Abstract
Genome-wide functional linkages among proteins in cellular complexes and metabolic pathways can be inferred from high throughput experimentation, such as DNA microarrays, or from bioinformatic analyses. Here we describe a method for the visualization and interpretation of genome-wide functional linkages inferred by the Rosetta Stone, Phylogenetic Profile, Operon and Conserved Gene Neighbor computational methods. This method involves the construction of a genome-wide functional linkage map, where each significant functional linkage between a pair of proteins is displayed on a two-dimensional scatter-plot, organized according to the order of genes along the chromosome. Subsequent hierarchical clustering of the map reveals clusters of genes with similar functional linkage profiles and facilitates the inference of protein function and the discovery of functionally linked gene clusters throughout the genome. We illustrate this method by applying it to the genome of the pathogenic bacterium Mycobacterium tuberculosis, assigning cellular functions to previously uncharacterized proteins involved in cell wall biosynthesis, signal transduction, chaperone activity, energy metabolism and polysaccharide biosynthesis.
INTRODUCTION
With the development of high throughput experimental and computational procedures, the identification of functionally linked proteins has progressed at a brisk pace, while methods for the visual interpretation of these large datasets have developed more slowly. Experimental methods such as high throughput yeast two-hybrid experiments (1) and whole genome microarray experiments (2) have yielded a plethora of information about functionally related genes and proteins, both in the form of protein–protein interaction data as well as information regarding protein function. Extensive databases, such as the Database of Interacting Proteins (3), the Biomolecular Interaction Network Database (4) and the MIPS Comprehensive Yeast Genome Database (5), have also been created to catalog thousands of protein–protein interactions identified by both large and small scale experiments.
In addition to high throughput experimental procedures, several computational methods have been developed to identify functionally linked genes and proteins. Among these are the Rosetta Stone method (6), which identifies individual proteins that exist as a single fusion protein in another organism (6,7), the Phylogenetic Profile method (8), which examines the correlated occurrence of proteins in various genomes, the Operon method (9,10), which functionally links genes likely to belong to common operons based on the distance between genes in the same orientation, and the Conserved Gene Neighbor method (11,12), which identifies genes that are located in close chromosomal proximity in multiple genomes. These methods provide non-homology based approaches for identifying functionally related proteins throughout a particular genome and complement the traditional homology based tools such as BLAST (13) for the identification of protein function. In addition, these methods permit a protein’s function to be defined in the context of its cellular interactions (14).
These computational methods are useful not only for the identification of protein function, as we have demonstrated previously in the genome of Mycobacterium tuberculosis (9), but can also be employed for the reconstruction of protein networks. Traditionally, these networks have been displayed as two-dimensional graphs of nodes and edges, where edges represent functional linkages between pairs of protein nodes (1,15). Although this classical representation has its merits, we have found that an alternative representation has advantages for depicting genome-wide functional linkages in prokaryotic genomes. Specifically, we employ a single scatter plot, with both axes organized according to the order of genes along the prokaryotic chromosome. This representation has allowed us to identify characteristics of protein network architecture that have previously eluded identification with the classical node and edge representation.
Andrei Grigoriev first proposed the use of a two- dimensional matrix to indicate experimentally identified protein–protein interactions in the 56 gene genome of bacteriophage T7 (16). Here we build on this idea and plot computationally derived protein functional linkages on individual scatter plots. Thousands of functional linkages in the M.tuberculosis genome are plotted on a single scatter plot, comprising what we describe as a genome-wide functional linkage map. These maps reveal global and local features of prokaryotic genome organization and suggest protein relationships on a genome-wide basis.
MATERIALS AND METHODS
Genome-wide functional linkage maps
Functional linkage maps were generated by first identifying M.tuberculosis protein pairs that are functionally linked by the Rosetta Stone, Phylogenetic Profile, Operon (distance threshold 100 bp) and Conserved Gene Neighbor methods. Pairs of proteins that are functionally linked by two or more computational methods were then identified. Protein pairs were converted to corresponding integer values (i.e. protein pair Rv0005 and Rv0006 was converted to integer pair 5,6), and a list of integer pairs was input into the graphing program SigmaPlot 2000 to create a scatter plot representing genome-wide functional linkages.
Hierarchical clustering
Bit vectors for each gene were created. The presence of a functional linkage was indicated by a bit entry of 1 in the vector at the position corresponding to the functionally linked gene. The absence of a functional linkage was indicated by a bit entry of 0. The program Cluster (17) was employed to cluster genes based on the similarity of their functional linkage profiles using a centered correlation coefficient as the comparison metric. The hierarchical clustering algorithm used was the average linkage algorithm. The program Treeview (17) was employed to visualize the clustering results. The original data table was represented graphically by coloring each cell according to the bit entry. Cells with a bit entry of 1 (corresponding to a functional linkage) were colored black, while cells with a bit entry of 0 (corresponding to absence of a functional linkage) were colored white.
Rosetta Stone method
Proteins were functionally linked by the Rosetta Stone method if individual proteins were found to be present as a single fused protein in another organism, as described by Marcotte et al. (6,18). In this case, if individual M.tuberculosis proteins have significant homology to distinct regions of a single ‘fusion’ protein in another organism then they are indicated as functionally linked by this method. A probabilistic score is calculated by estimating the likelihood of observing Rosetta Stone proteins given the number of homologs each protein has.
Phylogenetic profile method
Phylogenetic profiles were used to identify proteins that occur in a correlated fashion in numerous genomes, as described by Pellegrini et al. (8). A phylogenetic profile for each M.tuberculosis protein was created in the form of a bit vector, by searching for the presence or absence of homologs in each of the available fully sequenced genomes. The presence of an identifiable homolog in a particular genome was indicated by the integer 1 in the bit vector at the position corresponding to that genome, while the absence of a homolog was indicated by the integer 0. Phylogenetic profiles were then clustered based on the similarity of profiles, resulting in clusters of genes with similar profiles and likely related functions.
