Abstract
While genome and transcriptome sequencing has revealed a large number and diversity of Anopheles gambiae predicted proteins, identifying their functions and biosynthetic pathways remains challenging. Applied mass spectrometry based proteomics in conjunction with mosquito genome and transcriptome databases were used to identify 44 proteins as putative components of the eggshell. Among the identified molecules are two vitelline membrane proteins and a group of seven putative chorion proteins. Enzymes with peroxidase, laccase and phenoloxidase activities, likely involved in cross-linking reactions that stabilize the eggshell structure, also were identified. Seven odorant binding proteins were found in association with the mosquito eggshell, although their role has yet to be demonstrated. This analysis fills a considerable gap of knowledge about proteins that build the eggshell of anopheline mosquitoes.
Keywords: chorion, vitelline membrane, OBPs, phenoloxidase, peroxidase, mosquito
Introduction
Insect eggshells provide the embryo protection from physical and biological insults and insure their survival. Most current knowledge of insect eggshell morphology and composition comes from studies of Drosophila melanogaster (Margaritis, 1985; Waring, 2000; Cavaliere et al., 2008). The fruit fly eggshell is divided into two major and structurally-distinct layers, the inner vitelline membrane (VM) and the outer chorion (Trougakos and Margaritis, 2002). Proteins participating in eggshell assembling include abundant structural components of the vitelline membrane (VM26A.1, VM26A.2, VM34C, VM32E) and those involved in building the chorion layers (s15, s16, s18, s19, s36, s389) (Cavaliere et al., 2008; Fakhouri et al., 2006; Swever et al., 2005). These molecules become stabilized in the final eggshell structure through enzyme-catalyzed protein cross-linking. The D. melanogaster Pxd gene product (Pxd peroxidase) is an essential enzyme in a process that involves secretion of hydrogen peroxide by the follicle cells and the formation of di- and tri-tyrosyl cross-links among the eggshell proteins (Konstandi et al. 2005).
Although the primary function of eggshells in protecting embryos is conserved throughout the evolution of insects, differences in ecology, and in particular, ovipositional substrates are strong forces that drive the evolution of eggshell morphology, organization and composition (Jagadeeshan and Singh, 2007). Mosquito (Diptera, Culicidae) eggshells show remarkable diversity in physical properties and structure, presumably resulting from adaptation to the large variety of environments exploited by these insects. For example, the eggs of the yellow fever mosquito, Aedes aegypti, are highly-resistant to desiccation allowing embryos to survive for months in harsh and dry conditions (Christophers, 1960; Sota and Mogi, 1992). In contrast, the eggshells of the human malaria vector, Anopheles gambiae, are more permeable, restricting their survival and development to humid environments (Deane and Causey, 1943, Beier et al., 1990). Greater knowledge of the proteins that comprise eggshells is required to understand these differences and how they contribute to successful mosquito reproductive strategies. Aedes aegypti eggshell components have been characterized (Li and Li, 2006, Li, 1994), however there is considerable lack of information of the proteins that constitute the eggshells of anopheline mosquitoes. Here we use a mass spectrometry/proteomics approach to identify An. gambiae eggshell proteins.
Materials and Methods
Egg collection and eggshell isolation
Egg collection and eggshell isolation were carried out as described by Li and Li (2006) with modifications. Ovaries were dissected from females at 72 hours post blood meal (hPBM) in Ringer’s solution and frozen at −70°C. Approximately 500 eggs were homogenized in a HEPES buffer, pH8.0 (10 mM HEPES, 130 mM NaCl, 4.7 mM KCl, 0.5 mM phenyl thiocarbamide, 0.1mM p-nitrophenyl-p′-guanidinobenzoate, 5 mM ethylene diamine tetraacetic acid, 1% Triton X-100, 0.1 mM diethyldithiocarbamic acid, 1 mM phenylmethylsulfonyl fluoride). Eggshells were collected by sedimentation on ice for 15 minutes and the supernatant decanted. The sediment was washed five times for 15 min each with homogenization buffer. Eggshells then were sonicated for 30 seconds and centrifuged (100 g, 10 min, 4°C). The eggshell pellet was washed 5–6 times and centrifuged (100 g, 10 min, 4°C) to remove yolk and other cellular contaminants.
SDS-PAGE
The eggshell pellet was solubilized in SDS-sample buffer (Biorad) containing DTT or urea and DTT. The samples were boiled at 95°C for 3 min and resolved by 4–15% SDS-PAGE. Electrophoresis was carried out at 70V using Biorad Mini-protean II electrophoresis system. Proteins were stained with Coomassie blue R. Precision Plus (BioRad) protein standards were used to estimate the Mr of the resolved proteins.
