Abstract
The base pairing properties of oligonucleotide duplexes containing 8-aza-7-deaza-2′-deoxyisoguanosine, its 7-bromo or its 7-iodo derivative are described. The nucleosides were synthesized on a convergent route, protected and converted into phosphoramidites. Oligonucleotides were prepared on a solid-phase and were hybridized to yield duplexes with parallel (ps) or antiparallel (aps) chain orientation. The 8-aza-7-deaza-2′-deoxyisoguanosine-containing duplexes show almost identical base pairing stability as those containing 2′-deoxyisoguanosine, while the 7-substituted derivatives induce a significant duplex stabilization both in ps and aps DNA. Self-complementary duplexes with parallel chain orientation are exceptionally stable due to the presence of 5′-overhangs. The bulky halogen substituents were found to be well accommodated in the grooves both of aps and ps DNA.
INTRODUCTION
Parallel stranded (ps) DNA can be constructed from oligonucleotides incorporating isoguanine-cytosine and/or guanine-isocytosine pairs instead of the canonical guanine-cytosine pair (1–4). The dA–dT pair shows ambiguous base pairing properties and is therefore accepted in antiparallel (aps) as well as in ps DNA. Usually ps DNA is less stable than aps DNA (5). Modified nucleosides can be used to increase the stability of ps DNA to the level of aps DNA (6).
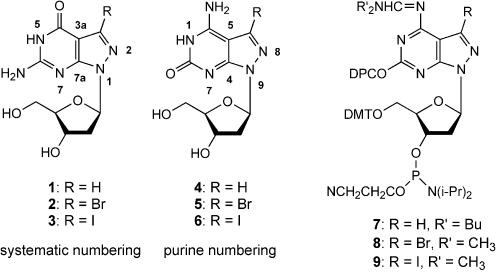
Earlier, it was shown that the introduction of 7-substituents into 7-deazapurine (7-deazapurine = pyrrolo[2,3-d]pyrimidine) or 8-aza-7-deazapurine (8-aza-7-deazapurine = pyrazolo [3,4-d]pyrimidine) nucleosides results in a stabilization of oligonucleotide duplexes with antiparallel chain orientation (7–12) (purine numbering is used throughout the Results and Discussion section). This was demonstrated on oligonucleotide duplexes containing 8-aza-7-deaza-2′-deoxyguanosine (1), 7-bromo-8-aza-7-deaza-2′-deoxyguanosine (2) or 7-iodo-8-aza-7-deaza-2′-deoxyguanosine (3) (13,14). Investiga tions with regard to quartet or pentaplex formation have also shown that 8-aza-7-deaza-2′-deoxyisoguanosine (4) forms supramolecular assemblies while the 7-bromo substituted derivative 5 as well as compound 1 prevent the molecules from aggregation (15,16).
Different from aps DNA, which forms a major and a minor groove, the grooves of ps DNA have a uniform size (5,17–20). As the 7-substituents of aps DNA are protruding into the major groove, the smaller groove of ps DNA might not provide enough space for the bulky 7-substituents. As little is known about the influence of 7-substituents on the duplex stability of ps DNA, this manuscript reports on the synthesis of corresponding oligonucleotides containing pyrazolo[3,4-d]pyrimidine analogs of isoguanine related nucleosides such as compounds 4–6. Oligonucleotides were prepared by solid-phase synthesis employing the phosphoramidites 7–9. The base pairing properties of the modified oligonucleotides are studied in duplexes with parallel and antiparallel chain orientation (Scheme 1).
Scheme 1.
MATERIALS AND METHODS
General
Thin-layer chromatography (TLC) was performed on TLC aluminum sheets covered with silica gel 60 F254 (0.2 mm, VWR International, Germany). Reverse-phase HPLC was carried out on a 4 × 250 mm RP-18 (10 µm) LiChrosorb column (VWR International) with a Merck-Hitachi HPLC pump (model 655 A-12) connected with a variable wavelength monitor (model 655-A), a controller (model L-5000) and an integrator (model D-2000). UV-spectra were recorded on a U-3200 spectrophotometer (Hitachi, Japan), λmax. in nm, ε in dm3 mol–1 cm–1. Half-life values (τ) were measured on a U-3200 spectrophotometer (Hitachi, Japan) connected with a temperature controller (Lauda, Germany). NMR spectra were measured on Avance DPX 250 or AMX 500 spectrometers (Bruker, Germany); chemical shifts (δ) are in p.p.m. downfield from internal TMS (1H, 13C) or external 85% H3PO4 (31P). The J-values are given in Hz. The solvents were purified and dried according to standard procedures. MALDI-TOF mass spectra were recorded on a Biflex-III spectrometer (Bruker, Leipzig, Germany) in the reflector mode. The average power of the nitrogen laser (337.1 nm) at 20 Hz was 3–4 mW (150–200 µJ/pulse) with a delay time of 600 ns. The enzymatic hydrolysis of the oligomers was performed as described below. Snake-venom phosphodiesterase (EC 3.1.15.1, Crotallus adamanteus) and alkaline phosphatase (EC 3.1.3.1, Escherichia coli) were generous gifts from Roche Diagnostics GmbH, Germany.
Oligodeoxyribonucleotides
The oligonucleotide syntheses were carried out on an ABI 392-08 DNA synthesizer (Applied Biosystems, Weiterstadt, Germany) in a 1 µmol scale using the phosphoramidites of the regular 2′-deoxynucleosides (Proligo, Hamburg, Germany), the 5′-O-DMT-N2-[(dimethylamino)methylidene]-2′-deoxy-5-methylisocytidine 3′-(β-cyanoethyl)phosphoramidite (4) together with the DPC- and butylamidine-protected phosphoramidite of 8-aza-7-deaza-2′-deoxyisoguanosine 7 (16). After cleavage of the oligonucleotides from the solid support, the former were deprotected in 25% aqueous ammonia solution for 12–16 h at 60°C. Purification of the 5′-dimethoxytrityl-oligomers was performed by reversed-phase HPLC (RP-18) with the following solvent gradient system [A: 0.1 M (Et3NH)OAc (pH 7.0)/MeCN 95:5; B: MeCN; 3 min 20% B in A with a flow rate of 1.0 ml min–1, 12 min 20–40% B in A with a flow rate of 1.0 ml min–1]. To remove the 4,4′-dimethoxytrityl residues the oligonucleotides were treated with 2.5% CHCl2COOH/CH2Cl2 for 5 min at room temperature. The detritylated oligomers were purified by reversed-phase HPLC with the gradient: 20 min 0–20% B in A with a flow rate of 1 ml min–1. Oligonucleotides synthesized in the trityl-off mode were deprotected in 25% aq. ammonia solution at 60°C for 16 h followed by purification by ion exchange chromatography (NucleoPac™-column, 4 × 250 mm) with the following solvent system [C: 1.5 M LiCl in aq. NaOH (pH 12.0)/D: aq. NaOH (pH 12.0); 5 min 5% D in E, 25 min 5–30% D in E, 10 min 30–5% D in E with a flow rate of 1.0 ml min–1]. The oligonucleotides were desalted and lyophil ized on a Speed-Vac evaporator to yield colorless solids. MALDI-TOF mass spectra are used for characterization (Supplementary Material).
