Abstract
Nineteen canonical and two Knirps-like family nuclear receptors (NRs) were identified in the genome of Tribolium castaneum. The current study was conducted to determine the function of these NRs in regulation of female reproduction and embryogenesis. RNA interference (RNAi)-aided knock-down in the expression of genes coding for 21 NRs showed that seven NRs E75, hormone receptor 3 (HR3), ecdysone receptor (EcR), ultraspiracle (USP), seven-up (SVP), FTZ transcription factor 1 (FTZ-F1) and hormone receptor 4 (HR4) are required for successful vitellogenesis and oogenesis. Knocking down the expression of genes coding for these seven NRs affected egg production by reducing the levels of vitellogenin mRNAs as well as by affecting the oocyte maturation. Expression of seven additional NRs hormone receptor 96 (HR96), hormone receptor 51 (HR51), hormone receptor 38 (HR38), hormone receptor 39 (HR39), Tailless (Tll), Dissatisfaction (Dsf) and Knirps-like is required for successful embryogenesis. The knock-down in the expression of genes coding for three other NRs (E78, hepatocyte nuclear factor 4, HNF4 and Eagle) partially blocked embryogenesis. This study showed that at least 17 out of the 21 NRs identified in T. castaneum play key roles in female reproduction and embryogenesis.
Keywords: vitellogenesis, oogenesis, embryogenesis, ecdysone receptor, RNAi
INTRODUCTION
Nuclear receptors (NRs) are a group of transcription factors that play key roles in the major biological processes including development (King-Jones and Thummel, 2005), cell growth (Parthasarathy and Palli, 2008), metabolic homeostasis (Alenghat et al., 2008; Palanker et al., 2009), reproduction and embryogenesis (Allen and Spradling, 2008; Moran and Jimenez, 2006; Niakan et al., 2006). All the NRs share common domain architecture, including a highly conserved DNA-binding domain (DBD) and a structurally conserved ligand-binding domain (LBD) (Bain et al., 2007). Due to this unique structure, NRs harbor a receptor function with DNA-binding capacity, ligand-binding ability and a transcriptional activation all combined in the same molecule, allowing a one-step response to a signal that results in a direct regulation of transcription of the target genes (King-Jones and Thummel, 2005). Based on the structures and the functions they carried out, the nuclear receptor super family is subdivided into three classes, the steroid receptor family including the progesterone receptor (PR), estrogen receptor (ER), glucocorticoid receptor (GR), androgen receptor (AR), and mineralocorticoid receptor; the thyroid/retinoid family including the thyroid receptor and the orphan receptor family, most of which can function as a ligand-independent fashion (Bain et al., 2007).
Nuclear receptors are switched on and off by small molecule ligands with properties similar to drugs. Therefore, NRs are attractive targets for developing new drugs against diseases like diabetes, cancer and heart disease (Chawla et al., 2001; Tsuchida et al., 2005). Nuclear receptors could also serve as great targets for development of insecticides. Four insecticides that target ecdysone receptor (EcR) have been commercialized to date (Palli et al., 2005). However, the other 20 NRs, which also harbor important functions, have not been exploited for development of insecticides. Several NRs including EcR, PR, GR and ER are being used for development of gene switches for use in medicine and agriculture (Palli et al, 2005). Knowledge in function of NRs in insects will aid in development of both insecticides and gene switches.
Some insect nuclear receptors are known to regulate female reproduction and embryogenesis. Nuclear receptor, HR39 was shown to regulate the female reproductive tract development and function in Drosophila melanogaster (Allen and Spradling, 2008). The NR, ultraspiracle (USP), is required for development of germ line tissues after fertilization, eggshell morphogenesis and embryonic development in D. melanogaster (Oro et al., 1992); some NRs affect sexual or reproduction behaviors. For example, estrogen receptor-α plays a key role in reproduction-related behaviors in female mice (Ogawa et al., 1998). The dissatisfaction (Dsf) gene of D. melanogaster codes for a tailless –like NR that regulates sexual behavior (Finley et al., 1998).