LC-MALDI TOF/TOF mass spectrometry
Eggshell preparations were rinsed several times using de-ionized, distilled water (ddH2O) to remove homogenization buffer, followed by treatment with either cyanogen bromide (CNBr) followed by trypsin, or trypsin alone. For CNBr/trypsin treatment, samples were vortexed and sonicated in a mixture of 5 μl ddH2O, 50 μl 5M CNBr in acetonitrile and 150 μl of trifloroacetic acid (TFA). Once dissolved, the mixture was capped with argon and incubated in the dark at 4°C overnight. To remove excess CNBr and TFA, the resulting solution was diluted with ddH2O and then vacuum-dried. This process was repeated twice followed by reconstitution with trypsin-digestion buffer (50mM NH4HCO3 with 30% acetonitrile). For treatment with trypsin alone, the eggshell pellet was suspended in trypsin-digest buffer.
Both CNBr-trypsin and trypsin-alone samples were treated with 5mM tris 2-carboxyethyl phosphine (TCEP, 5 mM) at room temperature (RT) for 20 min, and alkylated with iodoacetamide (10mM final concentration) in the dark at RT for 20 min. Reduced and alkylated samples were digested with trypsin gold (Promega) at a substrate-to-enzyme ratio of 1:50 (w/w) at 37°C overnight. Samples were centrifuged to remove insoluble material and the supernatant was vacuum-dried to remove acetonitrile and acidified with TFA (1% of final volume) prior to RP-HPLC fractionation.
Peptides were trapped for LC-MALDI analysis on an Acclaim PepMap100 Trap 0.3 × 5 mm C18 (Dionex) followed by washing with solvent A (2% acetonitrile, 0.1% TFA in water), then fractionated over a 0.075 × 150 mm C18 column (Michrom Inc.) by nanocapillary, reverse-phase HPLC using a linear gradient of 0 – 50% solvent B (85% acetonitrile, 5% isopropanol, 0.1% TFA) over 120 min at a flow rate of 0.2 μl/min, with online UV detection (LC Packings). Eluted material was mixed online, in a micromixing tee, with a solution of 75 mg/ml α-cyano-4-hydroxy cinnamic acid matrix in 10mM sodium monobasic phosphate, 50:50 CH3CN:water, driven at a rate of 0.4 μl/min. The mixture was deposited robotically on a stainless-steel plate in 288 spots., MS spectra were acquired from all spots using a 4700 MALDI-TOF/TOF (AB Inc.). All peaks with S:N >30 or cluster area >65 were subjected to MALDI TOF/TOF MS/MS.
MS/MS-based peptide and protein hits were discovered and scored using Mascot 2.2.04 (Matrix Science, London, UK), followed by Peptide Prophet (Nesvizhskii et al., 2003). Peptide identifications were accepted if they could be established with a Peptide Prophet error ≤ 0.05. Protein identifications were then accepted if they could be established with MASCOT score > 30, or contained at least two identified peptides. Peptides derived from the cleavage of vitelline/vitellogenin (AAF82131, AAF82132, AGAP004203) were excluded from analyses. Annotations and known or putative functions for the identified proteins were retrieved from Vectorbase, Anopheles gambiae gene set AgamP3.5, (http://www.vectorbase.org) and AnoXcel, Sep15/2009 release, (http://exon.niaid.nih.gov/transcriptome/AnoXcel-Sep09/AnoXcel-Sep-2009-Web.zip).
Transcription product amplification
Total RNA was extracted from ovaries dissected at 24, 48 and 72 hPBM using Trizol reagent and treated subsequently with DNase (Promega). Total RNA was extracted from the mosquito carcasses remaining after ovary dissections and treated similarly. Samples of carcass and ovary RNA (100 ng) were reverse-transcribed and cDNAs amplified using one-step RT-PCR (Invitrogen) and oligonucleotide primers designed for selected genes identified by mass spectrometry (Supplementary Table 1). To account for variation in mRNA levels in reverse-transcription reactions among samples, the products of amplification of the An. gambiae ribosomal S60 gene (AGAP002122-RA) were used for normalization. In all cases, the one-step RT-PCR reactions consisted of a 30 min reverse-transcription step at 50°C, followed by 15 minutes at 95°C and 34 cycles of 30 seconds at 94°C, 30 seconds at 65°C and 1 minute extension at 72°C. A final extension consisted of 10 minutes at 72°C. Amplified products were resolved on 1% agarose gels and visualized following ethidium-bromide staining. Products were cloned into TOPO TA vector and sequenced to confirm their identity.
