HB. Extended production of the Mullerian duct regressor in the American alligator. Gen Comp Endocrinol. 1994;96:122–128. doi: 10.1006/gcen.1994.1164. [DOI] [PubMed] [Google Scholar]
- Bansal M, Gatta GD, di Bernardo D. Inference of gene regulatory networks and compound mode of action from time course gene expression profiles. Bioinformatics. 2006;22:815–822. doi: 10.1093/bioinformatics/btl003. [DOI] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, Di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske LA, Capel B. Blurring the edges in vertebrate sex determination. Curr Opin Genet Dev. 2008;18:499–505. doi: 10.1016/j.gde.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JM, Gahr M, Horan K, Wibbels T, Crews D. Cloning and in situ hybridization analysis of estrogen receptor in the developing gonad of the red-eared slider turtle, a species with temperature-dependent sex determination. Dev Growth Differ. 1998;40:243–254. doi: 10.1046/j.1440-169x.1998.00013.x. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Modeling transcriptional regulatory networks. BioEssays. 2002;24:1118–1129. doi: 10.1002/bies.10189. [DOI] [PubMed] [Google Scholar]
- Brennan J, Tilmann C, Capel B. Pdgfr-α mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Sex determination in reptiles. Q Rev Biol. 1980;55:3–21. [Google Scholar]
- Bull JJ. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin-Cummings; 1983. [Google Scholar]
- Bull JJ, Vogt RC, Bulmer MG. Heritability of sex ratio in turtles with environmental sex determination. Evolution. 1982;36:333–341. doi: 10.1111/j.1558-5646.1982.tb05049.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Hillis DM, O'Steen S. Mammalian ZFY sequences exist in reptiles regardless of sex-determining mechanism. Science. 1988;242:567–569. doi: 10.1126/science.3140382. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Kornblihtt AR. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- Callebaut M, Van Nassauw L, Harrisson F. Comparison between oogenesis and related ovarian structures in a reptile, Pseudemys scripta elegans (turtle) and in a bird Coturnix coturnix japonica (quail) Reprod Nutr Dev. 1997;37:233–252. doi: 10.1051/rnd:19970301. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Sreenivasulu K, Raman R. Involvement of androgen receptor gene in male gonad differentiation in Indian garden lizard, Calotes versicolor. Mol Cell Endocrinol. 2009;303:100–106. doi: 10.1016/j.mce.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Cline TW. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. Vive la différence: males vs females in flies vs worms. Ann Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Crews D. Temperature-dependent sex determination: the interplay of steroid hormones and temperature. Zool Sci. 1996;13:1–13. doi: 10.2108/zsj.13.1. [DOI] [PubMed] [Google Scholar]
- Crews D, Bull JJ, Wibbels T. Estrogen and sex reversal in turtles: a dose-dependent phenomenon. Gen Comp Endocrinol. 1991;81:357–364. doi: 10.1016/0016-6480(91)90162-y. [DOI] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Deeming DC. Prevalence of TSD in crocodilians. In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 33–41. [Google Scholar]
- Desvages G, Girondot M, Pieau C. Sensitive stages for the effects of temperature on gonadal aromatase activity in embryos of the marine turtle Dermochelys coriacea. Gen Comp Endocrinol. 1993;92:54–61. doi: 10.1006/gcen.1993.1142. [DOI] [PubMed] [Google Scholar]
- Dinapoli L, Capel B. Germ cell depletion does not alter the morphogenesis of the fetal testis or ovary in the red-eared slider turtle (Trachemys scripta) J Exp Zool B Mol Dev Evol. 2007;308:236–241. doi: 10.1002/jez.b.21136. [DOI] [PubMed] [Google Scholar]
- Ditewig AC, Yao HHC. Organogenesis of the ovary: A comparative review on vertebrate ovary formation. Organogenesis. 2005;2:36–41. doi: 10.4161/org.2.2.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddamani LS. Differentiation and development of testis in the oviparous lizard, Calotes versicolor (Daud.) J Exp Zool A: Comp Exp Biol. 2006;305:299–308. doi: 10.1002/jez.a.265. [DOI] [PubMed] [Google Scholar]