The composition of oligonucleotides was proven by enzymatic hydrolysis: the oligonucleotides were dissolved in 0.1 M Tris–HCl buffer (pH 8.3, 200 µl), and treated with snake-venom phosphodiesterase (3 µl) at 37°C for 45 min and alkaline phosphatase (3 µl) at 37°C for another 30 min. The reaction mixtures were analyzed by HPLC (RP-18, at 260 nm, solvent system A, 0.7 ml min–1).
4-Amino-3-bromo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-1H-pyrazolo[3,4-d]-pyrimidin-6-one (5)
To a suspension of 7-bromo-8-aza-7-deazapurin-2,6-diamine 2′-deoxyribonucleoside 10 (21) (280 mg, 0.81 mmol) in water (15 ml), acetic acid (15 ml) and a solution of sodium nitrite (560 mg, 8.1 mmol) in water (2 ml) were added drop-wise at 60°C under stirring. After 10 min the pH of the solution was adjusted to 8 (25% aq. NH3). The crude product was applied to a Serdolit AD-4 column (4 × 20 cm, resin 0.1–0.2 mm, Serva, Germany). The column was washed with water (500 ml), and the product was eluted with H2O-PriOH (1:1). The solution was evaporated yielding 5 as a slight yellow powder after precipitation from H2O-PriOH (9:1) (185 mg, 66%). UV, λmax. (MeOH)/nm (ε/dm3mol–1cm–1) 233 (24 300); 291 (5800); 1H-NMR [D6 (DMSO)] 2.14, 2.65 [m, H2-C(2′)]; 3.44 [m, H2-C(5′)]; 3.75 [m, H-C(4′)]; 4.36 [s, H-C(3′)]; 5.02 [s, HO-C(3′), HO-C(5′)]; 6.31 [‘t’, J = 6.4, H-C(1′)]; 8.23 (NH2). Anal. calc. for C10H12Br N5O4: C, 34.70; H, 3.49; N, 20.23; found: C, 34.85; H, 3.68; N, 20.64.
4-Amino-3-iodo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-1H-pyrazolo[3,4-d]-pyrimidin-6-one (6)
To a suspension of 7-iodo-8-aza-7-deazapurin-2,6-diamine 2′-deoxyribonucleoside 11 (12) (100 mg, 0.26 mmol) in water (3 ml), acetic acid (300 µl) and a solution of sodium nitrite (80 mg, 1.2 mmol) in water (3 ml) were added drop-wise at 50°C under stirring. After 30 min the pH of the solution was adjusted to 8 (25% aq. NH3). The crude product was applied to a Serdolit AD-4 column (4 × 20 cm, resin 0.1–0.2 mm, Serva, Germany). The column was washed with water (500 ml), and the product was eluted with H2O-PriOH (1:1). The solution was evaporated yielding 6 as a slight yellow powder (70 mg, 70%). UV, λmax. (MeOH)/nm (ε/dm3mol–1cm–1) 237 (24 300); 293 (4600); 1H-NMR [D6 (DMSO)] 2.13, 2.67 [m, H2-C(2′)]; 3.47 [m, H2-C(5′)]; 3.75 [m, H-C(4′)]; 4.35 [s, H-C(3′)]; 4.88 [s, HO-C(5′)]; 5.28 [s, HO-C(3′)]; 6.33 [‘t’, J = 6.4, H-C(1′)]; 8.18 (NH2). Anal. calc. for C10H12 I N5O4: C, 30.55; H, 3.08; found: C, 30.90; H, 3.37.
3-Bromo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-4-(dimethylaminomethylidene)-1H-pyrazolo[3,4-d]pyrimidin-6-one (12)
To a solution of 5 (230 mg, 0.66 mmol) in methanol (2 ml) N,N-dimethylformamide dimethylacetal (300 µl, 2.24 mmol) was added. The mixture was stirred at room temperature for 30 min and evaporated to dryness. The residue was applied to FC (column 2 × 10 cm, CH2Cl2-MeOH 9:1 to 4:1), yielding 12 as a colorless foam (161 mg, 60%).
Rf (CH2Cl2-MeOH 4:1) 0.5; λmax. (MeOH)/nm (ε/dm3 mol–1cm–1) 230 (24 000); 249 (6600); 289 (4900); 1H-NMR [D6 (DMSO)] 2.14, 2.63 [m, H2-C(2′)]; 3.16, 3.21 [m, N(CH3)2]; 3.39 [m, H2-C(5′)]; 3.76 [m, H-C(4′)]; 4.33 [s, H-C(3′)]; 5.24 [br. s, HO-C(5′)]; 5.76 [br. s, HO-C(3′)]; 6.26 [‘t’, J = 6.5, H-C(1′)]; 8.57 (s, N = CH); 11.33 (br. s, NH). Anal. calc. for C13H17N6O4Br: C, 38.92; H, 4.27; N, 20.95; found: C, 38.60; H, 4.37; N, 20.82.
3-Iodo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-4-(dimethylaminomethylidene)-1H-pyrazolo[3,4-d]pyrimidin-6-one (13)
To a suspension of 6 (100 mg, 0.25 mmol) in methanol (2 ml) N,N-dimethylformamide dimethylacetal (400 µl, 2.99 mmol) was added. The mixture was stirred at 30°C for 12 h and evaporated to dryness. The residue was applied to FC (column 2 × 10 cm, CH2Cl2-MeOH 95:5 to 4:1), yielding 13 as an amorphous solid (71 mg, 62%). Rf (CH2Cl2-MeOH 9:1) 0.15; λmax. (MeOH)/nm (ε/dm3mol–1cm–1) 235 (18 700); 254 sh (7100); 291 (5000); 1H-NMR [D6 (DMSO)] 2.13, 2.64 [m, H2-C(2′)]; 3.15–3.48 [m, N(CH3)2, m, H2-C(5′)]; 3.76 [m, H-C(4′)]; 4.36 [s, H-C(3′)]; 4.88 [br. s, HO-C(5′)]; 5.22 [d, J = 4.4, HO-C(3′)]; 6.23 [‘t’, J = 6.6, H-C(1′)]; 8.59 (s, N = CH); 11.25 (br. s, NH). Anal. calc. for C13H17N6O4I: C, 34.84; H, 3.82; N, 18.75; found: C, 34.69; H, 3.58; N, 18.70.