Results and Discussion
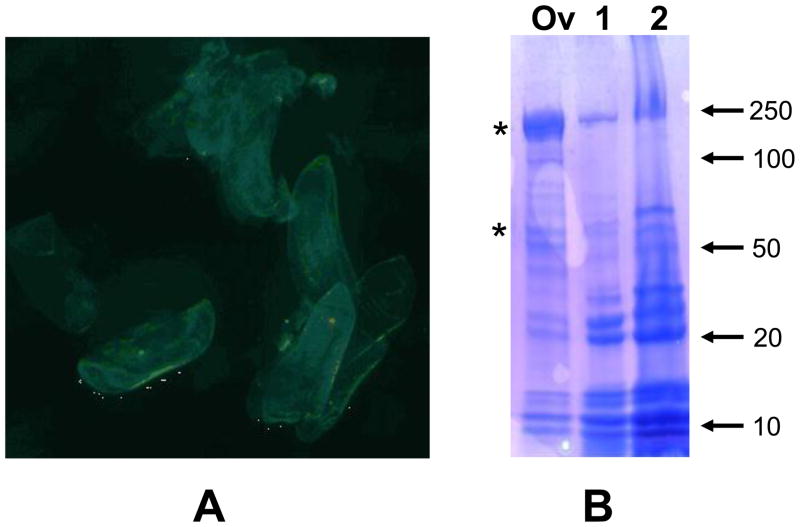
Eggshell proteins dissolved in mixtures of either SDS/DTT or SDS/urea/DTT and resolved by SDS-PAGE displayed similar patterns following staining (Figure 1). Two major groups of proteins are enriched during purification, one comprised of proteins with Mrs between 10 and 15 kDa and another with proteins between 20 and 30 kDa. Although microscopic examination of the eggshells preparations revealed no evident contamination with vitelline granules or other cellular structures, the 200 and 66 kDa vitelline subunits were never completely eliminated, even after extensive washing steps. Mass spectrometry-based proteomics in conjunction with mosquito genome and transcriptome databases were utilized to identify and assign 44 proteins from these preparations as putative components of the An. gambiae eggshell (Table 1). Prior to RP-HPLC fractionation of preparations treated with either CNBr or trypsin, or trypsin, alone the samples were centrifuged revealing that a small fraction of the eggshell material remained insoluble. The material contained in the pellets may represent non-proteinaceous components of the eggshell, such as lipids and carbohydrates, as well as insoluble proteins. Therefore we do not argue that in this study we have identified all of the proteins comprising the An. gambiae eggshell structure.
Figure 1.
Anopheles gambiae eggshell purification and protein analysis. A) Eggshells purified by differential sedimentation from ovaries dissected at 72 hours post blood meal and visualized by light microscopy. B) Ovary (Ov), and eggshell protein preparations dissolved in buffers containing SDS/DTT (1) or SDS/urea/DTT (2) were run on a 4–15% SDS-PAGE and stained with Coomassie blue R. The electrophoretic migrations of molecular weight markers are indicated at the right of the figure and represented in kDa. Vitellin subunits are indicated with asterisks.
Table I.
Anopheles gambiae eggshell proteins identified by LC-MSMS
| Gene ID | Protein score | Known/putative function/comments |
|---|---|---|
| Odorant binding proteins | ||
| AGAP000641 | 1516 | OBP34/37 |
| AGAP000642 | 1048 | OBP35 |
| AGAP000643 | 1014 | OBP36 |
| AGAP002025 | 499 | OBP11 |
| AGAP011647 | 152 | OBP1 |
| AGAP010648 | 96 | OBP44 |
| AGAP002189 | 138 | OBP13 |
| Enzymes | ||
| AGAP004038 | 415 | Chorion peroxidase |
| AGAP006176 | 104 | Laccase-2 |
| AGAP005959 | 97 | Yellowg2-Dopachrome conversion enzyme |
| AGAP004978 | 55 | prophenoloxidase 9 |
| AGAP003233 | 54 | peroxidase |
| AGAP007020 | 34 | thioredoxin |
| Vitelline membrane components | ||
| AGAP002134 | 645 | Vitelline membrane component |
| AGAP008696 | 560 | Vitelline membrane component |
| Putative chorion components | ||
| AGAP006551 | 125 | Putative chorion protein |
| AGAP006555 | 123 | Putative chorion protein |
| AGAP006553 | 118 | Putative chorion protein |
| AGAP006554 | 112 | Putative chorion protein |
| AGAP006549 | 72 | Putative chorion protein |
| AGAP006556 | 65 | Putative chorion protein |
| AGAP006550 | 45 | Putative chorion protein |
| Others | ||
| AGAP006524 | 197 | UNKNOWN |
| AGAP006563 | 140 | UNKNOWN |
| AGAP003149 | 94 | UNKNOWN |
| AGAP010147 | 77 | UNKNOWN |
| AGAP004182 | 70 | UNKNOWN |
| AGAP006527 | 70 | UNKNOWN |
| AGAP003047 | 64 | Schistosoma mansoni egg protein |
| AGAP004969 | 51 | Ionotropic glutamate receptor |
| AGAP010252 | 50 | Ribosomal protein L14 |
| AGAP002306 | 50 | Ribosomal protein L4 |
| AGAP002830 | 47 | C-1-tetrahydrofolate synthase |
| AGAP012261 | 46 | UNKNOWN |
| AGAP005802 | 45 | UNKNOWN |
| AGAP005061 | 42 | UNKNOWN |
| AGAP000547 | 37 | UNKNOWN |
| AGAP006584 | 35 | UNKNOWN |
| AGAP005338 | 35 | UNKNOWN |
| AGAP003911 | 34 | UNKNOWN |
| AGAP004887 | 33 | Ribosomal protein S17 |
| AGAP000672 | 33 | UNKNOWN |
| AGAP007758 | 32 | UNKNOWN |
| AGAP006805 | 28 | UNKNOWN |
Protein score is the sum of the unique (excluding duplicate matches) ions scores that made the significant homology cutoff in MASCOT (http://www.matrixscience.com/help/interpretation_help.html). The mass spectra of selected peptides are displayed in Supplementary Figure 1.