Operon method
A series of genes are considered functionally linked by the Operon method if the nucleotide distance between genes in the same orientation was less than or equal to a specified distance threshold (9). Multiple genes were linked if a series of genes in the same orientation all had intergenic distances less than or equal to the defined distance threshold. In the case of our genome-wide functional linkage maps, a distance threshold of 100 bp was employed.
Conserved Gene Neighbor method
Functional links were established by the Conserved Gene Neighbor method where genes appear as chromosomal neighbors in multiple genomes, as described by Overbeek et al. (11) and Dandekar et al. (12). For all possible pairs of M.tuberculosis genes, the nucleotide distance between homologs of these genes in all available sequenced genomes was calculated. Genes that were in close proximity in multiple genomes were indicated as functionally linked by this method. A probabilistic score reflects the likelihood of observing the intergenic distance between a pair of genes across all sequenced genomes.
Sanger Institute functional annotations
Sanger Institute M.tuberculosis H37Rv functional annotations were obtained from the Sanger M.tuberculosis web server at http://www.sanger.ac.uk/Projects/M_tuberculosis/Gene_list/.
Evaluation of functional linkages
The method of keyword recovery (9) was used to evaluate functional linkages represented in the genome-wide functional linkage map. The method of keyword recovery compares links between Swiss-Prot annotated proteins.
For each pair of functionally linked proteins we define the first protein of the pair ‘query protein A’ and the second protein of the pair ‘linked protein B’. All protein pairs are reciprocal, since a link from gene 1 to gene 2 is also represented as a link from gene 2 to gene 1. The keyword recovery of all linkages was calculated as:
where X is the total number of keywords in all query proteins, Y is the total number of linked gene pairs, x is the number of Swiss-Prot keywords of query protein A and nij is the number of times the query protein keyword j occurs in the annotation of the linked protein B.
Signal-to-noise was calculated as:
signal-to-noise = keyword recovery/random keyword recovery
where random keyword recovery was calculated for the same number of random pairwise Swiss-Prot annotated genes as exist computationally inferred links (mean of 100 random trials).
The maximum false-positive fraction (9) was calculated as the fraction of pairwise links that do not have any Swiss-Prot keywords in common (ignoring the keywords ‘hypothetical protein’, ‘3D structure’, ‘transmembrane’ and ‘complete proteome’).
RESULTS
Validation of functional linkages
The 9766 functional linkages represented in the genome-wide functional linkage map (Fig. 1B) are inferred by two or more computational methods. Previous studies have shown that functional linkages inferred by multiple methods are likely to share related functions (9,15). In order to validate the 9766 functional linkages of our genome-wide functional linkage map we employed a keyword recovery (9) scheme to compare links between Swiss-Prot annotated proteins. Keyword recovery means that identical keywords are found in the annotations for both proteins connected by the link.
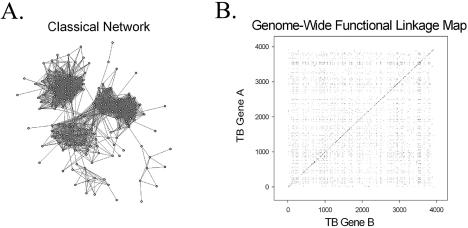
Figure 1.
A comparison of two methods for illustrating inferred protein functional linkages in the M.tuberculosis genome. (A) Classical representation of a protein network, consisting of nodes and edges. Each node represents a particular protein and each edge represents a functional linkage between two linked proteins. (B) Genome-wide functional linkage map representing all 9766 functional linkages inferred by two or more computational methods. Each of the functional linkages is depicted as a single point on this graph, where both the x- and y-axes are organized according to the order of genes along the M.tuberculosis chromosome. The dense clustering of functionally linked genes near the diagonal reflects both bacterial operon organization and close chromosomal proximity of genes of related function.
The method of keyword recovery allows us to evaluate a set of linkages based on known functional annotation. By comparing the Swiss-Prot keywords we can quantitatively evaluate the functional linkages represented in our genome-wide functional linkage map. The keyword recovery for linkages inferred by two or more methods is 0.55. Compared to the keyword recovery of randomly paired proteins (0.056) we have a signal-to-noise ratio of 9.8.
The maximum false-positive fraction (9) also reflects the functional similarity among linked proteins. The maximum false-positive fraction is the fraction of functionally linked proteins that do not share any keywords in common. The quantity 1 – maximum false-positive fraction indicates the fraction of pairwise links that have one or more keywords in common, and therefore some function in common. The maximum false-positive fraction for the 9766 linkages inferred by two or more methods is 0.20, demonstrating that 80% [100% × (1 – maximum false-positive fraction)] of linked pairs share some function in common. This percentage (80%) may in fact represent the lower boundary, since some proteins may have incomplete annotation or may employ different vocabularies to describe similar functions.
The linkage of genes by more than one method is expected in a number of cases since the computational methods we employ are inherently complementary. For example, Moreno-Hagelsieb et al. have shown that genes of common operons are more likely to occur as fusion genes (Rosetta Stone linkage), are more likely to occur as Conserved Gene Neighbors and are more likely to share similar Phylogenetic profiles than genes of different operons (19). Yanai et al. have also noted numerous instances where individual genes that constitute a fusion gene in one organism occur as operon members in another organism (20).
Linkages that are inferred by two or more methods have been shown to have a higher keyword recovery score and lower maximum false-positive score than linkages inferred by only a single method (9,15). The resulting lists of functionally linked genes, inferred by overlapping methods, therefore represent higher confidence functional linkages (15) and are well suited for the construction of our genome-wide functional linkage maps.