We recently identified nineteen canonical and two Knirps-like family NRs in the genome of T. castaneum (Tan and Palli, 2008). RNAi analysis showed that 10 out of the 19 canonical NRs, (E75, hormone receptor 3 (HR3), EcR, USP, seven-up (SVP), FTZ transcription factor 1 (FTZ-F1) and hormone receptor 4 (HR4) hormone receptor 51 (HR51), hormone receptor 38 (HR38), hormone receptor 39 (HR39) are important for metamorphosis. In addition, knocking down the expression of four NRs, tailless (Tll), Dsf, hepatocyte nuclear factor 4 (HNF4) and hormone receptor 78 (HR78) did not affect metamorphosis but caused defects in production of offspring. Also, knocking down the expression of non-canonical NR Knirps-like affected adults and caused reduction in egg production. To further understand the function of NRs in female reproduction, we used RNAi to knock-down the expression of gene coding for each NR during the adult stage and monitored progression in oogenesis, vitellogenesis, oviposition, embryogenesis and hatching. Seven NRs E75, HR3, EcR, USP, SVP, FTZ-F1 and hormone receptor 4 (HR4) are required for vitellogenesis and oogenesis because these seven NRs affected egg production by reducing the expression of vitellogenin genes, as well as by affecting the oocyte maturation. An additional seven NRs (hormone receptor 96 (HR96), HR51, HR38, HR39, Tll, Dsf and Knirps-like) are required for successful embryogenesis and hatching.
MATERIALS AND METHODS
Tribolium castaneum rearing and staging
Strain GA-1 of T. castaneum was reared on organic wheat flour containing 10% yeast at 30±1 °C under standard conditions (Beeman and Stuat, 1990). New adults were separated within 6 h post emergence.
RNA isolation and cDNA synthesis
Total RNA was isolated using the TRI reagent (Molecular Research Center Inc., Cincinnati, OH). The RNA was treated with DNase I (Ambion Inc., Austin, TX) and cDNA synthesis was performed using 2 μg of total RNA.
Double-stranded RNA (dsRNA) synthesis
Genomic DNA was used as a template to amplify regions of NRs and the PCR product was used for dsRNA synthesis. Genomic DNA was extracted from T. castaneum adults and purified using the DNeasy Tissue Kit (QIAGEN). All the primers used for dsRNA synthesis were reported by Tan and Palli (2008). The sequences for the rest of the primers used in real time PCR are shown in Table 1. The MEGA script RNAi Kit (Ambion Inc., Austin, TX) was used for dsRNA synthesis. For annealing dsRNA, the reaction products were incubated at 75°C for 5 min and cooled to room temperature over a period of 60 min. After the treatment with DNase, dsRNA was purified by phenol/chloroform extraction followed by ethanol precipitation and the dsRNA concentration was determined using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA).
Table 1.
The primers used in qRT-PCR
| gene | Forward primer 5′-3′ | Reverse primer 3′-5′ |
|---|---|---|
| Knirps-like | CCGACGTTTCTACCTCCTCA | TTCAACTCCAGGGTTGTGGT |
| E75 | CGGTCCTCAATGGAAGAAAA | TGTGTGGTTTGTAGGCTTCG |
| HR3 | CCGTGCAAAGTATGTGG | GTCGGCAGTATTGACATC |
| EcR | GATGGATGGCGAAGATCAGT | ACTTCGCTGGAACATGCTTT |
| USP | GATGCAAGCACAGGATGCTA | CCGACTTTATCCCTCGAACA |
| Tll | CACCAGGAAACGTCCTCAAT | GACCTGTGGCAGTGGTAGGT |
| HR51 | CTCAACCCACGAAACCATCT | GCTTTCAAGCAGGCAAATTC |
| Seven up | TATCGACCAACACCACAGGA | TGTCCGTAAACTGGGGAGTC |
| HR38 | CGCACCATTACGACTACCAA | TCTTGGGAGATGAGGGTGTC |
| FTZ-F1 | ACATTCGTGGTCGGATATGCTGGT | AGTTCTTGCAGTTTGGCGGTGATG |
| HR39 | CGACCGTCGACTGTACAAAA | AGTCGACATGGAACGGAAAC |
| HR4 | ACATTCGTGGTCGGATATGCTGG | TCTTGCCGGAGCTGTTCTATCGTT |
Microinjection
The newly hatched adult (within 6 h after emergence) were anesthetized with ether vapor for 4–5 min and lined on a glass slide covered with 2-sided tape. The dsRNA was injected into the dorsal side of the first or second abdominal segment using an injection needle pulled out from a glass capillary tube using a needle puller (Idaho technology). 0.8–1 μg (0.1 μl) dsRNA was injected into each new adult (within 6 h post emergence). The dsRNA prepared with 800 bp bacterial malE gene as a template was employed as a control. The injected beetles were removed from the slide and raised in whole wheat flour at 30±1 °C.