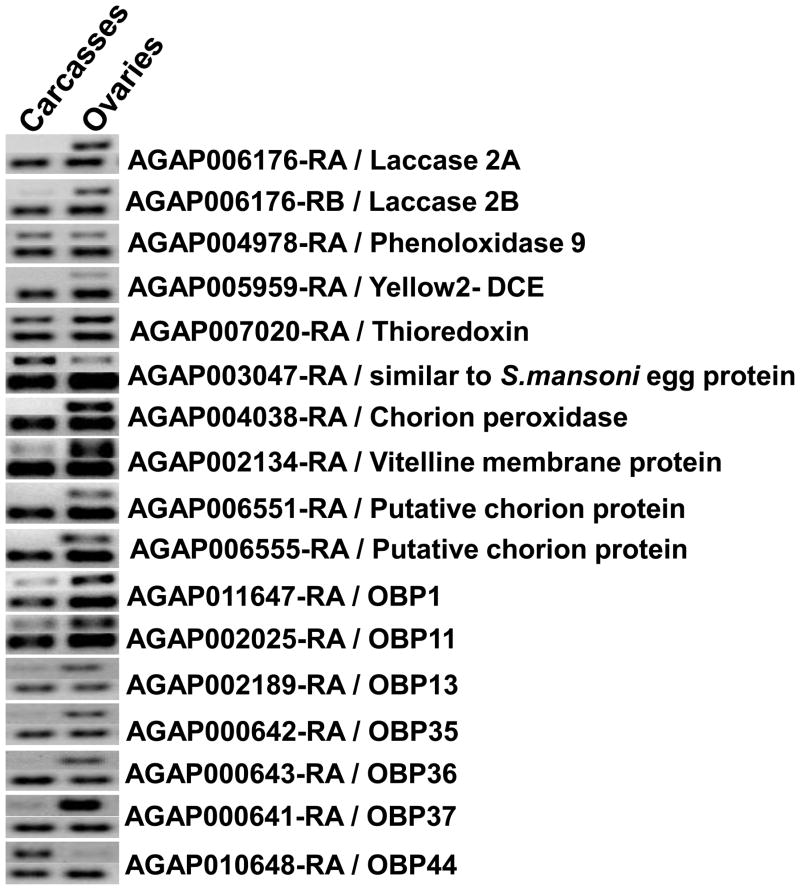
Organ-specific accumulations of a set of transcripts encoding the identified eggshell proteins were determined by RT-PCR (Figure 2). Transcripts encoding two Laccase 2 isoforms had similar pattern of mRNA accumulation and were detected preferentially in ovaries. Chorion peroxidase and dopachrome-conversion enzyme-encoding transcripts (AGAP004038-RA and AGAP005959-RA, respectively) were detected only in ovaries. AGAP003047, a gene that encodes a product similar to a Schistosoma mansoni egg protein, and AGAP007020 (thioredoxin) are expressed in both ovaries and carcasses. The transcript of a putative vitelline membrane component, AGAP002134-RA, is accumulated principally in the ovaries with a faint signal detected in carcass RNA.
Figure 2.
Accumulation of eggshell-related transcripts in Anopheles gambiae ovaries. Total RNA samples extracted from dissected ovaries or carcasses (body without ovaries) were utilized as templates in RT-PCR reactions. Amplified products were resolved by electrophoresis in 1% agarose gels and stained by ethidium bromide. Two parallel reactions were performed for each target transcript, one with eggshell-transcript specific primers (top signal pair) and the second with primers that amplify a fragment of the An. gambiae S60 ribosomal protein transcript (bottom signal pair).