Genome-wide functional linkage maps
Figure 1 illustrates the differences between the classical representation of functionally linked proteins, consisting of nodes and edges (Fig. 1A), and our alternative representation (Fig. 1B) of functional linkages in the genome of M.tuberculosis. Both Figure 1A and B represent M.tuberculosis functional linkages inferred by two or more computational methods. In Figure 1A we have graphed only a portion of the functional linkages to demonstrate the characteristic differences between the two methods of illustration. Although Figure 1A is able to capture certain general features of protein connectivity, in this case the presence of three large interconnected groups, it is difficult to examine and interpret the more subtle protein subgroups within these larger interconnected groups.
Figure 1B, in contrast, displays functional linkages predicted throughout the entire genome of M.tuberculosis. 9766 functional linkages, involving 1381 unique genes, are represented in the genome-wide functional linkage map in Figure 1B. Here we have depicted each functional linkage as a single point on this two-dimensional scatter plot. Both the x- and y-axes are a monotonically ordered list of genes, giving the order of genes along the M.tuberculosis H37Rv chromosome. Each point on this plot represents a functional linkage between the two genes at the corresponding chromosomal positions. For example, the point at coordinate 1,5 corresponds to the computationally assigned functional linkage between gene Rv0001(dnaA) and gene Rv0005(gyrB), both involved in DNA replication. Using these genome-wide functional linkage maps, we can begin to visualize both global and local features of protein network architecture that have been more difficult to visualize with the traditional node and edge graphs.
Notice that Figure 1B and other such functional linkage maps have mirror symmetry about the diagonal, because a linkage from protein m to n is the same as one from n to m. Certain global characteristics of protein connectivity are readily apparent in Figure 1B, such as the dense clustering of functionally linked genes near the diagonal. This clustering reflects both bacterial operon organization and close chromosomal proximity of genes of related function. We would not expect to identify this global trend using the standard node and edge graphs, because information regarding chromosomal location and proximity are not directly represented in these graphs. The central heavy band consists of points just off the diagonal, because none of the four methods detect homotypic functional linkages.
Our analysis of genome-wide functional linkage maps reveals certain chromosomal regions that have genes with many functional linkages and other regions that have few predicted linkages. This non-random connectivity may relate to the scale-free nature of protein network topology (21,22), and may enable detection of clusters of both promiscuous proteins as well as highly specialized proteins throughout the M.tuberculosis genome. A downloadable file of the genome-wide functional linkage map is provided at http://www.doe-mbi.ulca.edu/~strong/map. This file enables the identification of each point of the genome-wide functional linkage map in an interactive manner.
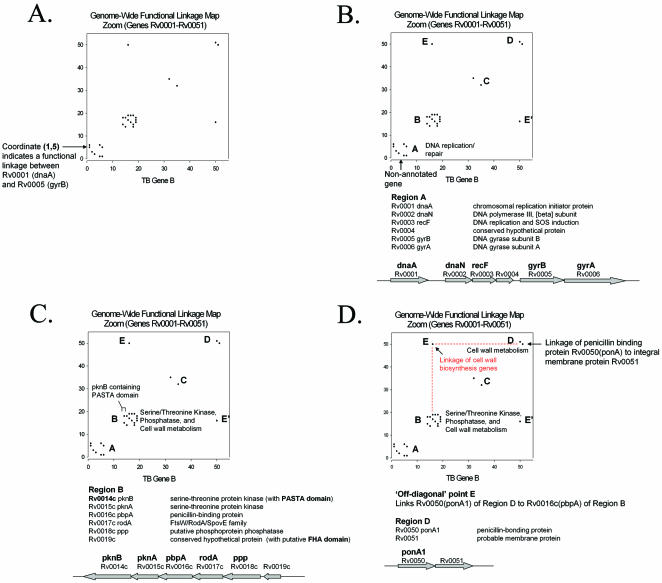
In Figure 2A we demonstrate how our genome-wide functional linkage maps can reveal important characteristics of protein network topology that may be missed by conventional methods. The functional linkages depicted in Figure 2A are identical to that of the genome-wide functional linkage map in Figure 1B, but cover only the portion of the map that contains the first 51 genes. Using this enlarged representation we again see the clustering of functionally linked genes near the diagonal. In Region A of the functional linkage map in Figure 2B we see a cluster of functionally linked genes all involved in DNA replication or repair. None of the proteins of Region A are homologous to any of the others in this region. This observation emphasizes the ability of our computational methods to identify relevant biological relationships among non-homologous proteins. Interestingly, the five annotated genes of this cluster flank the non-annotated gene Rv0004 (Fig. 2B), suggesting a possible cellular role for this gene in DNA replication or repair. The Rv0004 open reading frame is in the same orientation and slightly overlaps the open reading frame of Rv0003(recF), linking it by the Operon method. Additionally, although Rv0004 is similar in sequence only to other hypothetical and hypothetical conserved proteins, it is similar to a putative conserved domain (COG5512) (23) containing a zinc-ribbon module. Zinc-ribbon modules have been implicated in both DNA and RNA binding (24), adding support to our hypothesis of a cellular role for Rv0004 in DNA metabolism.
Figure 2.
An expanded view of the genome-wide functional linkage map of Figure 1B, showing the portion of the map that contains the first 51 genes. (A) Notice the clustering of functionally linked genes along the diagonal. Twenty-eight functional linkages in this region are represented as single points corresponding to each functionally linked pair of genes. (B) Region A contains a cluster of functionally linked genes involved in DNA replication or repair as well as the uncharacterized gene Rv0004. (C) Region B contains a cluster of genes involved in serine-threonine kinase/phosphatase activity, cell wall biosynthesis and the uncharacterized gene Rv0019c. The genes of this cluster are linked by 14 functional linkages, and we infer that all of the proteins of this cluster function together in a common pathway involving the regulation and synthesis of cell wall components. This inference is further supported by the study of Yeats et al. (25), in which they identified a protein domain that directly links eukaryotic-like serine-threonine protein kinases to cell wall biosynthesis proteins (25). (D) Here we use the ‘off-diagonal’ point E to link Cluster B to Cluster D. Point E links gene Rv0016c(ponA1) of Cluster B to gene Rv0050(ponA1) of Cluster D. Notice also that point E and point E′ are symmetry related and represent the same functional linkage.