3-Bromo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine (14)
To a suspension of compound 12 (170 mg, 0.42 mmol) in dry pyridine (3 ml) diphenylcarbamoyl chloride (200 mg, 0.86 mmol) and N,N-diisopropylethylamine (100 µl, 0.57 mmol) were added and stirred for 1 h at room temperature. Then, the mixture was poured into 5% aq. NaHCO3 (7 ml) and extracted with CH2Cl2 (3 × 5 ml). The CH2Cl2 layers were combined, dried over Na2SO4 and filtered. After evaporation of the solvent the residue was applied to FC (silica gel, column 2 × 15 cm). Elution with CH2Cl2 followed by CH2Cl2-acetone (step-wise gradient from 9:1 to 4:1) gave 14 as a colorless foam (204 mg, 81%).
Rf (CH2Cl2-acetone 4:1) 0.28; λmax (MeOH)/nm (ε/dm3mol–1cm–1) 238 (20 700); 322 (32 000); 1H-NMR [D6(DMSO)] 2.26, 2.74 [m, H2-C(2′)]; 3.23–3.83 [m, N(CH3)2; m, H2-C(5′)]; 3.81 [m, H-C(4′)]; 4.41 [m, H-C(3′)]; 4.77 [t, J = 5.6, HO-C(5′)]; 5.31 [d, J = 4.6, HO-C(3′)]; 6.45 [‘t’, J = 6.3, H-C(1′)]; 7.27–7.44 (m, arom. H); 8.87 (s, N = CH). Anal. calc. for C26H26N7O5Br: C, 52.36; H, 4.39; N, 16.44; found C, 52.14; H, 4.24; N, 16.19.
3-Iodo-1-(2-deoxy-β-d-erythro-pentofuranosyl)-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine (15)
As described for 14, with 13 (206 mg, 0.46 mmol) in dry pyridine (3 ml), diphenylcarbamoyl chloride (200 mg, 0.86 mmol) and N,N-diisopropylethylamine (100 µl, 0.57 mmol) gave 15 as a colorless foam (224 mg, 76%). Rf (CH2Cl2-acetone 4:1) 0.27; λmax (MeOH)/nm (ε/dm3mol–1cm–1) 235 (27 200), 323 (27 100); 1H-NMR [D6 (DMSO)] 2.26, 2.77 [m, H2-C(2′)]; 3.27–3.54 [m, N(CH3)2, m, H2-C(5′)]; 3.81 [m, H-C(4′)]; 4.42 [m, H-C(3′)]; 4.76 [t, J = 5.7, HO-C(5′)]; 5.76 [d, J = 4.6, HO-C(3′)]; 6.26 [‘t’, J = 6.4, H-C(1′)]; 8.88 (s, N = CH). Anal. calc. for C26H26N7O5I: C, 48.53; H, 4.07; N, 15.24; found: C, 48.73; H, 4.17; N, 14.89.
3-Bromo-1-[2-deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine (16)
Compound 14 (200 mg, 0.34 mmol) was dried by repeated co-evaporation from anhydrous pyridine and dissolved in anhydrous pyridine (1 ml). The solution was treated with dimethoxytrityl chloride (233 mg, 0.69 mmol) at room temperature under stirring (45 min). Methanol (1 ml) was introduced, and the stirring was continued for 5 min. The mixture was poured into 5% aq. NaHCO3 solution (5 ml) and extracted with CH2Cl2 (3 × 5 ml). The organic layers were combined and dried over Na2SO4. After evaporation of the solvent, the residue was applied to FC (silica gel, column 2 × 15 cm), which was pre-washed with CH2Cl2 and eluted with CH2Cl2-acetone (step-wise gradient from 98:2 to 9:1) to give 16 as a colorless foam (254 mg, 84%).
Rf (CH2Cl2-acetone 9:1) 0.48; λmax (MeOH)/nm (ε/dm3mol–1cm–1) 235 (47 700); 322 (29 700); 1H-NMR [D6(DMSO)] 2.32, 2.77 [m, H2-C(2′)]; 3.04 [m, H2-C(5′)]; 3.22, 3.27 [m, N(CH3)2]; 3.68 (m, 2 CH3O); 3.93 [m, H-C(4′)]; 4.50 [t, J = 4.9, H-C(3′)]; 5.35 [d, J = 4.9, HO-C(3′)]; 6.51 [m, H-C(1′)]; 6.71–7.45 (m, arom. H); 8.87 (s, N = CH). Anal. calc. for C47H44N7O7Br: C, 62.81; H, 4.93; N, 10.91; found: C, 62.80; H, 4.99; N, 10.96.
3-Iodo-1-[2-deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine (17)
As described for 16, with 15 (200 mg, 0.31 mmol) in dry pyridine (3 ml) and dimethoxytrityl chloride (122 mg, 0.36 mmol) gave 17 as a colorless foam (230 mg, 78%). Rf (CH2Cl2-acetone 9:1) 0.50; λmax (MeOH)/nm (ε/dm3mol–1cm–1) 234 (30 500); 322 (17 700); 1H-NMR [D6(DMSO)] 2.30, 2.75 [m, H2-C(2′)]; 3.02 [m, H2-C(5′)]; 3.28 [m, N(CH3)2]; 3.68 (m, 2 CH3O); 3.91 [m, H-C(4′)]; 4.47 [t, J = 4.9, H-C(3′)]; 5.33 [d, J = 4.9, HO-C(3′)]; 6.48 [m, H-C(1′)]; 6.71–7.44 (m, arom. H); 8.88 (s, N = CH). Anal. calc. for C47H44N7O7I: C, 59.69; H, 4.69; N, 10.37; found: C, 59.46; H, 4.53; N, 10.22.