- Ehrmann DA, Barnes RB, Rosenfield RL. Polycystic ovary syndrome as a form of functional ovarian hyperandrogenism due to dysregulation of androgen secretion. Endocr Rev. 1995;16:322–353. doi: 10.1210/edrv-16-3-322. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. 2004;27:328–334. doi: 10.1111/j.1365-2605.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Endo D, Kanaho Y, Park MK. Expression of sex steroid hormone-related genes in the embryo of the leopard gecko. Gen Comp Endocrinol. 2008;155:70–78. doi: 10.1016/j.ygcen.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O'Brien MJ. Development in vitro of mouse oocytes from primordial follicles. Biol Reprod. 1996;54:197–207. doi: 10.1095/biolreprod54.1.197. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Quinn AE, Sarre SD, O'Meally D, Georges A, Marshall Graves JA. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 2009;17:91–98. doi: 10.1007/s10577-008-9019-5. [DOI] [PubMed] [Google Scholar]
- Fahrioglu U, Murphy MW, Zarkower D, Bardwell VJ. mRNA expression analysis and the molecular basis of neonatal testis defects in Dmrt1 mutant mice. Sex Dev. 2006;1:42–58. doi: 10.1159/000096238. [DOI] [PubMed] [Google Scholar]
- Fleming A, Crews D. Estradiol and incubation temperature modulate regulation of steroidogenic factor 1 in the developing gonad of the red-eared slider turtle. Endocrinology. 2001;142:1403–1411. doi: 10.1210/endo.142.4.8043. [DOI] [PubMed] [Google Scholar]
- Fleming A, Wibbels T, Skipper JK, Crews D. Developmental expression of steroidogenic factor 1 in a turtle with temperature-dependent sex determination. Gen Comp Endocrinol. 1999;116:336–346. doi: 10.1006/gcen.1999.7360. [DOI] [PubMed] [Google Scholar]
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Ukeshima A, Miyayama Y. Observations of primordial germ cells in the turtle embryo (Caretta caretta): light and electron microscopic studies. Dev Growth Differ. 1979;21:3–10. doi: 10.1111/j.1440-169X.1979.00003.x. [DOI] [PubMed] [Google Scholar]
- Girling JE. The reptilian oviduct: A review of structure and function and directions for future research. J Exp Zool. 2002;293:141–170. doi: 10.1002/jez.10105. [DOI] [PubMed] [Google Scholar]
- Gnessi L, Basciani S, Mariani S, Arizzi M, Spera G, et al. Leydig cell loss and spermatogenic arrest in platelet-derived growth factor (PDGF)-A-deficient mice. J Cell Biol. 2000;149:1019–1025. doi: 10.1083/jcb.149.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum E, Carr JL. Sexual differentiation in the spiny softshell turtle (Apalone spinifera), a species with genetic sex determination. J Exp Zool. 2001;290:190–200. doi: 10.1002/jez.1049. [DOI] [PubMed] [Google Scholar]
- Harlow PS. Temperature-dependent sex determination in lizards. In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 42–52. [Google Scholar]
- Harrison DA. Sex determination: controlling the master. Curr Biol. 2007;17:R328–R330. doi: 10.1016/j.cub.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Harry JL, Williams KL, Briscoe DA. Sex determination in loggerhead turtles: differential expression of two hnRNP proteins. Development. 1990;109:305–312. doi: 10.1242/dev.109.2.305. [DOI] [PubMed] [Google Scholar]
- Hu YC, Wang PH, Yeh S, Wang RS, Xie C, et al. Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc Natl Acad Sci USA. 2004;101:11209–11214. doi: 10.1073/pnas.0404372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert J. Localisation extra-embryonnaire des gonocytes chez l'embryon d'orvet (Anguis fragilis L.) Arch Anat Microsc Morphol Exp. 1971;60:261–268. [PubMed] [Google Scholar]
- Hubert J. Ultrastructure des ebauches gonadiques du lezard vivipare (Lacerta vivipara Jacquin) pendant le periode de colonization par les gonocytes. Arch Anat Microsc Morphol Exp. 1976;65:241–254. [PubMed] [Google Scholar]
- Janes DE, Wayne ML. Evidence for a genotype X environment interaction in sex-determining response to incubation temperature in the leopard gecko, Eublepharis macularius. Herpetologica. 2006;62:56–62. [Google Scholar]