Region B of Figure 2C illustrates the power of functional linkages to detect unexpected metabolic relationships. In this region we find that three of the six genes in this functionally linked cluster have functions related to serine/threonine kinase or serine/threonine phosphatase activity [Rv0014c(pknB), Rv0015c(pknA) and Rv0018c(ppp)], two genes are involved in cell wall biosynthesis [Rv0016c(pbpA) and Rv0017c(rodA)] and one gene is uncharacterized (Rv0019c). While at first glance the relationship among all six genes is not obvious, this cluster of computationally linked genes exemplifies an important aspect of our computational methods, specifically that these methods excel at identifying protein function at the cellular level of metabolism as opposed to exclusively at the biochemical level. Recently, a protein domain named the PASTA (penicillin-binding protein and serine/threonine kinase associated) domain (25) was identified, linking eukaryotic-like serine/threonine kinases, such as pknB (Rv0014c), directly to cell wall biosynthesis (25). Yeats et al. suggested that pknB-like serine/threonine protein kinases, which contain tandem repeats of the PASTA domain, may be important regulators of cell wall synthesis, by sensing unlinked peptidoglycans in the extracellular environment (25). The functional linkages of Region B are consistent with this proposal of Yeats et al.
All six of the genes in cluster B occur next to each other in the same genomic orientation (Fig. 2C), and five of the six genes occur as a contiguous string of genes with a 4 bp overlap between each adjacent gene pair. The overlap of genes in the same orientation by 4 bp is a common feature of operon structure in Escherichia coli (26) and we therefore hypothesize that these M.tuberculosis genes are not only functionally linked but also form a common operon. It is well known that genes of common operons often encode proteins of related cellular function (27), ranging from physically interacting proteins to proteins involved in common biochemical pathways. In this case, pknB, pknA and ppp may play a role in the regulation of M.tuberculosis cell wall biosynthesis. The final gene in this functionally linked cluster, Rv0019c, may also play a role in this proposed pathway. Although Rv0019c has been annotated as a conserved hypothetical protein, it does show similarity to a putative Forkhead-associated (FHA) domain (COG1716). FHA domains have been found to mediate phosphorylation- dependent protein–protein interactions (28), and are often involved in signal transduction mechanisms.
Figure 2D illustrates an example of the use of off-diagonal points to identify functionally linked gene clusters in the M.tuberculosis genome. Notice that off-diagonal points E and E′ are symmetry related and represent the same linkage. Point E links gene Rv0016c (pbpA) of Cluster B to gene Rv0050 (ponA1) of Cluster D. Cluster D consists of two genes, Rv0050(ponA1) and Rv0051. Rv0050(ponA1) encodes a penicillin-binding protein involved in cell wall biosynthesis and Rv0051 is predicted to be an integral membrane protein. These genes are linked by both the Operon and Conserved Gene Neighbor methods and may serve related functions in cell wall biosynthesis. In this example, the off-diagonal point directly links Cluster B to Cluster D, and we infer that the genes of these two clusters have related cellular functions associated with M.tuberculosis cell wall biosynthesis.
Hierarchical clustering
Although the off-diagonal points can identify functionally linked genes and gene clusters throughout the genome, we adopt a complementary method to facilitate this process. We apply hierarchical clustering to group the genes of our genome-wide functional linkage maps based on the similarity of their functional linkage profiles. A functional linkage profile for a particular gene is equivalent to a single row in our genome-wide functional linkage map and consists of a record of all the genes to which a particular gene is functionally linked.
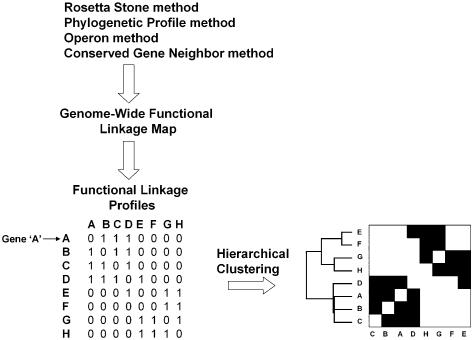
Figure 3 provides an overview of the strategy. First, the Rosetta Stone, Phylogenetic profile, Operon and Conserved Gene Neighbor computational methods are employed to identify functionally linked genes throughout the genome. Second, a genome-wide functional linkage map is constructed, representing each of the computationally assigned functional linkages. Next, each row of the functional linkage map is converted to a binary vector (functional linkage profile), where genes that are functionally linked to our query gene have a bit entry of 1 and genes that are not linked have a bit entry of 0. In the simplified example depicted in Figure 3, the functional linkage profile for gene ‘A’ indicates that gene A is functionally linked to genes B, C and D (as indicated by the bit score of 1). Lastly, hierarchical clustering of the profiles results in a re-ordering of the genes, clustering together genes that share similar functional linkage profiles.
Figure 3.
Schematic of the methods used in this study. The Rosetta Stone, Phylogenetic Profile, Operon and Conserved Gene Neighbor methods were employed to identify functionally linked proteins throughout the M.tuberculosis genome. Functional linkages inferred by two or more computational methods were used to construct a genome-wide functional linkage map. The map was then converted into separate bit vectors representing the functional linkage profile of each gene in the map. Hierarchical clustering resulted in a reordering of the genes, clustering genes with similar functional linkage profiles.