3-Bromo-1-[2-deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine 3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (8)
A solution of compound 16 (150 mg, 0.17 mmol) in dry THF (1 ml) was preflushed with argon and kept under argon atmosphere. Then, N,N-diisopropylethylamine (80 µl, 0.46 mmol) and chloro(2-cyanoethoxy)(diisopropylamino)phosphine (96 µl, 0.43 mmol) were added under stirring at room temperature. The stirring was continued for 30 min and the reaction mixture quenched with 5 ml CH2Cl2. An aq. solution of 5% NaHCO3 (10 ml) was added, the layers separated, and the aqueous layer extracted with CH2Cl2 (3 × 15 ml). The combined organic extracts were dried over Na2SO4, filtered, evaporated and applied to FC (silica gel, column 2 × 10 cm, CH2Cl2-acetone 9:1). Evaporation of the main zone yielded compound 8 as a colorless foam (142 mg, 77%). Rf (CH2Cl2-acetone 9:1) 0.9. δP(CDCl3) 149.4, 149.6.
3-Iodo-1-[2-deoxy-5-O-(4,4′-dimethoxytriphenylmethyl)-β-d-erythro-pentofuranosyl]-4-(dimethylaminomethylidene)-6-[(diphenylcarbamoyl)oxy]-1H-pyrazolo[3,4-d]pyrimidine 3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (9)
As described for 8, with 17 (200 mg, 0.21 mmol) in dry dichloromethane (5 ml) with N,N-diisopropylethylamine (110 µl, 0.63 mmol) and chloro(2-cyanoethoxy)(diisopropylamino)phosphine (110 µl, 0.49 mmol) under argon gave 9 as a colorless foam (190 mg, 78%). Rf (CH2Cl2-acetone 9:1) 0.95. δP(CDCl3) 149.3, 149.5.
RESULTS AND DISCUSSION
Monomers
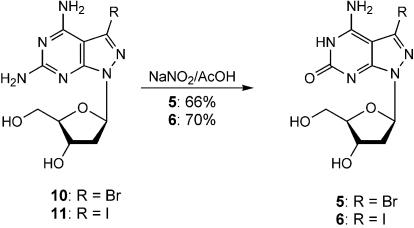
For the synthesis of the 7-substituted 8-aza-7-deaza-2′-deoxyisoguanosines 5 and 6 the corresponding 8-aza-7-deazapurin-2,6-diamine 2′-deoxyribonucleosides 10 and 11 (12,21) were used as precursors. Selective deamination of the 2-amino groups with sodium nitrite in acetic acid furnished the nucleosides 5 or 6 (Scheme 2).
Scheme 2.
As 2′-deoxyisoguanosine is sensitive to acidic conditions, which causes depurination during oligonucleotide synthesis the stability of the N-glycosylic bond of the base modified nucleosides was investigated. Already the substitution of the purine moiety by the 7-deaza- or 8-aza-7-deazapurine results in an increase of the glycosylic bond stability (22,23). The presence of 7-halogen substituents at the 8-aza-7- deaza-2′-deoxyadenosine led to a further stabilization of the glycosylic bond accompanied by an increased duplex stability. On the other hand the 7-deazapurine (pyrrolo[2,3-d]pyrimidine) nucleosides, which showed the highest glycosylic bond stability, exhibit the lowest duplex stability (23). Thus, the hydrolysis of the 7-halogenated isoguanine related nucleosides 5 and 6 was studied. It was followed UV-spectrophotometrically in 0.5 M HCl at 60°C resulting in half-lives of 10 min for compound 5 and 14 min for 6 (Table 1). Comparison with the purine 2′-deoxyisoguanosine shows a significant glycosylic bond stabilization induced by the 7-subsituents.
Table 1. Half-life values of the glycosylic bond stability of 2′-deoxyisoguanosine derivatives.
| Compound | τ (min) | Conditions | τ (min) | Conditions |
|---|---|---|---|---|
| iGda | 8 | 0.1 M HCl, rt | ||
| 4 | 12.5 | 0.5 M HCl, rt | <2 | 0.5 M HCl, 40°C |
| 5 | 10 | 0.5 N HCl, 60°C | 46 | 0.5 N HCl, 40°C |
| 6 | 14 | 0.5 N HCl, 60°C | 52 | 0.5 N HCl, 40°C |
aSeela and Gabler (24).
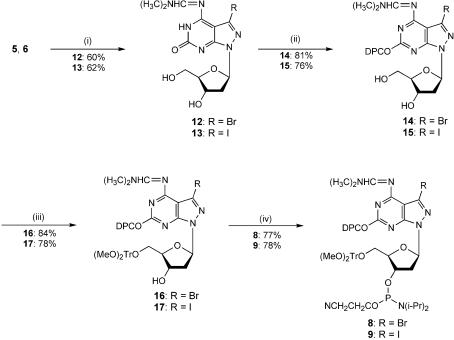
Next, the phosphoramidites 8 and 9 were prepared, which were later used for solid-phase oligonucleotide synthesis. The amino groups were protected with the N,N-dimethylaminomethylidene residue using dimethylformamide dimethylacetal/methanol thereby furnishing compound 12 in 60% and 13 in 62% yield with half-lives of 10 and 5 min in aq. ammonia solution. In the case of 2′-deoxyisoguanosine the diphenylcarbamoyl residue was chosen for protection of the 2-oxo-group (3). Subsequently, the amino- and oxo-protected compounds were converted into the 5′-O-DMT-derivatives 16 and 17 under standard conditions. Phosphitylation with chloro(2-cyanoethoxy)(diisopropylamino)phosphine furnished the phosphoramidites 8 (25) and 9 (Scheme 3).
Scheme 3. (i) Dimethylformamide dimethylacetal, methanol. (ii) Diphenylcarbamoyl chloride, pyridine, N,N-diisopropylethylamine. (iii) Dimethoxytrityl chloride, pyridine. (iv) Chloro(2-cyanoethoxy)(diisopropylamino)phosphine, N,N-diisopropylethylamine, THF.
The compounds were characterized by 1H- and 13C-NMR spectroscopy as well as by elemental analysis. 13C-NMR shift assignment was performed by gated decoupled spectra. As observed for other pyrazolo[3,4-d]pyrimidine and isoguanine derivatives a few signals are not appearing (26). Compared with the non-halogenated compound 4 the 7-bromo substituent (5) leads to a 15 p.p.m. up-field shift of C(7); an additional shift of 10 p.p.m. is observed for the iodinated compound 6. The assignment of C(1′) and C(4′) are based on the differences of the coupling constant 1J(C,H), which are larger for C(1′) than for C(4′) (27) (Table 2).