- Janzen FJ. Heritable variation for sex ratio under environmental sex determination in the common snapping turtle (Chelydra serpentina) Genetics. 1992;131:155–161. doi: 10.1093/genetics/131.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen FJ, Paukstis GL. Environmental sex determination in reptiles: Ecology, evolution, and experimental design. Q Rev Biol. 1991;66:149–179. doi: 10.1086/417143. [DOI] [PubMed] [Google Scholar]
- Janzen FJ, Krenz JG. Phylogenetics: Which was first, TSD or GSD? In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 121–130. [Google Scholar]
- Jeyasuria P, Place AR. Embryonic brain-gonadal axis in temperature-dependent sex determination of reptiles: A role for P450 aromatase (CYP19) J Exp Zool. 1998;281:428–449. [PubMed] [Google Scholar]
- Kettlewell JR, Raymond CS, Zarkower D. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis. 2000;26:174–178. [PubMed] [Google Scholar]
- Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Matsumoto T, Matsuyama R, Shiina H, Sato T, et al. Androgen receptor function in folliculogenesis and its clinical implication in premature ovarian failure. Trends Endocrinol Metab. 2007;18:183–189. doi: 10.1016/j.tem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Korpelainen H. Sex ratios and conditions required for environmental sex determination in animals. Biol Rev Camb Phil Soc. 1990;65:147–184. doi: 10.1111/j.1469-185x.1990.tb01187.x. [DOI] [PubMed] [Google Scholar]
- Lance VA. Is regulation of aromatase expression in reptiles the key to understanding temperature-dependent sex determination? J Exp Zool A Ecol Genet Physiol. 2009;311:314–322. doi: 10.1002/jez.465. [DOI] [PubMed] [Google Scholar]
- Lang JW, Andrews HV. Temperature-dependent sex determination in crocodilians. J Exp Zool. 1994;270:28–44. [Google Scholar]
- Livera G, Delbes G, Pairault C, Rouiller-Fabre V, Habert R. Organotypic culture, a powerful model for studying rat and mouse fetal testis development. Cell Tissue Res. 2006;324:507–521. doi: 10.1007/s00441-006-0167-7. [DOI] [PubMed] [Google Scholar]
- Lopez AJ. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- Lovern MB, Holmes MM, Wade J. The green anole (Anolis carolinensis): A reptilian model for laboratory studies of reproductive morphology and behavior. ILAR J. 2004;45:54–64. doi: 10.1093/ilar.45.1.54. [DOI] [PubMed] [Google Scholar]
- Marín I, Baker BS. The evolutionary dynamics of sex determination. Science. 1998;281:1990–1994. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, et al. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Merchant-Larios H, Ruiz-Ramirez S, Moreno-Mendoza N, Marmolejo-Valencia A. Correlation among thermosensitive period, estradiol response, and gonad differentiation in the sea turtle Lepidochelys olivacea. Gen Comp Endocrinol. 1997;107:373–385. doi: 10.1006/gcen.1997.6946. [DOI] [PubMed] [Google Scholar]
- Moore BC, Uribe-Aranzábal MC, Boggs ASP, Guillette LJ., Jr Developmental morphology of the neonatal alligator (Alligator mississippiensis) ovary. J Morphol. 2008;269:302–312. doi: 10.1002/jmor.10583. [DOI] [PubMed] [Google Scholar]
- Moore MC, Hews DK, Knapp R. Hormonal control and evolution of alternative male phenotypes: Generalizations of models for sexual differentiation. Am Zool. 1998;38:133–151. [Google Scholar]
- Moreno-Mendoza N, Harley VR, Merchant-Larios H. Temperature regulates SOX9 expression in cultured gonads of Lepidochelys olivacea, a species with temperature sex determination. Dev Biol. 2001;229:319–326. doi: 10.1006/dbio.2000.9952. [DOI] [PubMed] [Google Scholar]
- Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- Muller JK, Gross TS, Borgert CJ. Topical dose delivery in the reptilian egg treatment model. Env Toxicol Chem. 2007;26:914–919. doi: 10.1897/06-290r.1. [DOI] [PubMed] [Google Scholar]
- Murdock C, Wibbels T. Cloning and expression of aromatase in a turtle with temperature-dependent sex determination. Gen Comp Endocrinol. 2003a;130:109–119. doi: 10.1016/s0016-6480(02)00573-7. [DOI] [PubMed] [Google Scholar]
- Murdock C, Wibbels T. Expression of Dmrt1 in a turtle with temperature-dependent sex determination. Cytogenet Genome Res. 2003b;101:302–308. doi: 10.1159/000074353. [DOI] [PubMed] [Google Scholar]