NR effects on egg laying, embryogenesis and oocyte maturation
Healthy females developed from adults injected with dsRNA were mated with uninjected virgin males. The beetles were removed from the flour at two weeks after beginning of matting. The number of eggs laid and larvae developed from eggs laid by each pair were recorded after incubation of eggs for one additional week. The ovaries were dissected following the methods described in Trauner and Büning (2007) and processed for scanning electron microscopy. The dissected ovaries were dehydrated in a series of ethanol (25%, 50%, 75%, 90% and 100%) by incubating in each concentration for 1 h with shaking. The ovaries were then immersed in HDMS (1,1,1,3,3,3 hexadimethyldisilazane) for 5 min, and air dried at room temperature. The ovaries were mounted on stainless steel stubs with sticky tape under dust free conditions. The ovaries were then sputtered with gold using Hummer VI Sputtering System (Technics) at plasma discharge rate of 10 mA for 180 s. Scans were performed with a Hitachi S-800 Scanning Electron Microscope set at 10 kV and 10 mA. Images were documented using Evex Nanoanalysis and Digital Imaging software (Evex Analytical version 2.0.1192). The oocytes were staged as described previously (Ullmann, 1973).
Embryo collection, fixing and staining with DAPI
Eggs were collected within 24 h after egg laying and incubated at 28°C for an additional three days. Then the eggs were dechlorinated by placing the strainer containing the eggs for 2 min into a Petri dish filled with 25% bleach. After washing, the embryos were fixed and stained following the procedure described by Bitra and Palli (2010).
Quantitative real-time PCR (qRT-PCR)
Relative mRNA levels of selected NRs after injection of dsRNA were determined by qRT-PCR. cDNA prepared using RNA isolated from control malE dsRNA or NR dsRNA injected female adults, primers designed based on NR sequences and MyiQ single color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) were used to perform qRT-PCR. Some of the primers used in qRT-PCR were reported by Tan and Palli (2008) and the primers used for qRT-PCR of vitellogenin genes, vg1 and vg2 were reported by Parthasarathy et al. (2010b), and the rest of the primers used in qRT-PCR are shown in Table. 1. All the primers for qRT-PCR were designed based on the sequence from the NR regions that are outside the dsRNA target regions. qRT-PCR reactions were performed using a common program as follows: initial incubation of 95 °C for 3 min was followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, 72 °C for 30 s. Standard curves were obtained using a 10-fold serial dilution of pooled cDNA. Quantitative mRNA measurements were performed in triplicate and normalized to an internal control of T. castaneum ribosomal protein 49 (RP49) mRNA.
Statistical analysis
The following linear models were used for analyzing the expression of each NR, yij = μ + ti + sj + tsij + eijk where yijl is an observed expression level of each NR, μ is the overall mean, ti is the ith (i=1,2,3) tissues effect (ovary, fat body or the whole body), sk is the kth (k=1,2…7) days effect after emergence (from d0 to d7), tsij, is the interaction between tissues and days effect, and eijk is the residual error, as described previously by Xu et al (2009).
The number of eggs laid by each pair, larvae hatched from eggs laid and expression levels of Vg1 and Vg2 in beetles injected with dsRNA were analyzed using the one way ANOVA program. Knock-down efficiency was analyzed by student-t test on the expression level between RNAi beetles and control. Multiple comparisons for Knockdown specificity were tested by Bonferroni option. The analysis was performed using SPSS version 13.0.