Table 2. 13C-NMR chemical shifts of 8-aza-7-deaza-2′-deoxyisoguanosine derivatives (D6-DMSO, 25°C).
| a | C(3) | C(3a) | C(4)d | C(6)d | C(7a)d | C(1′) | C(2′ | C(3′) | C(4′) | C(5′) | HC = N | CO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b | C(7) | C(5) | C(6)d | C(2)d | C(4)d | |||||||
| 4(26) | 134.7 | 92.6 | 157.0 | 155.2 | e | 84.3 | c | 71.2 | 87.6 | 62.5 | ||
| 5 | 120.9 | 91.9 | 157.8 | 157.1 | e | 83.2 | c | 70.6 | 87.4 | 62.2 | ||
| 6 | 99.6 | 89.8 | 157.9 | 157.0 | e | 83.0 | c | 70.4 | 87.4 | 62.3 | ||
| 12 | 122.3 | 97.4 | 158.0 | 157.0 | e | 83.4 | 37.5 | 70.8 | 87.4 | 62.4 | 158.7 | |
| 13 | 100.8 | 94.7 | 158.0 | 157.0 | e | 83.3 | 37.6 | 70.7 | 87.3 | 62.3 | 158.6 | |
| 14 | 129.3 | 104.7 | 159.1 | 158.1 | 156.4 | 83.8 | 37.7 | 70.7 | 87.7 | 62.2 | 163.3 | 150.8 |
| 15 | 107.9 | 93.8 | 158.6 | 157.8 | 155.9 | 83.9 | c | 70.8 | 87.7 | 62.3 | 163.2 | 150.9 |
| 16 | 129.3 | 104.6 | 159.2 | 158.0 | 156.4 | 83.5 | 35.1 | 70.5 | 85.2 | 64.2 | 163.4 | 150.9 |
| 17 | 107.7 | 93.7 | 158.7 | 157.9 | 155.9 | 83.5 | c | 70.7 | 85.6 | 64.3 | 163.2 | 150.9 |
aSystematic numbering.
bPurine numbering.
cSuperimposed by DMSO.
dTentative assignment.
eNot detected.
Oligodeoxyribonucleotides
Synthesis. The synthesis of the oligonucleotides using the protocol of phosphoramidite chemistry was performed on an ABI 392-08 synthesizer employing the standard conditions (28). The building blocks 7, 8 and 9 as well as standard phosphoramidites were used. Oligonucleotides were purified according to the synthesis mode (trityl-on; trityl-off) either by reversed-phase HPLC or by ion-exchange chromatography. The nucleoside composition of oligonucleotides was determined by tandem hydrolysis with snake-venom phosphodiesterase and alkaline phosphatase or by MALDI-TOF spectroscopy. The stability of oligonucleotide duplexes was determined by temperature-dependent measurements of UV-spectra (Cary 1E, Varian). The calculation of the thermodynamic data was performed with the program MeltWin 3.0 (29).
Base-pairing properties of the halogenated 2′-deoxyisoguanosine analogs 5 and 6. The base pairing properties of 2′-deoxyisoguanosine and its 7-deazapurine analog have already been investigated by our laboratory. Oligonucleotide duplexes with ps and aps strand orientation were studied (22). In both series of duplexes it was observed that base pairing is selective for either cytosine in ps-duplexes or isocytosine in aps DNA with the advantage that the 2′-deoxyisoguanosine containing the purine heterocycle shows a higher duplex stability than the pyrrolo[2,3-d]pyrimidine derivative. On the other hand, a lower N-glycosylic bond stability was found for the purine compound. As we want to combine the advantages of the high duplex stability with a high glycosylic bond stability, compounds 4–6 were incorporated into oligonucleotide duplexes and their base pair stability was studied.
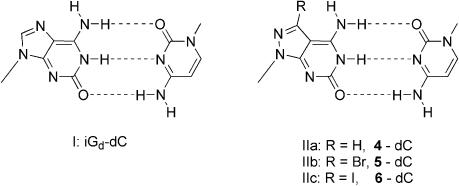
Self-complementary duplexes with parallel chain orientation. The incorporation of compound 4 in the alternating self-complementary duplex 5′-d(4-C)3 (19) already led to a small but significant duplex stabilization when compared with 2′-deoxyisoguanosine, while the replacement of iGd (duplex 18) by 7-deaza-2′-deoxyisoguanosine (duplex 22) led to a strong destabilization of the duplex structure (23). Next the hexanucleotides 5′-d(5-C)3 (20) and 5′-d(6-C)3 (21), containing 7-bromo- and 7-iodo-8-aza-7-deaza-2′-deoxyisoguanosine were investigated (Table 3). As shown in Table 3 the 8-aza-7-deaza-2′-deoxyisoguanosine 4 (duplex 19) showed a stabilizing effect (ΔTm = 1.6°C per modification). This effect increased significantly when halogen substituents were introduced in position-7 with the highest Tm value for the 7-iodinated duplex 21 (ΔTm = 5.8°C per modification). According to the chain orientation the self-complementary duplexes contain only 5 bp as well as two 5′-overhangs. Apart from the modified residues within the duplex chain the overhanging residues also stabilize the duplex structure (30). This is due to terminal stacking of the dangling ends. Nevertheless, the base pair stabilization by the 7-substituents within the ps-duplex is already strong. Analogously to the base pair of iGd–dC (motif I) the base pair motifs IIa–c are suggested for the duplexes 19–21 (Scheme 4).
Table 3. Tm-values and thermodynamic data of duplex formation of ps self-complementary oligonucleotidesa.
| Tm(°C) | ΔTm/mod (°C) | ΔH° (kcal/mol) | ΔS° [cal/(mol K)] | ΔG°310 (kcal/mol) | ||
|---|---|---|---|---|---|---|
| 5′-d(iG-C-iG-C-iG-C)b | 18 | 33 | –34 | –88 | –6.2 | |
| 5′-d(iG-C-iG-C-iG-C) | ||||||
| 5′-d(4-C-4-C-4-C)b | 19 | 41 | +1.6 | –47 | –129 | –7.8 |
| 5′-d(4-C-4-C-4-C) | ||||||
| 5′-d(5-C-5-C-5-C) | 20 | 57 | +4.8 | –64 | –162 | –10.2 |
| 5′d(5-C-5-C-5-C) | ||||||
| 5′d(6-C-6-C-6-C) | 21 | 62 | +5.8 | –46 | –116 | –9.9 |
| 5′d(6-C-6-C-6-C) | ||||||
| 5′-d(c7iG-C-c7iG-C-c7iG-C)b,c | 22 | 22 | –2.2 | –40 | –112 | –5.3 |
| 5′-d(c7iG-C-c7iG-C-c7iG-C) |
aMeasured in 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate buffer, pH 7.
bSee Seela et al. (23).