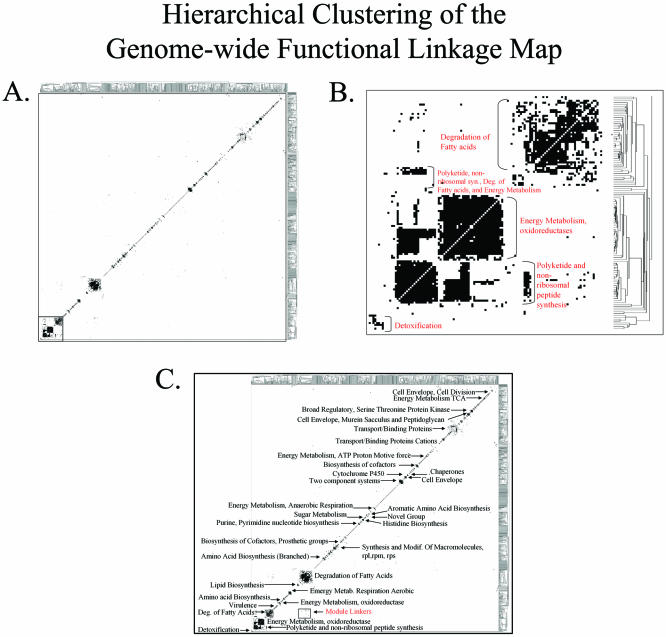
Figure 4A shows the result of clustering the genes of the M.tuberculosis genome-wide functional linkage map depicted in Figure 1B. 1381 genes have functional linkages to other genes inferred by two or more computational methods and are included in the clustered map of Figure 4A. After hierarchical clustering, we see that ∼80% of these genes cluster into distinct groups, most of which contain genes of related cellular function (http://www.doe-mbi.ucla.edu/~strong/map/). These clusters range from small clusters with 2–12 genes (more than 100 of these) to large clusters containing more than 12 genes (about 6 of these). For example the three large clusters in Figure 4B contain 23, 35 and 48 genes, respectively.
Figure 4.
Hierarchical Clustering of the genome-wide functional linkage map. (A) Hierarchical Clustering groups the genes of our genome-wide functional linkage map based on the similarity of functional linkage profiles. 1381 M.tuberculosis genes have functional linkages inferred by two or more computational methods and are depicted in this figure. Approximately 80% of the genes represented in this map cluster into distinct modules. (B) An expanded version of the area bordered by the box in (A). Notice that many of the modules contain genes of related cellular function, as judged by their annotations. Here we see modules involved in detoxification, polyketide synthesis, energy metabolism and degradation of fatty acids. (C) Cellular function can be assigned to most modules of our hierarchical clustered map, including those shown here. Modules are present for a number of important M.tuberculosis pathways, including fatty acid degradation, energy metabolism, cell envelope biosynthesis and virulence factors. In addition, a few modules contain a high frequency of non-annotated proteins, suggesting previously uncharacterized M.tuberculosis pathways or complexes.
Although in our original functional linkage map the dense clustering along the diagonal reflects operon organization and genomic clustering of functionally related genes, the diagonal in Figure 4A arises from two factors. The first is that our hierarchical clustering algorithm clusters genes along both axes. Since our original genome-wide functional linkage map is symmetric, the resulting clustered map places identical genes at the equivalent position on both axes. The second contributing factor is that the functional linkage profiles of clustered genes tend to contain linkages to other genes of the same cluster.
These resulting gene clusters are analogous to the functional modules coined by Snel et al. (29), in which they used the Conserved Gene Neighbor method to construct traditional node and edge protein networks to identify groups of proteins involved in related cellular functions. While we employ a different method than Snel et al. for the visualization and examination of protein networks, we do see related observations, such as the presence of distinct functional modules consisting of proteins involved in related cellular functions, as well as the presence of linker proteins (29), which link functional modules of varied function. These observations also correspond well with the observations of Rives et al. (30) and Ravasz et al. (31), who employed analogous methods of network clustering to investigate the modular organization of yeast protein interaction networks (30) and E.coli metabolic networks (31), respectively.
Our method of hierarchical clustering tends to cluster genes of related biological function, but we see relatively little overlap among separate clusters, as seen by the modest number of off-diagonal functional linkages connecting the varying clusters. All off-diagonal functional linkages in Figure 4A that occur to the right of the diagonal have been indicated in bold so that they can be more easily observed at this scale. Although these off-diagonal linkages are the minority, they may in fact represent an important group of genes that link functional modules.
Figure 4B illustrates an expanded view of the region bordered by the box in Figure 4A. In this region we observe modules involved in detoxification, polyketide sythesis, energy metabolism and degradation of fatty acids. Cellular function can be assigned to most modules throughout our clustered genome-wide functional linkage map, some of which are indicated in Figure 4C. We observe functional modules involved in a wide variety of cellular functions, ranging from lipid biosynthesis to energy metabolism. Additionally, we observe a number of modules containing a high frequency of non-annotated proteins, possibly indicating previously uncharacterized pathways in M.tuberculosis. Some functional categories, such as amino acid biosynthesis, are partitioned into separate modules corresponding to specific pathways within the general amino acid biosynthesis category. Separate modules are observed for branched amino acid biosynthesis, histidine biosynthesis, cysteine biosynthesis, chorismate and tyrosine biosynthesis and one module corresponding to arginine and tryptophan synthesis (http://www.doe-mbi.ucla.edu/~strong/map/).