Amplification products corresponding to the transcripts of genes encoding OBPs (AGAP000641- OBP34/37, AGAP000642- OBP35, AGAP000643- 0BP36, AGAP002025- OBP11, AGAP002189- OBP13, AGAP010648- OBP44, AGAP011647- OBP1) were detected in both ovaries and carcasses, and with exception of OBP 44, mRNA accumulation was higher in the ovaries. Insect OBPs are reported to be expressed mainly in antennae, and are proposed to bind small hydrophobic odorant molecules and carry and present them to the olfactory receptors (Biessmann et al., 2002). Structurally-related proteins also have been identified in non-sensory organs of insects, supporting the hypothesis that OBPs and OBP-like proteins could be utilized in tasks unrelated to olfaction (Biessmann et al., 2002). For example, D7-related (D7r) proteins similar in structure to OBPs are abundant components in the saliva of hematophagic Diptera. The An. gambiae D7r proteins have been shown to bind the biogenic amines serotonin, norepinephrine, and histamine with high affinity thereby reducing the concentrations of these effectors at the feeding site and resulting in an anti-hemostatic activity (Mans et al., 2007, Calvo et al., 2009). The Tenebrio molitor THP12 gene encodes a member of the OBP family that is accumulated in the hemolymph and has a strong capacity to bind fatty acids, a finding consistent with a role in the transport of small hydrophobic molecules (Rothemund et al., 1999). OBPs similarly could carry substrates for eggshell assembling reactions. Furthermore, studies support the conclusion that odorant molecules act as chemo-attractants for sperm (Fukuda et al., 2004), and the OBPs identified in this study could enrich specific attractant molecules at the surface of the mosquito eggshell. The Ae. aegypti OBP22 was found to be expressed in both the sensory organs and male reproductive apparatus, providing additional support for this hypothesis (Li et al., 2008).
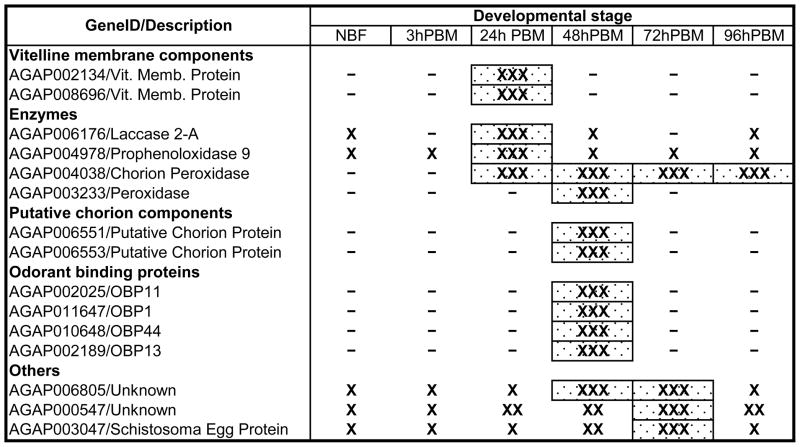
Temporal patterns of gene expression during oogenesis were retrieved from http://www.angaged.bio.uci.edu/(Marinotti et al., 2006). Although these data describe RNA accumulation in whole mosquitoes, a number of genes identified as encoding eggshell proteins increase in representation following a blood meal (Table 2, Supplementary figure 1). Furthermore, a sequential pattern of RNA accumulation is evident, with transcripts encoding vitelline membrane proteins and three enzymes displaying a maximum accumulation at 24 hPBM. The transcripts of one of the peroxidases (AGAP003233-RA) and those encoding OBPs and the putative chorion proteins have maximal abundance at 48 hPBM. Three other transcripts encoding proteins with unknown function are up-regulated during oogenesis with maximum accumulation at 48 or 72 hPBM.
TABLE 2.
Timing of Eggshell Gene Expression During Oogenesis
 |
Two proteins corresponding to the transcripts AGAP002134-RA, and AGAP008696-RA, contain 25–26 amino acid sequences similar to a hydrophobic vitelline membrane (VM) domain identified previously in D. melanogaster (Andrenacci et al., 2001, Figure 3). This domain is necessary for the assembly of the VM proteins in the vitelline membrane. The deletion of ten amino acids within this domain impairs the integration of VM proteins into the vitelline membrane and their cross-linking at the final stages of oogenesis. This domain is present in all the D. melanogaster vitelline membrane proteins and homologous proteins of the mosquito Ae. aegypti (Edwards et al., 1998) and An. gambiae (this study). In all three species, this domain contains three precisely spaced cysteine residues, suggesting that it performs the general function of holding together the various vitelline membrane proteins by disulfide cross-links. AGAP002134-PA contains one of these conserved domains. AGAP008696-RA encodes a protein with two of the hydrophobic domains in an open reading frame (+2) not displayed currently in Vectorbase. Support for this modified annotation is provided by the peptide sequence, EYHHAPAPAYHHAPAYEAKKEYAAPAYEAPKKEYGHHAAPPMHHGYEAKKPA YGGYGKR, identified in the LC-MS/MS analyses and also encoded in the AGAP008696-RA +2-frame. A comparison of the full length sequences of the identified An. gambiae VM proteins with those of the D. melanogaster and Ae. aegypti homologues (Andrenacci et al., 2001; Edwards et al., 1998), reveals a low overall primary sequence conservation (not shown) indicating that these genes diverged significantly in the course of evolution and that the extensive amino acid substitutions external to the conserved VM motif do not affect their established structural function.
Figure 3.