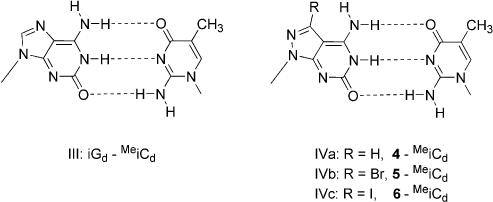
cd(c7iG) = 7-Deaza-2′-deoxyisoguanosine 4 = c7z8iGd = 8-Aza-7-deaza-2′-desoxyisoguanosine. 5 = Br7c7z8iGd = 7-Bromo-8-aza-7-deaza-2′-desoxyisoguanosine. 6 = I7c7z8iGd = 7-Iodo-8-aza-7-deaza-2′-desoxyisoguanosine.
Scheme 4.
Non self-complementary duplexes with parallel chain orientation. Next, non self-complementary duplexes were studied (Table 4). In order to induce parallel chain orientation the oligonucleotides contain two ‘iGd’–dC and two MeiCd–dG base pairs. The duplex 23·24 was used as reference compound. The duplex stability increased in the order: iGd ≌ 4 < 5 < 6. The most stable duplex was found in the case of the 7-iodo compound (27·24) with a Tm-enhancement of 2.5°C per modification, while that of compound 5 showed a ΔTm of 1.5°C. These effects are smaller than those observed for the self-complementary duplexes discussed above. This results from the absence of the dangling ends present in the self-complementary ps-duplexes shown in Table 4.
Table 4. Tm-values and thermodynamic data of non self-complementary ps duplexesa,b.
| Tm (°C) | ΔTm/mod (°C) | ΔH° (kcal/mol) | ΔS° [cal/(mol K)] | ΔG°310 (kcal/mol) | ||
|---|---|---|---|---|---|---|
| 5′-d(TiCATAAiCTiG iGAT)c | 23 | 44 | –85 | –242 | –10.3 | |
| 5′-d(A GTATTGA C CTA) | 24 | |||||
| 5′-d(TiCATAAiCT4 4 AT) | 25 | 43 | –0.5 | –73 | –206 | –9.0 |
| 5′-d(AGTATTGA CC TA) | 24 | |||||
| 5′-d(TiCATAAiCT 5 5 AT) | 26 | 47 | +1.5 | –76 | –211 | –10.0 |
| 5′-d(AGTATTGA CC TA) | 24 | |||||
| 5′-d(TiCATAAiCT 6 6 AT) | 27 | 49 | +2.5 | –77 | –214 | –10.7 |
| 5′d(AGTATTGA CC TA) | 24 |
aIn 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate, pH 7; oligonucleotide concentration: 5 µM (single strand).
bd(iC) = MeiCd = 5-Methyl-2′-deoxyisocytidine.
cSee Seela and Wei (22).
Tables 5 and 6 summarize the pairing properties of the ‘isoguanine’ nucleosides located opposite the four canonical nucleosides. The mismatch discrimination was strong but dependent on the position of incorporation. As expected a stronger discriminatory effect occurs when the modification is located in the centre of the duplex (Table 5) while it is less pronounced near the termini (Table 6). Rather, sequence independent discriminatory effects are observed when the nucleosides 4–6 are located opposite to 2′-deoxythymidine or 2′-deoxyadenosine.
Table 5. Tm-values and thermodynamic data of non self-complementary ps duplexes of the sequence 5′-d(ATiCiCAXTTATXA)·5′-d(TAGGTYAATACT)a,b.
| X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| iG·Cc | 28·29 | 39 | |||||||||
| 4·C | 30·29 | 39 | ±0 | 5·C | 35·29 | 43 | +4 | 6·C | 36·29 | 44 | +5 |
| 4·G | 30·31 | – | >–20 | 5·G | 35·31 | – | >–20 | 6·G | 36·31 | – | >–20 |
| 4·T | 30·32 | 27 | –12 | 5·T | 35·32 | 29 | –10 | 6·T | 36·32 | 31 | –7 |
| 4·A | 30·33 | 28 | –11 | 5·A | 35·33 | 28 | –11 | 6·A | 36·33 | 28 | –11 |
| 4·iC | 30·34 | 26 | –13 | 5·iC | 35·34 | 35 | –4 | 6·iC | 36·34 | 36 | –3 |
aIn 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate, pH 7; oligonucleotide concentration: 5 µM (single strand).
bd(iC) = MeiCd = 5-Methyl-2′-deoxyisocytidine.
cSee Seela and Wei (22).
Table 6. Tm-values and thermodynamic data of non self-complementary ps duplexes of the sequence 5′-d(TiCATAAiCTXXAT)·5′-d(AGTATTGAYCTA)a,b.
| X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| iG·Cc | 23·24 | 44 | |||||||||
| 4·G | 25·35 | 32 | –12 | 5·G | 26·35 | 38 | –6 | 6·G | 27·35 | 39 | –5 |
| 4·T | 25·36 | 29 | –15 | 5·T | 26·36 | 34 | –10 | 6·T | 27·36 | 34 | –10 |
| 4·A | 25·37 | 30 | –14 | 5·A | 26·37 | 33 | –11 | 6·A | 27·37 | 32 | –12 |
| 4·iC | 25·38 | 40 | –4 | 5·iC | 26·38 | 45 | +1 | 6·iC | 27·38 | 46 | +2 |
aIn 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate, pH 7; oligonucleotide concentration: 5 µM (single strand).
bd(iC) = MeiCd = 5-Methyl-2′-deoxyisocytidine.
cSee Seela and Wei (22).
Non self-complementary duplexes with antiparallel chain orientation. As shown in Tables 7 and 8, aps-duplexes containing iGd–MeiCd base pairs and standard dA–dT pairs are more stable than the ones with ps chains. It is apparent that the duplex with 7-iodo-8-aza-7-deaza-2′-deoxyisoguanosine (6) opposite 2′-deoxy-5-methylisocytidine (27·36 and 34·42) forms the most stable base pair followed by that of the bromo derivative 5 (26·35 and 34·41). Their stability increase (ΔTm) is higher than that of the parent duplexes as well as the duplex containing the base pair 4–MeiCd (25·30 and 34·40). The duplex stability increased in the order iGd ≤ 4 < 5 < 6, which is the same as in parallel DNA. However, the base pair stabilization induced by the 7-halogeno substituents is more pronounced in aps than in ps DNA.