The two largest modules of Figure 4C contain genes involved in the degradation of fatty acids. Although both modules are involved in fatty acid degradation, the two modules correspond to distinct steps within the fatty acid degradation pathway. The smaller of these two modules contains 34 acyl-CoA synthetase fadD family members, involved in the first step of fatty acid degradation, while the larger module contains members of the fadA (6 genes), fadB (3 genes), fadE (34 genes) and echA (21 genes) families, representing all four steps of the downstream β-oxidation of fatty acids. While these two modules are separate, a number of off-diagonal module linkers link the two modules. The module linkers link acyl-CoA synthetase fadD members to acyl-CoA dehydrogenase fadE family members. These proteins catalyze consecutive steps in the fatty acid degradation pathway.
Other examples of module linkers include a linkage between a serine-threonine kinase/phosphatase module and a cell envelope module. This linkage is mediated by the functional linkage between Rv0018c (ppp) and Rv0019c. In addition, we observe a linkage between two related ribosomal modules, mediated by the functional linkage between Rv0717 (rpsN1) and Rv0702 (rplD).
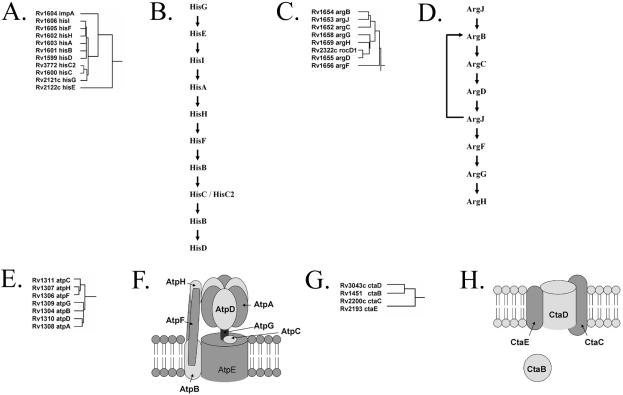
Figure 5 illustrates the ability of our hierarchical clustering methods to identify genes that are involved in common biochemical pathways as well as protein complexes. In Figure 5A we see that all nine genes of the histidine biosynthesis pathway (Fig. 5B) cluster together along with the hisC2 paralog, enabling complete reconstruction of the M.tuberculosis histidine biosynthesis pathway. Similarly, as demonstrated in Figure 5C, all seven genes of the arginine biosynthesis pathway (Fig. 5D) cluster together, yielding complete reconstruction of the arginine biosynthesis pathway. We also see an additional gene, rocD1, embedded in this cluster of genes. rocD1 encodes an ornithine aminotransferase which is involved in the arginine degradation pathway.
Figure 5.
Reconstruction of known biochemical pathways and protein complexes in M.tuberculosis. Panels (A), (C), (E) and (G) each depict the clustering pattern of a subset of genes of our hierarchical clustered functional linkage map. Panels (B), (D), (F) and (H) represent known biochemical pathways and known protein complexes. (A) All nine genes of the histidine biosynthesis pathway are clustered together, along with the hisC2 paralog, yielding complete enumeration of the catalytic elements of the histidine biosynthesis pathway. (B) Histidine biosynthesis pathway. (C) All seven genes of the arginine biosynthesis pathway cluster together, giving a complete enumeration of the arginine biosynthesis enzymes. (D) Arginine biosynthesis pathway. (E) Seven of the eight genes involved in the mycobacterial ATP synthase complex cluster together. (F) ATP synthase multiprotein complex. (G) All three proteins of the bacterial cytochrome c oxidase complex cluster together along with the cytochrome c oxidase assembly factor, ctaB. (H) Cytochrome c oxidase.
Figure 5E demonstrates the clustering of seven of the eight mycobacterial ATP synthase genes. Each of these genes encodes individual protein subunits of the M.tuberculosis ATP synthase complex (Fig. 5F). Although these proteins share little sequence similarity, they do share similar functional linkage profiles and are therefore clustered together during hierarchical clustering. The eighth gene of the ATP synthase multiprotein complex, Rv1305(atpE), is functionally linked to the other proteins only by the Operon method and therefore is not present in this cluster, which requires linkages by at least two methods. Our last example involves the prokaryotic cytochrome c oxidase protein complex (Fig. 5H). In Figure 5G we see that in addition to all three members of the cytochrome c oxidase complex clustering together, a fourth gene, ctaB, encoding the cytochrome c oxidase assembly factor, is also included. Notice also that all four genes of this cluster are from very different chromosomal locations. These examples exemplify the ability of our methods to cluster genes of related cellular function, corresponding to both biochemical pathways as well as protein complexes.
Inference of protein function
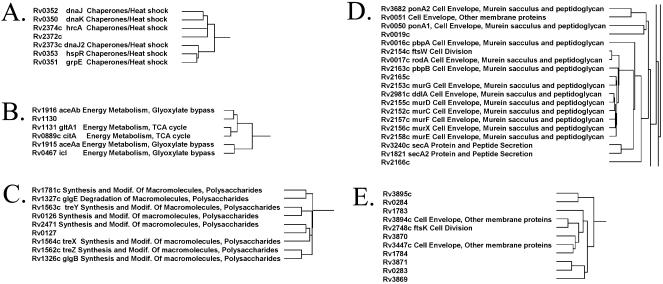
We can also use these clusters to infer function for previously uncharacterized proteins. Figure 6A–E demonstrates examples of gene clusters that contain a mixture of annotated and non-annotated genes. In Figure 6A we show a cluster of genes encoding proteins involved in chaperone activity, along with a single non-annotated gene, Rv2372c. Because of the clustering pattern we infer that Rv2372c has a function related to that of chaperone/heat shock proteins. In Figure 6B we have five genes involved in the TCA cycle and the closely associated glyoxylate bypass pathway. Although Rv1130 is annotated as a conserved hypothetical protein with no known function, we infer that Rv1130 has a cellular function related to that of the TCA and glyoxylate bypass pathway. Although no function has been assigned to Rv1130, it is homologous to a putative prpD domain (COG2079). Horswill et al. have shown that a Salmonella enterica PrpD enzyme is involved in the conversion of propionate to pyruvate (32), a substrate destined for the TCA cycle. This added evidence enables further refinement of our hypothesis and we infer that Rv1130 is involved in the conversion of propionate to pyruvate, which after oxidative decarboxylation is fed into the TCA cycle.