Conservation of a hydrophobic amino acid motif among Anopheles gambiae, Aedes aegypti and Drosophila melanogaster vitelline membrane proteins. This conserved motif is composed of 25–27 amino acids and is required for the proper assembly of the eggshell. Protein IDs: AGAP002134, AGAP008696, An. gambiae vitelline membrane identified in this study, with a and b corresponding to the two domains discussed in the text; AAB51284, AAB51282 and AAB1283 correspond to Ae. aegypti vitelline membrane proteins; and CG9046, CG9271, CG9048 and CG16874 are D. melanogaster vitelline membrane proteins. The alignment was performed at PRALINE multiple sequence alignment, http://www.ibi.vu.nl/programs/pralinewww/.
Enzymes with phenoloxidase (AGAP004978-PA), laccase (AGAP006176-PA), dopachrome-conversion (AGAP005959-PA and AGAP000879-PA), and peroxidase (AGAP004038-PA, AGAP003233-PA, and AGAP007020-PA) activities were identified. The product of AGAP004978-PA is the An. gambiae prophenoloxidase 9 (AgPPO9), which is the orthologue of Culex quinquefasciatus CPIJ016564 and Ae. aegypti AAEL013492 (AePPO5, Kim et al., 2005). Despite conservation of sequence and temporal expression profiles, (AgPPO9 and AePPO5 transcripts are accumulated in female mosquitoes following a blood meal), AgPPO9 transcripts are present in ovaries of vitellogenic females while AePPO5 is not expressed in the ovaries (Kim et al., 2005; Marinotti et al., 2006). AgPPO9 transcripts are found in tissues other than the ovaries, indicating that the enzyme is involved in processes in addition to eggshell formation. AePPO5 (Kim et al. 2005) and AgPPO9 lack a secretory signal peptide, and the mechanism involved in the transport of these enzymes to the eggshell is unknown. Extracellular proteins, such as fibroblast growth factors, interleukin-1 and galectins found in the extracellular matrix of other organisms are exported without a classical N-terminal signal peptide by the non-classical secretory pathway that works independently of the ER Golgi network (Bendtsen et al., 2004, Marie et al., 2008). However, it is also possible that the mosquito proteins are being synthesized locally by the follicle cells and deposited on the surface of the eggshell following apoptosis of the cells.
Laccases have p-diphenol oxidase enzymatic activity and participate in cuticular tanning in insects. Pan et al. (2009) reported that Cx. pipiens pallens laccase 2 (CpLac2) is expressed abundantly in the egg developmental stages and postulated a role in eggshell tanning. The presence of the An. gambiae Laccase 2 transcripts (AGAP006176-RA and -RB) in ovaries provides additional support for this hypothesis. Furthermore, the peptides identified by mass spectrometry verified that the AGAP006176-PA isoform is present in the eggshell. The accumulation of AGAP006176-PB in the eggshell remains to be demonstrated unambiguously.
The translation products of AGAP005959 and AGAP000879 contain motifs similar to those found in the Major Royal Jelly protein family. This protein family includes the products of the Drosophila yellow genes that encode dopachrome-conversion enzymes important for cuticle melanization (Claycomb, 2004). The D. melanogaster yellow-g2 gene product participates in the cross-linking of the chorion and/or underlying vitelline membrane proteins.
Three peroxidases, corresponding to the products of AGAP004038 (HPX8), AGAP003233, and AGAP007020, have temporally-distinct expression patterns. HPX8, the orthologue of the D. melanogaster (Dpxt), and Ae. aegypti (AAT27427; AAEL004386) chorion peroxidases, has transcripts that accumulate following a blood meal. The An. gambiae transcript level remains high even after oviposition, an unexpected phenotype for a gene involved in eggshell production. AGAP003233 encodes a protein with a haem-peroxidase domain, is orthologous to the D. melanogaster chorion peroxidase, pxt (Konstandi et al., 2005), and its transcript is accumulated at 48 hPBM in a pattern similar to other putative chorion components. Similar to AgPPO9, the AGAP003233 product does not have a typical secretory signal peptide, again supporting the hypothesis that follicle cells utilize a non-classical transport pathway for the secretion of these enzymes. Covalent cross-linking of the chorion proteins involves endogenous hydrogen peroxide (Margaritis, 1985) and AGAP007020 encodes the TPX5 thioredoxin-dependent peroxidase, an enzyme believed to be associated intimately with the regulation of intracellular peroxide concentration (Salinas et al., 2004).