- Murdock C, Wibbels T. Dmrt1 expression in response to estrogen treatment in a reptile with temperature-dependent sex determination. J Exp Zool B Mol Dev Evol. 2006;306:134–139. doi: 10.1002/jez.b.21076. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Moriya M, Baba T, Michikawa Y, Yamanobe T, et al. An endoplasmic reticulum protein, calreticulin, is transported into the acrosome of rat sperm. Exp Cell Res. 1993;205:101–110. doi: 10.1006/excr.1993.1063. [DOI] [PubMed] [Google Scholar]
- Nelson NJ, Cree A, Thompson MB, Keall SN, Daucherty CH. Temperature-dependent sex determination in tuatara. In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 53–58. [Google Scholar]
- Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, Janes DE. Evolution of sex chromosomes in Sauropsida. Int Comp Biol. 2008;48:512–519. doi: 10.1093/icb/icn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Lee DG, Yu JR, Jung SK, Choi K, et al. Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2835–2845. doi: 10.1091/mbc.12.9.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieau C, Dorizzi M. Oestrogens and temperature-dependent sex determination in reptiles: all is in the gonads. J Endocrinol. 2004;181:367–377. doi: 10.1677/joe.0.1810367. [DOI] [PubMed] [Google Scholar]
- Place AR, Lance VA. The temperature-dependent sex determination drama: same cast, different stars. In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 99–110. [Google Scholar]
- Place AR, Lang J, Gavasso S, Jeyasuria P. Expression of P450arom in Malaclemys terrapin and Chelydra serpentina: A tale of two sites. J Exp Zool. 2001;290:673–690. doi: 10.1002/jez.1118. [DOI] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: Are sex chromosomes an evolutionary trap? Zool J Linnean Soc. 2009;156:168–183. [Google Scholar]
- Pudney J. Spermatogenesis in nonmammalian vertebrates. Microsc Res Techn. 1995;32:459–497. doi: 10.1002/jemt.1070320602. [DOI] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JAM. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Quinn AE, Radder RS, Sarre SD, Georges A, Ezaz T, Shine R. Isolation and development of a molecular sex marker for Bassiana duperreyi, a lizard with XX/XY sex chromosomes and temperature-induced sex reversal. Mol Gen Genomics. 2009;281:665–672. doi: 10.1007/s00438-009-0437-7. [DOI] [PubMed] [Google Scholar]
- Radder RS, Quinn AE, Georges A, Sarre SD, Shine R. Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol Lett. 2008;4:176–178. doi: 10.1098/rsbl.2007.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey M, Crews D. Adrenal-kidney-gonad complex measurements may not predict gonad-specific changes in gene expression patterns during temperature-dependent sex determination in the red-eared slider turtle (Trachemys scripta elegans) J Exp Zool Part A. 2007;307:463–470. doi: 10.1002/jez.399. [DOI] [PubMed] [Google Scholar]
- Ramsey M, Crews D. Steroid signaling and temperature-dependent sex determination – reviewing the evidence for early action of estrogen during ovarian determination in turtles. Sem Cell Dev Biol. 2009;20:283–292. doi: 10.1016/j.semcdb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey M, Shoemaker C, Crews D. Gonadal expression of Sf1 and aromatase during sex determination in the red-eared slider turtle (Trachemys scripta), a reptile with temperature-dependent sex determination. Differentiation. 2007;75:978–991. doi: 10.1111/j.1432-0436.2007.00182.x. [DOI] [PubMed] [Google Scholar]
- Raynaud A, Pieau C. Embryonic development of the genital system. In: Billett F, Gans C, editors. Biology of the Reptilia, Volume 15: Development B. New York: Wiley and Sons; 1985. pp. 149–300. [Google Scholar]
- Rhen T, Crews D. Variation in reproductive behavior within a sex: neural systems and endocrine activation. J Neuroendocrinol. 2002;14:517–532. doi: 10.1046/j.1365-2826.2002.00820.x. [DOI] [PubMed] [Google Scholar]
- Rhen T, Lang JW. Temperature-dependent sex determination in the snapping turtle: manipulation of the embryonic sex steroid environment. Gen Comp Endocrinol. 1994;96:243–254. doi: 10.1006/gcen.1994.1179. [DOI] [PubMed] [Google Scholar]
- Rhen T, Lang JW. Among-family variation for environmental sex determination in reptiles. Evolution. 1998;52:1514–1520. doi: 10.1111/j.1558-5646.1998.tb02034.x. [DOI] [PubMed] [Google Scholar]