Table 7. Tm-values and thermodynamic data of non self-complementary aps duplexesa,b.
| Tm (°C) | ΔTm/mod (°C) | ΔH° (kcal/mol) | ΔS° [cal/(mol K)] | ΔG°310 (kcal/mol) | ||
|---|---|---|---|---|---|---|
| 3′-d(ATCCAGTTATGA) | 24 | 51 | –85 | –237 | –11.6 | |
| 5′-d(TAGGTCAATACT) | 29 | |||||
| 3′-d(TAGGTCAATACT) | 29a | 49 | –97 | –275 | –11.5 | |
| 5′-d(ATCCAGTTATGA) | 24a | |||||
| 3′-d(TAiG iGTiCAATAiCT)c | 23 | 60 | –94 | –257 | –14.8 | |
| 5′-d(AT iC iCAiG TTATiGA) | 28 | |||||
| 3′-d(TA 4 4T iCAATA iCT) | 25 | 61 | +0.25 | –96 | –263 | –14.6 |
| 5′-d(ATiCiCA 4 TTAT 4 A) | 30 | |||||
| 3′-d(TA 5 5 T iCAATAiCT) | 26 | 69 | +2.25 | –106 | –283 | –17.6 |
| 5′-d(ATiCiCA 5 TTAT 5 A) | 35 | |||||
| 3′-d(TA 6 6 T iCAATAiCT) | 27 | 73 | +3.25 | –119 | –321 | –20.0 |
| 5′-d(ATiCiCA 6 TTAT 6 A) | 36 |
aMeasured in 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate, pH 7; oligonucleotide concentration: 5 µM (single strand).
bd(iC) = MeiCd = 5-Methyl-2′-deoxyisocytidine.
cSee Seela et al. (4).
Table 8. Tm-values and thermodynamic data of non self-complementary aps duplexes of the sequence 5′-d(TAGGTXAATACT)·3′-d(ATCCAYTTATGA)a,b.
| X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | X·Y | Tm (°C) | ΔTm/mod (°C) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| iC·iGc | 34·39 | 54 | |||||||||
| iC·4 | 34·40 | 56 | +2 | iC·5 | 34·41 | 57 | +3 | iC·6 | 34·42 | 58 | +4 |
| C·4 | 29·40 | 42 | –12 | C·5 | 29·41 | 47 | –7 | C·6 | 29·42 | 48 | –6 |
| G·4 | 31·40 | 42 | –12 | G·5 | 31·41 | 47 | –7 | G·6 | 31·42 | 47 | –7 |
| T·4 | 32·40 | 41 | –13 | T·5 | 32·41 | 42 | –12 | T·6 | 32·42 | 42 | –12 |
| A·4 | 33·40 | 28 | –26 | A·5 | 33·41 | 27 | –27 | A·6 | 33·42 | 28 | –26 |
aIn 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate, pH 7; oligonucleotide concentration: 5 µM (single strand).
bd(iC) = MeiCd = 5-Methyl-2′-deoxyisocytidine.
cSee Seela and Wei (22).
Regarding the various sizes of the 7-substituents the iodo-compound 6 makes a stronger contribution to the duplex stabilization than the bromo-nucleoside 5. An explanation for this behavior might result from the differences of the polarizabilities (αm) of the nucleobases. We have calculated values of 14.06 (αm/10–24 cm3) for 4, 17.05 (αm/10–24 cm3) for 5 and 19.07 (αm/10–24 cm3) for 6 (30). The size of the substituents, even in the case of the large iodo-substituent, does not interfere with the limited size of the grooves.
The nucleosides 4–6 opposite the four canonical 2′-deoxyribonucleosides show comparable Tm decreases, particularly for halogenated nucleosides 5 and 6. Comparison with the unsubstituted compound 4 discloses a lower discriminatory effect. The position of incorporation does not affect the duplex stabilities severely as was found for parallel stranded non-self-complementary duplexes. A related base pair motif as reported for the 2′-deoxyisoguanosine–2′-deoxy-5-methylisocytidine pair (motif III) is also suggested for that of the pyrazolo[3,4-d]pyrimidine nucleosides (motifs IVa–IVc) (Scheme 5).
Scheme 5.
CONCLUSIONS
Replacement of 2′-deoxyisoguanosine by 8-aza-7-deaza-2′-deoxyisoguanosine (4) results in parallel stranded duplexes showing a similar base pair stability. The base pair is further stabilized when halogen substituents, such as bromine or iodine, are introduced at the 7-position of the modified base (5,6). It is most pronounced in self-complementary parallel stranded duplexes containing overhangs.
Similar to parallel stranded duplexes the halogenated 8-aza-7-deaza-2′-deoxyisoguanosine derivatives 5 and 6 strongly stabilize duplexes with antiparallel chain orientation. The stabilizing effect of the substituents is more pronounced in the aps DNA than in those with parallel chain orientation. Although the size of the grooves is different in ps and aps DNA, the bulky halogen substituents are well accommodated in both duplex structures. As the 7-halogenated derivatives of 8-aza-7-deaza-2′-deoxyisoguanosine are rather resistant to ‘depurination’ they are favorable mimics of 2′-deoxyisoguanosine. They are currently used as probes for hybridization experiments performed on microarrays.
SUPPLEMENTARY MATERIAL
Supplementary Material containing MALDI–TOF data is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Yang He and Dr Helmut Rosemeyer for the NMR spectra and Mrs Elisabeth Feiling for the oligonucleotide synthesis. Financial support by the Roche Diagnostics GmbH is gratefully acknowledged.