Seven small proteins (~11 kDa) with secretory signal peptides at their amino terminal ends are encoded by the genes, AGAP006549, AGAP006550, AGAP006551, AGAP006553, AGAP006554, AGAP006555 and AGAP006556. These proteins do not have any known functional motifs and are encoded by genes clustered in an ~15.5kb region on Chromosome 2L (location: 34,113,357 and 34,128,909). Eggshell genes of Drosophilids and the silk moth, Bombyx mori, are organized in clusters (Hibner et al., 1991; Vlachou and Komitopoulou, 2001), consistent with the interpretation that chorion genes duplication and divergence is a frequent event in insect evolution. Despite conservation in genomic organization, rapid evolution and sequence diversion of sex- and reproduction-related genes is a common observation. Thus, chorion proteins as a group evolve rapidly as an adaptation to divergences in ovipositional substrate and different microhabitats, providing adaptation to stresses such as desiccation, plant toxins, pathogen invasions and predation (Jagadeeshan and Singh, 2007). Anopheles and Drosophila lineages diverged ~250 million years ago (Gaunt and Miles, 2002) and their females lay eggs in different substrates. The strong selective pressures on chorion components resulting from the distinct life histories likely resulted in extensive mutations, precluding identification of a common ancestry based on amino acid sequence conservation.
Proteins with similarity to cytoplasmic or cell membrane components, for example, ribosomal proteins, tropomyosin, c-1-tetrahydrofolate synthase and glutamate receptor, also were identified and could represent either contamination of the eggshell preparations with cellular fragments or indications of novel function for these molecules. Seventeen additional proteins identified in these analyses have no associated function or description in the existing databases. The transcripts encoding these proteins, with three exceptions (Table 2), are not induced during oogenesis, further supporting the interpretation that they encode constitutively-expressed cellular components.
In summary, associations among proteomics data and EST databases helps establish the identity of large numbers of proteins and supports the development of hypotheses for their function. The present study provides data that 44 predicted mosquito genes and their corresponding transcripts and proteins likely are involved in eggshell formation. We do not expect that we have identified all of the proteins comprising this structure, but we have set the foundation for future molecular, biochemical and genetic studies. For example, RNAi-based gene-silencing techniques offer an effective way to validate the function of the identified proteins in eggshell assembling. Furthermore, the annotation of the An. gambiae genome is a dynamic process, with gene boundaries and corresponding transcripts and proteins re-assigned constantly (Gomez et al., 2005; Li et al., 2006). We identified one instance of either alternative or incorrect protein prediction (AGAP008696-PA), and a new reading frame for the corresponding transcript was validated. The identification of two vitelline membrane proteins, a group of seven putative chorion proteins, peroxidase, laccase and phenoloxidase enzymes, and odorant binding proteins, as components of the An. gambiae eggshell fills a considerable knowledge gap for this important vector species.
Supplementary Material
Acknowledgments
Lynn Olson assisted in preparing the manuscript. DA and GY were supported in part by NIH-Fogarty D43 TW01505, WC and PG by the Chao Comprehensive Cancer Center, OM and AAJ by NIH NIAID AI29746.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrenacci D, Cernilogar FM, Taddel C, Rotoli D, Cavaliere V, Graziani F, Gargiulo G. Specific domains drive VM32E protein distribution and integration in Drosophila eggshell layers. Journal of Cell Science. 2001;114:2819–2829. doi: 10.1242/jcs.114.15.2819. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Engineering Design and Selection. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Walter MF, Dimitratos S, Woods D. Isolation of cDNA clones encoding putative odourant binding proteins from the antennae of the malaria-transmitting mosquito, Anopheles gambiae. Insect Molecular Biology. 2002;11:123–132. doi: 10.1046/j.1365-2583.2002.00316.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito anti-inflammatory protein. Proceedings of the National Academy of Sciences. 2009;106:3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere V, Bernardi F, Romani P, Duchi S, Gargiulo G. Building up the Drosophila eggshell: First of all the eggshell genes must be transcribed. Developmental Dynamics. 2008;237:2061–2072. doi: 10.1002/dvdy.21625. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Developmental Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Deane MP, Causey OR. Viability of Anopheles gambiae eggs and morphology of unusual types found in Brazil. American Journal of Tropical Medicine and Hygiene. 1943;23:95–102. [Google Scholar]
- Edwards MJ, Severson DW, Hagedorn HH. Vitelline envelope genes of the yellow fever mosquito, Aedes aegypti. Insect Biochemistry and Molecular Biology. 1998;28:915–925. doi: 10.1016/s0965-1748(98)00083-6. [DOI] [PubMed] [Google Scholar]