Figure 6.
Inference of protein function based on hierarchical clustering of genes with similar functional linkage profiles. (A) Chaperone/heat shock activity. Notice that six genes involved in chaperone/heat shock activity cluster together with the uncharacterized gene Rv2372c. We infer that Rv2372c has a function associated with chaperone/heat shock activity. (B) Energy metabolism, TCA cycle and glyoxylate bypass. Five genes involved in the TCA cycle or the closely associated glyoxylate bypass pathway cluster along with the conserved hypothetical protein Rv1130. We infer that Rv1130 has a function related to these proteins based on the clustering pattern. (C) Polysaccharide metabolism. Eight genes involved in polysaccharide metabolism cluster along with the conserved hypothetical protein Rv0127. We infer that Rv0127 has a function related to that of the genes in this cluster (polysaccharide metabolism). (D) Cell envelope biosynthesis. A group of proteins involved in cell envelope metabolism, as well as three non-annotated genes, cluster together in this region. We infer that the uncharacterized genes Rv0019c, Rv2165c and Rv2166c have functions related to cell envelope biosynthesis. (E) Numerous non-annotated genes clustered with a few cell envelope/division genes. In this case, the majority of the clustered proteins are uncharacterized. Even so, of the annotated proteins involved in this cluster, all three share a common function associated with either cell division or the cell envelope. We therefore infer that the other proteins of this cluster may have related functions.
Figure 6C shows a cluster of genes involved in polysaccharide metabolism, along with the conserved hypothetical protein Rv0127. Three of the genes in this cluster encode proteins involved in trehalose metabolism (treX, treY and treZ), one gene encodes the glycosyl hydrolase glgE and one gene encodes the glycan branching enzyme glgB. We infer that Rv0127 has a cellular function involved in polysaccharide metabolism. In Figure 6D we have a 19 gene cluster involved in cell envelope biosynthesis, in addition to three non-annotated genes, Rv0019c, Rv2165c and Rv2166c. Notice that again we see Rv0019c, previously encountered in Figure 2C, but this time we come upon it in the context of our clustered genes. While previously we inferred that Rv0019c has a function related to cell wall/envelope metabolism based on its genomic organization, here our hierarchical clustering methods support that inference. In addition, we also infer that Rv2165c and Rv2166c have functions related to cell envelope metabolism.
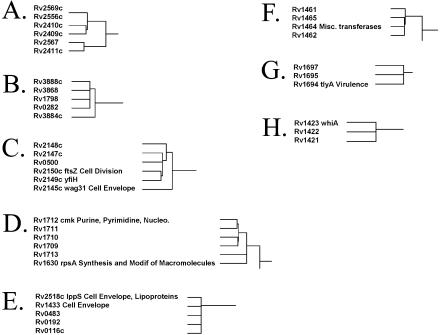
Finally, in Figure 6E we focus on a cluster containing a majority of non-annotated genes interspersed with three genes involved in either the cell envelope or cell division. We hypothesize that this cluster may represent a previously uncharacterized group of M.tuberculosis genes involved in the cell envelope/cell division. Although each of these uncharacterized proteins is annotated as either a hypothetical protein or a conserved hypothetical protein, six out of the eight proteins have two or more computationally predicted transmembrane helices, a common characteristic of proteins involved in cell division and cell wall-related functions. This type of cluster is not unique in its composition. In fact, a number of clusters contain a high percentage of uncharacterized proteins (Fig. 7 and http://www.doe-mbi.ucla.edu/~strong/map/). Some of these clusters do not contain any annotated genes and we hypothesize that these clusters represent previously uncharacterized pathways and complexes.
Figure 7.
Functional linkage modules containing numerous uncharacterized genes. Some of the clusters contain very few annotated genes or do not contain any annotated genes at all. These clusters may represent previously uncharacterized M.tuberculosis pathways or complexes.
DISCUSSION
We find that the four computational methods, the Rosetta Stone, Phylogenetic profile, Operon and Conserved Gene Neighbor methods, can be applied not only to link pairs of proteins throughout a particular genome, but also to construct complex protein networks. These methods have varied applications, ranging from the analysis of functional linkages on a genome-wide scale to the inference of protein function. We propose that genome-wide functional linkage maps may provide a useful method for the visualization and interpretation of both experimentally derived (3–5) as well as computationally inferred (33,34) functional linkage datasets on a genome-wide basis.
Although the functional linkage maps we have discussed thus far have incorporated functional linkages inferred by at least two of our computational methods, we have also constructed genome maps using linkages established by each of the four computational methods alone (Supplementary Fig. 1). Although we expect the Operon method to link genes exclusively near the diagonal, we also observe that the other three methods have a tendency to identify functional linkages along the diagonal, corresponding to functionally linked genes in close chromosomal proximity.
The use of individual computational methods is also likely to aid in the inference of protein function. Since our original clustered map contains only linkages inferred by the overlap of two or more methods, examination of linkages established by individual methods may provide additional information and may aid in the identification of protein function involving clusters of previously uncharacterized genes. We envision that these functional linkages may suggest potential functional roles for these proteins and may indicate potential research directions or biochemical experiments in which to investigate these proteins.
Using a combination of our genome-wide functional linkage maps and hierarchical clustering, we have been able to elucidate features of protein network architecture that have previously eluded inference. These two types of graphical representation have allowed us to rapidly analyze genome-wide functional linkages and have enabled us to infer protein function and identify potential pathways involving previously uncharacterized proteins. While we have focused our attention here on the deadly bacterial pathogen M.tuberculosis, these methods can also be applied to any prokaryotic organism, and may even be extended to examine genome features of eukaryotes.