REFERENCES
- 1.Seela F., Gabler,B. and Kazimierczuk,Z. (1993) 2′-Deoxyisoguanosine: synthesis and incorporation into oligodeoxyribonucleotides. Collect. Czech. Chem. Commun., 58, 170–173. [Google Scholar]
- 2.Sugiyama H., Ikeda,S. and Saito,I. (1996) Remarkably stable parallel-stranded oligonucleotides containing 5-methylisocytosine and isoguanine. J. Am. Chem. Soc., 118, 9994–9995. [Google Scholar]
- 3.Seela F. and Wei,C. (1997) Oligonucleotides containing consecutive 2′-deoxyisoguanosine residues: synthesis, duplexes with parallel chain orientation, and aggregation. Helv. Chim. Acta, 80, 73–85. [Google Scholar]
- 4.Seela F., He,Y. and Wei,C. (1999) Parallel-stranded oligonucleotide duplexes containing 5-methylisocytosine-guanine and isoguanine-cytosine base pairs. Tetrahedron, 55, 9481–9500. [Google Scholar]
- 5.van de Sande J.H., Ramsing,N.B., Germann,M.W., Elhorst,W., Kalisch,B.W., von Kitzing,E., Pon,R.T., Clegg,R.C. and Jovin,T.M. (1988) Parallel stranded DNA. Science, 241, 551–557. [DOI] [PubMed] [Google Scholar]
- 6.Seela F., Wei,C., Becher,G., Zulauf,M. and Leonard,P. (2000) The influence of modified purine bases on the stability of parallel DNA. Bioorg. Med. Chem. Lett., 10, 289–292. [DOI] [PubMed] [Google Scholar]
- 7.Buhr C.A., Wagner,R.W., Grant,D. and Froehler,B.C. (1996) Oligodeoxynucleotides containing C-7 propyne analogs of 7-deaza-2′-deoxyguanosine and 7-deaza-2′-deoxyadenosine. Nucleic Acids Res., 24, 2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balow G., Mohan,V., Lesnik,E.A., Johnston,J.F., Monia,B.P. and Acevedo,O.L. (1998) Biophysical and antisense properties of oligodeoxynucleotides containing 7-propynyl-, 7-iodo- and 7-cyano-7-deaza-2-amino-2′-deoxyadenosines. Nucleic Acids Res., 26, 3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seela F. and Thomas,H. (1995) Duplex stabilization of DNA: oligonucleotides containing 7-substituted 7-deazaadenines. Helv. Chim. Acta, 78, 94–108. [Google Scholar]
- 10.Ramzaeva N. and Seela,F. (1996) Duplex stability of 7-deazapurine DNA: oligonucleotides containing 7-bromo- or 7-iodo-7-deazaguanine. Helv. Chim. Acta, 79, 1549–1558. [Google Scholar]
- 11.Seela F. and Chen,Y. (1996) Oligonucleotides containing 7- or 8-methyl-7-deazaguanine: steric requirements of major groove substituents on the DNA structure. Chem. Commun., 2263–2264. [Google Scholar]
- 12.Becher G., He,J. and Seela,F. (2001) Major-groove-halogenated DNA: the effects of bromo and iodo substituents replacing H-C(7) of 8-aza-7-deazapurine-2,6-diamine or H-C(5) of uracil residues. Helv. Chim. Acta, 84, 1048–1065. [Google Scholar]
- 13.Seela F. and Becher,G. (1999) Oligonucleotides containing pyrazolo[3,4-d]pyrimidines: the influence of 7-substituted 8-aza-7-deaza-2′-deoxyguanosines on the duplex structure and stability. Helv. Chim. Acta, 82, 1640–1655. [Google Scholar]
- 14.Seela F. and Becher,G. (1998) Stabilisation of duplex DNA by 7-halogenated 8-aza-7-deazaguanines. Chem. Commun., 2017–2018. [Google Scholar]
- 15.Seela F. and Kröschel,R. (2001) Quadruplex and pentaplex self-assemblies of oligonucleotides containing short runs of 8-aza-7-deaza-2′-deoxyisoguanosine or 2′-deoxyisoguanosine. Bioconjugate Chem., 12, 1043–1050. [DOI] [PubMed] [Google Scholar]
- 16.Seela F. and Kröschel,R. (2003) Oligonucleotide duplexes containing N8-glycosylated 8-aza-7-deazaguanine and self-assembly of 8-aza-7-deazapurines on the nucleoside and the oligomeric level. Org. Biomol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 17.Pattabiraman N. (1986) Can the double helix be parallel? Biopolymers, 25, 1603–1606. [DOI] [PubMed] [Google Scholar]
- 18.Germann M.W., Vogel,H.J., Pon,R.T. and van de Sande,J.H. (1989) Characterization of a parallel-stranded DNA hairpin. Biochemistry, 28, 6220–6228. [DOI] [PubMed] [Google Scholar]
- 19.Zhou N., Germann,M.W., van de Sande,J.H., Pattabiraman,N. and Vogel,H.J. (1993) Solution structure of the parallel-stranded hairpin d(T8<>C4A8) as determined by two-dimensional NMR. Biochemistry, 32, 646–656. [DOI] [PubMed] [Google Scholar]
- 20.Yang X.-L., Sugiyama,H., Ikeda,S., Saito,I. and Wang,A.H.-J. (1998) Structural studies of a stable parallel-stranded DNA duplex incorporating isoguanine: cytosine and isocytosine: guanine basepairs by nuclear magnetic resonance spectroscopy. Biophys. J., 75, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seela F. and Becher,G. (2001) Pyrazolo[3,4-d]pyrimidine nucleic acids: adjustment of dA-dT to dG-dC base pair stability. Nucleic Acids Res., 29, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seela F. and,Wei,C. (1999) The base-pairing properties of 7-deaza-2′-deoxyisoguanosine and 2′-deoxyisoguanosine in oligonucleotide duplexes with parallel and antiparallel chain orientation. Helv. Chim. Acta, 82, 726–745. [Google Scholar]
- 23.Seela F., Wei,C., Melenewski,A., He,Y., Kröschel,R. and Feiling,E. (1999) Parallel-stranded DNA formed by new base pairs related to the isoguanine-cytosine or isocytosine-guanine motifs. Nucl. Nucl., 18, 1543–1548. [Google Scholar]
- 24.Seela F. and Gabler,B. (1994) Facile synthesis of 2′-deoxyisoguanosine and related 2′,3′-dideoxyribonucleosides. Helv. Chim. Acta, 77, 622–630. [Google Scholar]
- 25.Seela F., Kröschel,R. and He,Y. (2001) Parallel DNA containing pyrazolo[3,4-d]pyrimidine analogues of isoguanine. Nucl. Nucl., 20, 1283–1286. [DOI] [PubMed] [Google Scholar]
- 26.Kazimierczuk Z., Mertens,R., Kawczynski,W. and Seela,F. (1991) 2′-Deoxyisoguanosine and base-modified analogues: chemical and photochemical synthesis. Helv. Chim. Acta, 74, 1742–1748. [Google Scholar]
- 27.Seela F. and Bussmann,W. (1985) Assignment of 13C chemical shifts of α-D-ribonucleoside sugar carbons by 1J(CH) coupling constants. Nucl. Nucl., 4, 391–394. [Google Scholar]
- 28.Users’ Manual of the DNA Synthesizer. Applied Biosystems, Weiterstadt, Germany, p. 392. [Google Scholar]
- 29.McDowell J.A. and Turner,D.H. (1996) Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAG GU CUC)2 by two-dimensional NMR and simulated annealing. Biochemistry, 35, 14077–14089. [DOI] [PubMed] [Google Scholar]
- 30.Rosemeyer H. and Seela,F. (2002) Modified purine nucleosides as dangling ends of DNA duplexes: the effect of the nucleobase polarizability on stacking interactions. J. Chem. Soc., Perkin Trans., 2, 746–750. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.