- Fakhouri M, Elalayli M, Sherling D, Hall JD, Miller E, Sun X, Wells L, LeMosy EK. Minor proteins and enzymes of the Drosophila eggshell matrix. Developmental Biology. 2006;293:127–41. doi: 10.1016/j.ydbio.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. Journal of Cell Science. 2004;15:5835–5845. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographical landmarks. Molecular Biology and Evolution. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Gomez SM, Eiglmeier K, Segurens B, Dehoux P, Couloux A, Scarpelli C, Wincker P, Weissenbach J, Brey PT, Roth CW. Pilot Anopheles gambiae full-length cDNA study: sequencing and initial characterization of 35,575 clones. Genome Biology. 2005;6:R39. doi: 10.1186/gb-2005-6-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibner BL, Burke WD, Eickbush TH. Sequence Identity in an Early Chorion Multigene Family Is the Result of Localized Gene Conversion. Genetics. 1991;128:595–606. doi: 10.1093/genetics/128.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeshan S, Singh RS. Rapid Evolution of Outer Egg Membrane Proteins in the Drosophila melanogaster Subgroup: A Case of Ecologically Driven Evolution of Female Reproductive Traits. Molecular Biology and Evolution. 2007;24:929–938. doi: 10.1093/molbev/msm009. [DOI] [PubMed] [Google Scholar]
- Kim SR, Yao Y, Han Q, Christensen BM, Li J. Identification and molecular characterization of a prophenoloxidase involved in Aedes aegypti chorion melanization. Insect Molecular Biology. 2005;14:185–194. doi: 10.1111/j.1365-2583.2004.00547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstandi OA, Papassideri IS, Stravopodis DJ, Kenoutis CA, Hasan Z, Katsorchis T, Wever R, Margaritis LH. The enzymatic component of Drosophila melanogaster chorion is the Pxd peroxidase. Insect Biochemistry and Molecular Biology. 2005;35:1043–1057. doi: 10.1016/j.ibmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Li J. Egg chorion tanning in Aedes aegypti mosquito. Comparative Biochemistry and Physiology. 1994;109A:835–843. doi: 10.1016/0300-9629(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Li J, Riehle MM, Zhang Y, Xu J, Oduol F, Gomez SM, Eiglmeier K, Ueberheide BM, Shabanowitz J, Hunt DF, Ribeiro JM, Vernick KD. Anopheles gambiae genome re-annotation through synthesis of ab initio and comparative gene prediction algorithms. Genome Biology. 2006;7:R24. doi: 10.1186/gb-2006-7-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JS, Li J. Major chorion proteins and their crosslinking during chorion hardening in Aedes aegypti mosquitoes. Insect Biochemistry and Molecular Biology. 2006;36:1195–1203. doi: 10.1016/j.ibmb.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Picimbon JF, Ji S, Kan Y, Chuanling Q, Zhou JJ, Pelosi P. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochemical and Biophysical Research Communications. 2008;372:464–468. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Calvo E, Ribeiro JM, Andersen JF. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. Journal of Biological Chemistry. 2007;282:36626–36633. doi: 10.1074/jbc.M706410200. [DOI] [PubMed] [Google Scholar]
- Margaritis LH. The egg-shell of Drosophila melanogaster.III. Covalent cross linking of the chorion proteins involves endogenous hydrogen peroxide. Tissue Cell. 1985;17:553–559. doi: 10.1016/0040-8166(85)90031-x. [DOI] [PubMed] [Google Scholar]
- Marie M, Sannerud R, Avsnes Dale H, Saraste J. Take the ‘A’ train: on fast tracks to the cell surface. Cellular and Molecular Life Sciences. 2008;65:2859–2874. doi: 10.1007/s00018-008-8355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Calvo E, Nguyen QK, Dissanayake S, Ribeiro JM, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Molecular Biology. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Pan C, Zhou Y, Mo J. The clone of laccase gene and its potential function in cuticular penetration resistance of Culex pipiens pallens to fenvalerate. Pesticide Biochemistry and Physiology. 2009;93:105–111. [Google Scholar]
- Rothemund S, Liou YC, Davies PL, Krause E, Sönnichsen FD. A new class of hexahelical insect proteins revealed as putative carriers of small hydrophobic ligands. Structure. 1999;15:1325–1332. doi: 10.1016/s0969-2126(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Salinas G, Selkirk ME, Chalar C, Maizels RM, Fernández C. Linked thioredoxin-glutathione systems in platyhelminths. Trends in Parasitology. 2004;20:340–346. doi: 10.1016/j.pt.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Sota T, Mogi M. Interspecific variation in desiccation survival time of Aedes (Stegomyia) mosquito eggs is correlated with habitat and egg size. Oecologia. 1992;90:353–358. doi: 10.1007/BF00317691. [DOI] [PubMed] [Google Scholar]
- Swevers L, Raikhel AS, Sappington TW, Shirk P, Iatrou K. Vitellogenesis and Post-Vitellogenic Maturation of the Insect Ovarian Follicle. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Elsevier Pergamon; Oxford, UK: 2005. pp. 87–156. [Google Scholar]
- Trougakos IP, Margaritis LH. Novel morphological and physiological aspects of insect eggs. In: Hilker M, Meiners T, editors. Chemoecology of insect eggs and egg deposition. Blackwell Wissenschaftsverlag; Berlin, Germany: 2002. pp. 3–36. [Google Scholar]
- Vlachou D, Komitopoulou The chorion genes of the medfly. II. DNA sequence evolution of the autosomal chorion genes s18, s15, s19 and s16 in Diptera. Gene. 2001;270:41–52. doi: 10.1016/s0378-1119(01)00482-6. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. International Reviews of Cytology. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.