RESULTS
NRs expression in the whole body, fat body and ovary
Quantitative real-time PCR was employed to determine mRNA levels of all 21 NRs in the whole body (Day 0 to Day 7), as well as in the fat body (Day 0 to Day 7) and the ovary tissues (Day 3 to Day 7). Six NRs (Eagle, HR78, Tll, HR51, HR83 and PNR) showed very low levels of mRNA in the whole body, as well as in the fat body and ovary tissues on all the days tested (data not shown). Therefore, the expression profiles of these six NRs were not included in further analyses. In the whole body samples, USP, EcR, HR96, FTZ-F1, E75, HR39, HR38, SVP, HR4, E78 and HR3 showed higher levels of expression when compared to expression of the other four NRs, Tll, Dsf, Knirps-like and HR51 (Fig. 1A). Fairly constant levels of HR96 and FTZ-F1 mRNA were detected in the whole body samples of adults between 0–7 days post adult emergence (PAE). The USP mRNA levels were lower in adults soon after emergence and then increased gradually between 1–7 days PAE. The mRNA levels of EcR, E75, HR39, HR38, HR4 and HR3 were higher in adult female beetles soon after emergence and then the mRNA levels gradually decreased during 1–7 days PAE. In contrast, Tll mRNA levels were lower soon after adult emergence then increased gradually during 1–7 days PAE.
Figure 1.
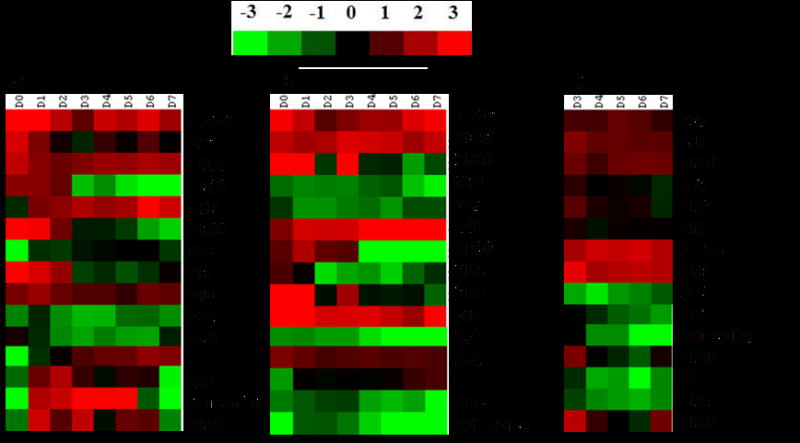
mRNA levels of NRs in the whole body (A), fat body (B) and ovary (C). Total RNA isolated from staged insects (whole body, fat body or ovary samples) and primers designed based on NR sequences were used in qRT-PCR to quantify mRNA levels of all 21 NRs. Relative mRNA levels were calculated by comparing the NR mRNA levels to internal standard RP49 mRNA levels. Three biological replicates were performed for each time point. The mean normalized values are displayed using Cluster 3.0 program.
Except for a few minor differences, the expression of patterns of NRs in the fat body are similar to those described above for the whole body suggesting that the mRNA levels in the fat body influence the mRNA levels in the whole body as fat body constitutes the majority of cells synthesizing RNA in the whole body (Fig 1B). One notable exception is the expression pattern of Tll, which is significantly different between the whole body and the fat body. While the Tll mRNA levels gradually decreased in the whole body samples of 1–7 day-old adults, the Tll mRNA levels gradually increased in the fat body dissected from 1–7 day-old adults. The mRNA levels of USP, EcR, HR96 and FTZ-F1 were high in the ovaries dissected from 3–7 day-old adult females (Fig. 1C). HR3, HR4 and HR38 mRNA levels were high on day 3 PAE, and then the mRNA levels decreased during days 4–7 PAE. Unlike in the whole body and fat body, the E75 mRNAs were present in the ovary during all days tested with slightly higher expression on days 5 and 6. Higher levels of HR39 were detected on day 3 and day 7 when compared to the other three days tested. These data showed that 15 NRs were expressed well in the adult females suggesting that they may play important roles in regulation of reproduction and embryogenesis.