We have been able to assign function to a number of previously uncharacterized genes, including genes involved in cell wall metabolism, chaperone/heat shock activity, energy metabolism and polysaccharide metabolism, and suggest a potential pathway involving serine/threonine kinases and cell wall metabolism genes. Some of the proteins we have assigned function to, in turn, may serve as potential drug targets since a number of the pathways to which they are linked have been previously proposed as drug targets (35–37).
Mycobacterium tuberculosis genome-wide functional linkage maps, dendrograms and functional linkages are available at http://www.doe-mbi.ucla.edu/~strong/map/.
SUPPLEMENTARY MATERIAL
Supplementary Material is avaliable at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
M.S. is supported by a USPHS National Research Service Award GM07185. This work was also supported by the National Institutes of Health under grant no. P01 GM31299-20.
REFERENCES
- 1.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 2.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 3.Xenarios I., Salwinski,L., Duan,X.J., Higney,P., Kim,S.M. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader G.D., Betel,D. and Hogue,C.W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mewes H.W., Frishman,D., Guldener,U., Mannhaupt,G., Mayer,K., Mokrejs,M., Morgenstern,B., Munsterkotter,M., Rudd,S. and Weil,B. (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Detecting protein function and protein–protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 7.Enright A.J., Iliopoulos,I., Kyrpides,N.C. and Ouzounis,C.A. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 8.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strong M., Mallick,P., Pellegrini,M., Thompson,M.J. and Eisenberg,D. (2003) Inference of protein function and protein linkages in M. tuberculosis based on prokaryotic genome organization: a combined computational approach. Genome Biol., 4, R59.1–R59.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellegrini M., Thompson,M., Fierro,J. and Bowers,P. (2001) Computational method to assign microbial genes to pathways. J. Cell. Biochem., 37, 106–109. [DOI] [PubMed] [Google Scholar]
- 11.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 15.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriev A. (2001) A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res., 29, 3513–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcotte C.J.V. and Marcotte,E.M. (2002) Predicting functional linkages from gene fusions with confidence. Appl. Bioinformatics, 1, 93–100. [PubMed] [Google Scholar]
- 19.Moreno-Hagelsieb G., Trevino,V., Perez-Rueda,E., Smith,T.F. and Collado Vides,J. (2002) Transcription unit conservation in the three domains of life: a perspective from Escherichia coli. Trends Genet., 17, 175–177. [DOI] [PubMed] [Google Scholar]
- 20.Yanai I., Wolf,Y.I. and Koonin,E.V. (2002) Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol., 3, 24.1–24.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuchty S. (2002) Interaction and domain networks of yeast. Proteomics, 2, 1715–1723. [DOI] [PubMed] [Google Scholar]
- 22.Jeong H., Tombor,B., Albert,R., Oltvai,Z.N. and Barabasi,A.L. (2000) The large-scale organization of metabolic networks. Nature, 407, 651–654. [DOI] [PubMed] [Google Scholar]
- 23.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravind L. and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeats C., Finn,R.D. and Bateman,A. (2002) The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci., 27, 438–440. [DOI] [PubMed] [Google Scholar]
- 26.Salgado H., Moreno-Haelsieb,G., Smith,T. and Collado-Vides,J. (2000) Operons in Escherichia coli: genomic analysis and predictions. Proc. Natl Acad. Sci. USA, 97, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodish H., Berk,A., Zipursky,S.L., Matsudaira,P., Darnell,J. and Baltimore,D. (1995) Molecular Cell Biology, 3rd Edn. Scientific American Books, New York, NY. [Google Scholar]
- 28.Pallen M., Chaudhuri,R. and Khan,A. (2002) Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends Microbiol., 10, 556–563. [DOI] [PubMed] [Google Scholar]
- 29.Snel B., Bork,P. and Huynen,M.A. (2002) The identification of functional modules from the genomic association of genes. Proc. Natl Acad. Sci. USA, 99, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rives A.W. and Galitski,T. (2003) Modular organization of cellular networks. Proc. Natl Acad. Sci. USA, 100, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravasz E., Somera,A.L., Mongru,D.A., Oltvai,Z.N. and Barabasi,A.L. (2002) Hierarchical organization of modularity in metabolic networks. Science, 297, 1551–1555. [DOI] [PubMed] [Google Scholar]
- 32.Horswill A.R. and Escalante-Semerena,J.C. (2001) In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry, 40, 4703–4713. [DOI] [PubMed] [Google Scholar]
- 33.von Mering C., Huynen,M., Jaeggi,D., Schmidt,S., Bork,P. and Snel,B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res., 31, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellor J.C., Yanai,I., Clodfelter,K.H., Mintseris,J. and DeLisi,C. (2002) Predictome: a database of putative functional links between proteins. Nucleic Acids Res., 30, 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose J.D., Maddry,J.A., Comber,R.N., Suling,W.J., Wilson,L.N. and Reynolds,R.C. (2002) Synthesis and biological evaluation of trehalose analogs as potential inhibitors of mycobacterial cell wall biosynthesis. Carbohydr. Res., 337, 105–120. [DOI] [PubMed] [Google Scholar]
- 36.McKinney J.D., Honer zu Bentrup,K., Munoz-Elias,E.J., Miczak,A., Chen,B., Chan,W.T., Swenson,D., Sacchettini,J.C., Jacobs,W.R.,Jr and Russell,D.G. (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature, 406, 735–738. [DOI] [PubMed] [Google Scholar]
- 37.Drews S.J., Hung,F. and Av-Gay,Y. (2001) A protein kinase inhibitor as an antimycobacterial agent. FEMS Microbiol. Lett., 205, 369–374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.