- Rhen T, Metzger K, Schroeder A, Woodward R. Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle, Chelydra serpentina. Sex Dev. 2007;1:255–270. doi: 10.1159/000104775. [DOI] [PubMed] [Google Scholar]
- Rhen T, Jangula A, Schroeder A, Woodward-Bosh R. The platelet-derived growth factor signaling system in snapping turtle embryos, Chelydra serpentina: potential role in temperature-dependent sex determination and testis development. Gen Comp Endocrinol. 2009;161:335–343. doi: 10.1016/j.ygcen.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett JC, Patrizio P, Schmid JE, Hecht NB, Dix DJ. Gene expression patterns associated with infertility in humans and rodent models. Mutat Res. 2004;549:225–240. doi: 10.1016/j.mrfmmm.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Rockman MV. Reverse engineering the genotype-phenotype map with natural genetic variation. Nature. 2008;456:738–744. doi: 10.1038/nature07633. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sarre SD, Georges A, Quinn A. The ends of a continuum: Genetic and temperature-dependent sex determination in reptiles. BioEssays. 2004;26:639–645. doi: 10.1002/bies.20050. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Yao HH, Pierucci-Alves F, Capel B. Colocalization of WT1 and cell proliferation reveals conserved mechanisms in temperature-dependent sex determination. Genesis. 2003;35:193–201. doi: 10.1002/gene.10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, et al. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131:933–942. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25:19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, et al. Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci USA. 2006;103:224–229. doi: 10.1073/pnas.0506736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine R, Elphick MJ, Donnellan S. Co-occurrence of multiple, supposedly incompatible modes of sex determination in a lizard population. Ecol Lett. 2002;5:486–489. [Google Scholar]
- Shoemaker CM, Crews D. Analyzing the coordinated gene network underlying temperature-dependent sex determination in reptiles. Sem Cell Dev Biol. 2009;20:293–303. doi: 10.1016/j.semcdb.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C, Ramsey M, Queen J, Crews D. Expression of Sox9, Mis, and Dmrt1 in the gonad of a species with temperature-dependent sex determination. Dev Dyn. 2007a;236:1055–1063. doi: 10.1002/dvdy.21096. [DOI] [PubMed] [Google Scholar]
- Shoemaker CM, Queen J, Crews D. Response of candidate sex-determining genes to changes in temperature reveals their involvement in the molecular network underlying temperature-dependent sex determination. Mol Endocrinol. 2007b;21:2750–2763. doi: 10.1210/me.2007-0263. [DOI] [PubMed] [Google Scholar]
- Smith CA, Joss JMP. Gonadal sex differentiation in Alligator mississippiensis, a species with temperature-dependent sex determination. Cell Tissue Res. 1993;273:149–162. [Google Scholar]
- Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- Smith CA, McClive PJ, Hudson Q, Sinclair AH. Male-specific cell migration into the developing gonad is a conserved process involving PDGF signaling. Dev Biol. 2005;284:337–350. doi: 10.1016/j.ydbio.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Smith CA, Shoemaker CM, Roeszler KN, Queen J, Crews D, Sinclair AH. Cloning and expression of R-Spondin1 in different vertebrates suggests a conserved role in ovarian development. BMC Dev Biol. 2008;8:72. doi: 10.1186/1471-213X-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CWJ, Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Swain A, Lovell-Badge R. Mammalian sex determination: A molecular drama. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- Takada S, DiNapoli L, Capel B, Koopman P. Sox8 is expressed at similar levels in gonads of both sexes during the sex determining period in turtles. Dev Dyn. 2004;231:387–395. doi: 10.1002/dvdy.20132. [DOI] [PubMed] [Google Scholar]
- Torres Maldonado LC, Landa Piedra A, Moreno Mendoza N, Marmolejo Valencia A, Meza Martinez A, Merchant Larios H. Expression profiles of Dax1, Dmrt1, and Sox9 during temperature sex determination in gonads of the sea turtle Lepidochelys olivacea. Gen Comp Endocrinol. 2003;129:20–26. doi: 10.1016/s0016-6480(02)00511-7. [DOI] [PubMed] [Google Scholar]