Function of NRs in reproduction
To determine the function of NRs in reproduction of T. castaneum female adults, we prepared dsRNAs for 19 canonical and 2 Knirps-family NRs and injected them into newly eclosed adult female beetles. Knock-down in the expression of genes coding for all the NRs except Tll caused a 14–100% decrease in egg production when compared to eggs produced by control females injected with malE dsRNA. Knock-down in the expression of genes coding for seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) caused complete block in egg production (Fig. 2A). The eggs laid by females injected with Knirps-like, HR96, Tll, HR51, Dsf, HR38 and HR39 dsRNA produced no offspring (Fig. 2B). In addition, eggs laid by females injected with E78, HNF4 and Eagle dsRNA showed 45–75% reduction in hatching (Fig. 2B). These data from RNAi experiments showed that seven NRs are required for reproduction and an additional seven NRs are required for embryogenesis.
Figure 2.
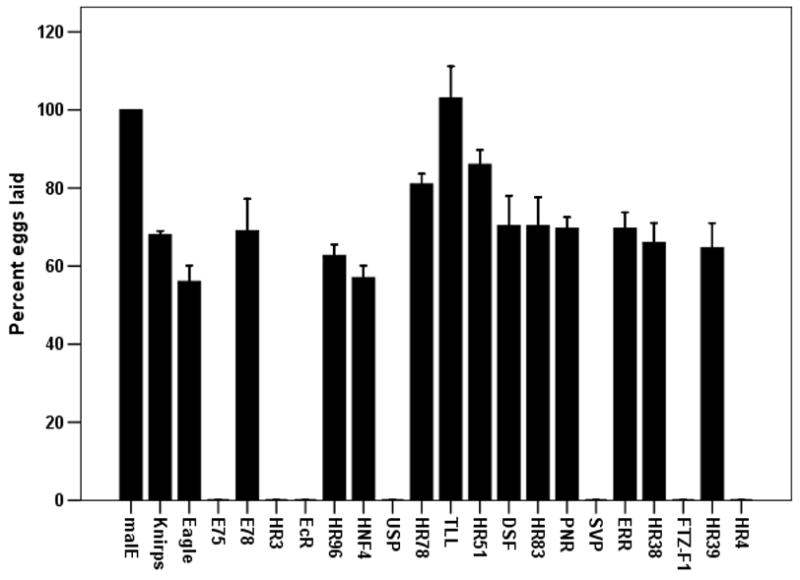
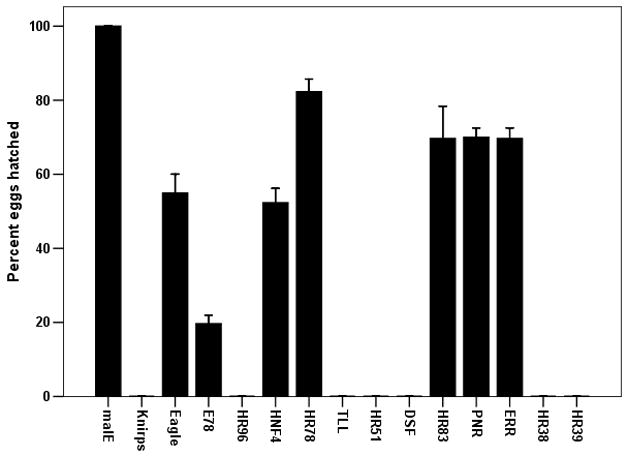
The effect of knock-down in the expression of genes coding for NRs on egg laying (A) and the production of offspring from the eggs laid (B). The dsRNA of NRs were injected into newly eclosed adult females and the injected female beetles were mated with virgin uninjected male beetles on the 4th day after injection. The eggs laid over a two week period by each pair were counted. The larvae hatched from the eggs laid were counted after incubating eggs for one additional week. The percent eggs laid and percent hatched were calculated by comparing the number of eggs laid and hatch rate from control beetles injected with malE dsRNA. Mean±S. E. (n=4) are shown.
Knock-down efficiency and specificity
Out of the all NRs tested, Eagle, HNF4, HR78, HR83, PNR and ERR showed the least effect on female reproduction and embryogenesis. To determine whether the lack of this effect on reproduction was due to inefficient knock-down in gene expression, we performed qRT-PCR to quantify mRNA levels of these genes in RNAi beetles. As shown in Figure 3A, the mRNA levels of these six NRs tested decreased by 44–81% in RNAi beetles when compared to their expression in malE dsRNA injected control beetles. The expression of genes coding for these NRs was reduced in dsRNA injected beetles yet no effect on reproduction and embryogenesis was observed suggesting that these NRs may not have a significant role in female reproduction or embryogenesis.
Figure 3.
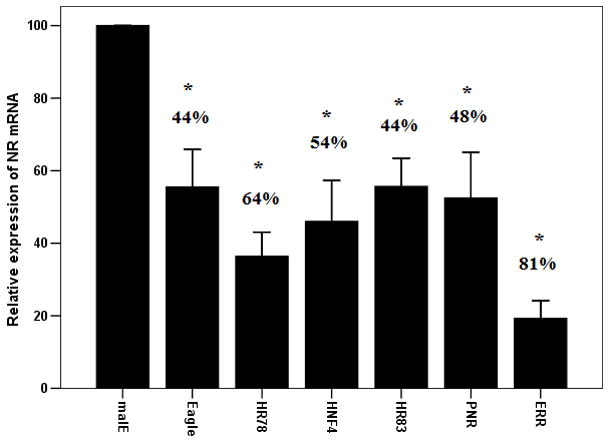
Efficiency (A) and specificity (B) of knock-down in expression of target genes in beetles injected with NR dsRNA. A. Knock-down efficiency test for 6 NRs including Eagle HR78, HNF4, ERR, PNR and HR83. Nuclear receptors or malE dsRNA were injected into the newly emerged adults. qRT-PCR was carried out to quantify mRNA levels at 96 h after injection. Relative expression was determined by comparing NR mRNA levels with the mRNA levels of internal standard RP49. Knock-down efficiency was calculated by comparing NR mRNA levels in NR dsRNA injected insects with the expression levels in malE dsRNA injected insects. Mean±S.E (n=3) are shown.
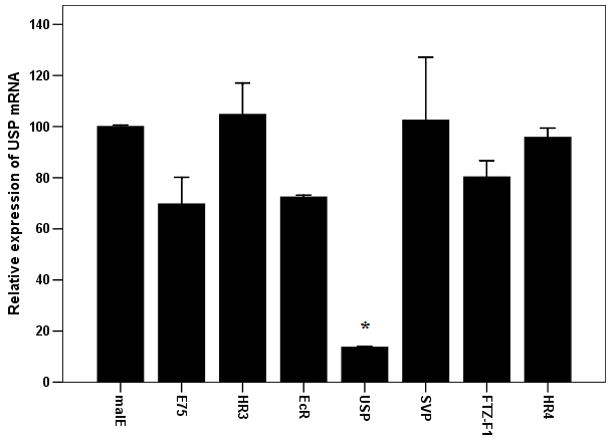
B: Knock-down specificity among seven NRs E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4. The dsRNAs for these seven NRs were injected into newly eclosed females and USP mRNA levels were quantified at 96 h after injection.
The dsRNA for each NR was designed in the region that is not well conserved among the 21 nuclear receptors. Therefore, the dsRNA prepared for each NR is expected to knock-down the expression of its own target NR but not the other receptors. As shown in Figure 1A, injection of dsRNA of seven nuclear receptors E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4 blocked egg production completely. To determine the specificity of dsRNA in knocking-down the expression of these seven NRs, we injected dsRNA of these seven NRs and quantified mRNA levels of USP in RNAi animals. As shown in Figure 3B, the mRNA levels of USP were significantly reduced in beetles injected with USP dsRNA but not in the beetles injected with dsRNA for the other six NRs. Also, more than 60% knock-down in the expression of target gene was achieved in beetles injected with each of the seven NRs (data not shown). These data demonstrate the specificity of dsRNA in knocking-down the expression of target NRs.
The effect of NR knock-down on Vg gene expression
As shown above, seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) are required for egg production. To determine whether these seven NRs block egg production by affecting Vg gene expressions, Vg1 and Vg2 mRNA levels were quantified in insects injected with dsRNA of these seven NRs. The mRNA levels of both Vg1 and Vg2 were significantly reduced in beetles injected with dsRNA for these seven NRs when compared to their expression in control insects injected with malE dsRNA (Figs 4A&B). The extent of the effect on Vg gene expression was variable among the NRs tested. Knock-down in the expression of gene coding for FTZ-F1 caused complete reduction in Vg gene expression. Knock-down in the expression of the genes coding for the other six NRs tested caused 50–75% reduction in Vg gene expressions. Some of the variability observed in the reduction of Vg gene expression in NR RNAi beetles could be due to variability in the knock-down efficiency among these seven NRs.