- Uribe MCA, Guillette LJ., Jr Oogenesis and ovarian histology of the American alligator Alligator mississippiensis. J Morphol. 2000;245:225–240. doi: 10.1002/1097-4687(200009)245:3<225::AID-JMOR4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. Introduction. In: Valenzuela N, Lance VA, editors. Temperature-Dependent Sex Determination in Vertebrates. Washington: Smithsonian Institution; 2004. pp. 1–4. [Google Scholar]
- Valenzuela N. Evolution of the gene network underlying gonadogenesis in turtles with temperature-dependent and genotypic sex determination. Integr Comp Biol. 2008a;48:476–485. doi: 10.1093/icb/icn031. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. Relic thermosensitive gene expression in a turtle with genotypic sex determination. Evolution. 2008b;62:234–240. doi: 10.1111/j.1558-5646.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela N, Shikano T. Embryological ontogeny of aromatase gene expression in Chrysemys picta and Apalone mutica turtles: Comparative patterns within and across temperature-dependent and genotypic sex-determining mechanisms. Dev Genes Evol. 2007;217:55–62. doi: 10.1007/s00427-006-0106-3. [DOI] [PubMed] [Google Scholar]
- Valenzuela N, LeClere A, Shikano T. Comparative gene expression of steroidogenic factor 1 in Chrysemys picta and Apalone mutica turtles with temperature-dependent and genotypic sex determination. Evol Dev. 2006;8:424–432. doi: 10.1111/j.1525-142X.2006.00116.x. [DOI] [PubMed] [Google Scholar]
- Valleley EMA, Muller U, Ferguson MWJ, Sharpe PT. Cloning and expression analysis of two ZFY-related zinc finger genes from Alligator mississippiensis, a species with temperature-dependent sex determination. Gene. 1992;119:221–228. doi: 10.1016/0378-1119(92)90275-t. [DOI] [PubMed] [Google Scholar]
- Valleley EMA, Cartwright EJ, Croft NJ, Markham AF, Coletta PL. Characterisation and expression of Sox9 in the leopard gecko, Eublepharis macularius. J Exp Zool. 2001;291:85–91. doi: 10.1002/jez.7. [DOI] [PubMed] [Google Scholar]
- Viets B, Ewert MA, Talent LG, Nelson C. Sex determining mechanisms in squamate reptiles. J Exp Zool. 1994;270:45–56. [Google Scholar]
- Wallis MC, Waters PD, Graves JAM. Sex determination in mammals – before and after the evolution of SRY. Cell Mol Life Sci. 2008;65:3182–3195. doi: 10.1007/s00018-008-8109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Allan CM, Jimenez M, Lim PR, Davey RA, et al. Female mice haploinsufficient for an inactivated androgen receptor (AR) exhibit age-dependent defects that resemble the AR null phenotype of dysfunctional late follicle development, ovulation, and fertility. Endocrinology. 2007;148:3674–3684. doi: 10.1210/en.2007-0248. [DOI] [PubMed] [Google Scholar]
- Western PS, Harry JL, Graves JAM, Sinclair AH. Temperature-dependent sex determination in the American alligator: AMH precedes SOX9 expression. Dev Dyn. 1999;216:411–419. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<411::AID-DVDY9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Western PS, Harry JL, Marshall Graves JA, Sinclair AH. Temperature-dependent sex determination in the American alligator: expression of SF1, WT1 and DAX1 during gonadogenesis. Gene. 2000;241:223–232. doi: 10.1016/s0378-1119(99)00466-7. [DOI] [PubMed] [Google Scholar]
- Wibbels T, Bull JJ, Crews D. Chronology and morphology of temperature-dependent sex determination. J Exp Zool. 1991;260:371–381. doi: 10.1002/jez.1402600311. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Davies DC. The control of sexual differentiation of the reproductive system and brain. Reproduction. 2007;133:331–359. doi: 10.1530/REP-06-0078. [DOI] [PubMed] [Google Scholar]
- Xita N, Tsatsoulis A. Review: fetal programming of polycystic ovary syndrome by androgen excess: evidence from experimental, clinical, and genetic association studies. J Clinical Endocrinol Metab. 2006;91:1660–1666. doi: 10.1210/jc.2005-2757. [DOI] [PubMed] [Google Scholar]
- Yao HHC, Dinapoli L, Capel B. Cellular mechanisms of sex determination in the red-eared slider turtle, Trachemys scripta. Mech Dev. 2004;121:1393–1401. doi: 10.1016/j.mod.2004.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]