Figure 4.
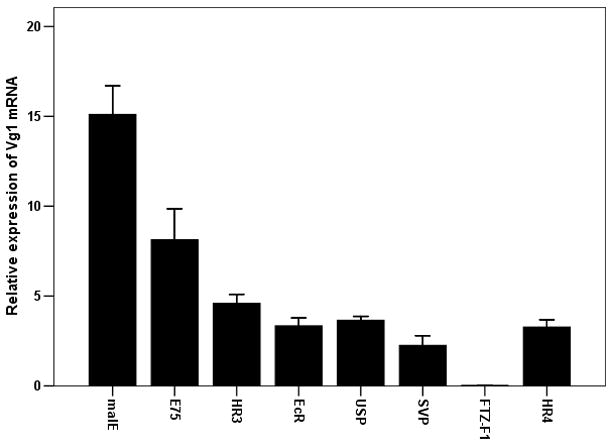
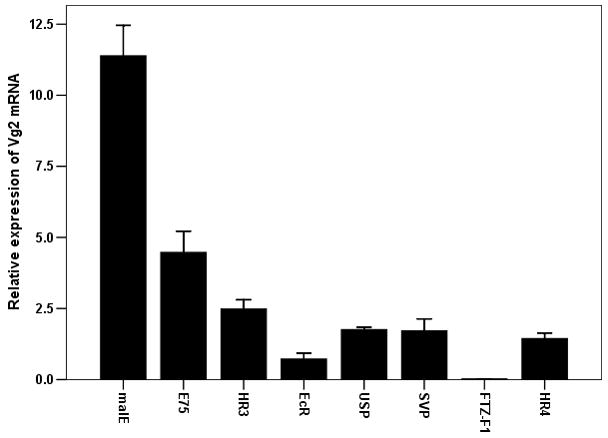
Effect of knock-down in the expression of genes coding for seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) on the expression of Vg1 (A) and Vg2 (B) genes. Seven NRs or malE dsRNA were injected into the newly emerged adults. qRT-PCR was carried out to quantify mRNA levels of Vg1 or Vg2 at 96 h after injection. The relative expression was determined by comparing NR mRNA levels with the mRNA levels of internal standard RP49. The mean relative expression compared to that in malE RNAi beetles was calculated by comparing Vg1 or Vg2 mRNA levels in NR dsRNA injected insects with the expression levels in malE dsRNA injected insects. Mean±S.E (n=3) are shown.
The effect of NRs knock-down on oocyte maturation
To determine whether the seven NRs block egg production by affecting the oocyte maturation, the ovaries were dissected from insects injected with dsRNA for seven NRs on the 4th day after injection and observed under a scanning electron microscope. The developing oocytes were classified as described by Ullmann (1973). The primary oocyte in the ovaries dissected from control beetles injected with malE dsRNA were well developed and reached stage 8 by 4 days PAE (Fig. 5). In contrast, the primary oocytes in the ovaries dissected from insects injected with EcR or USP were blocked at stages 2–3 (Fig. 5). The ovaries dissected from insects injected with HR4, E75, HR3, FTZ-F1 and SVP dsRNA contained primary oocytes that were blocked at stages 3–4 (Fig. 5).
Figure 5.

Effect of knock-down in the expression of genes coding for seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) on oocyte maturation. dsRNA of seven NRs or malE was injected into newly emerged females and ovaries were dissected at four days after injection, processed and images were captured using a scanning electron microscope as described in Materials and Methods section.
The effect of NR knock-down on embryonic development
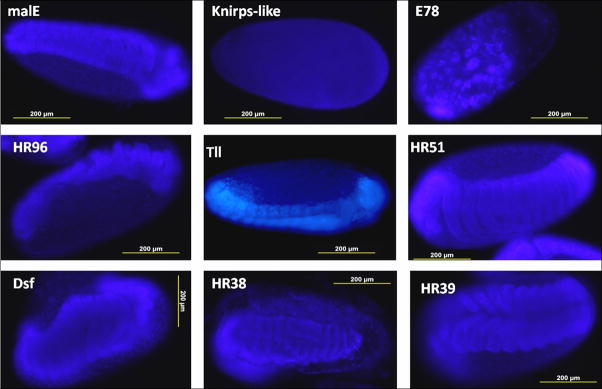
To determine the stages at which the embryonic development was blocked due to the lack of nuclear receptor proteins, the eggs were collected within 24 h of egg laying and on the fourth day after collection the embryos were fixed and stained with DAPI. Most of the eggs laid by control beetles injected with malE dsRNA successfully completed embryonic development and became first instar larvae. In contrast, more than 90% embryos in the eggs laid by beetles injected with Knirps-like dsRNA were arrested at the early blastoderm stage (Fig. 6). Similarly, the embryos developed from eggs laid by E78 dsRNA injected beetles were arrested at the beginning of germ band growth (Fig. 6). The embryos developed in eggs laid by beetles injected with HR96 dsRNA were blocked at the mid-stage of germ band growth. Knock-down in expression of Tll, Dsf, HR38 and HR39 also affected embryogenesis, these embryos were blocked near the stage of germ band growth completion. Embryos in the eggs laid by HR51 dsRNA injected beetles were arrested at a later stage of embryogenesis where in the formation of first instar larvae was completed but none of the larvae were unable to hatch and remained in the egg shell (Fig. 6).
Figure 6.
Nuclear staining of embryos of T. castaneum
Eggs were collected within 24 h after egg laying and stained after 4 days of egg laying to observe the stage of arrest of embryogenesis. All the pictures were taken at 10× magnification.
DISCUSSION
The first major contribution of this study is the identification of seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) that play critical roles in female reproduction of the red flour beetle, Tribolium castaneum. Out of the 21 NRs tested, knock-down in expression of seven NRs (E75, HR3, EcR, USP, SVP, FTZ-F1 and HR4) completely blocked egg laying. Further analysis showed that these seven NRs are required for both Vg production and oocyte maturation because knock-down in expression of these seven NR genes caused significant reduction in Vg synthesis, blocked oocyte maturation. It is interesting that all the seven NRs identified as essential genes for female reproduction are known to play key roles in 20E signal transduction (King-Jones and Thummel, 2005). Two of the identified NRs, EcR and USP heterodimerize and bind to 20E. E75 is an early gene, HR3 and HR4 are delayed-early genes and FTZ-F1 is a competent factor. All these five NRs are 20E induced transcription factors that play key roles in expression and/or repression of genes in 20E signal transduction cascade. These data suggest that 20E regulates female reproduction including oogenesis and vitellogenesis in T. castaneum. In a recent study Parthasarathy et al., (2010a) reported that 20E directly regulates oocyte maturation in T. castaneum. Initiation of oocyte maturation is a prerequisite for vitellogenin synthesis, hence 20E indirectly regulates vitellogenin synthesis in the fat body (Parthasarathy et al., 2010b). Taken together, these studies in T. castaneum suggest that the NRs that are involved in 20E action may directly regulate oogenesis.
E75 is an ecdysone induced transcription factor involved in the 20E pathway mediated by EcR and USP heterodimer (Segraves and Hogness, 1990). It acts as an early gene to regulate vitellogenesis and oogenesis. For example, in D. melanogaster it mediates egg chamber maturation during mid-oogenesis (Buszczak et al., 1999). In Aedes aegypti, all three E75 isoforms are induced at the onset of vitellogenesis by a blood meal-activated hormonal cascade, and highly expressed in the mosquito ovary (Pierceall et al., 1999). In the ovary of the honey bee queen, E75 is expressed preferentially in the follicle cells that surround the oocyte at the late stage (Paul et al., 2006).
HR3 and HR4 seem to have similar functions in regulation of development; they not only repress early genes induced by 20E but also induce the downstream genes, such as FTZ-F1 (Kageyama et al., 1997; King-Jones et al.,, 2005). The entry into metamorphism depends on the precise timing of both these functions (Gauhar et al., 2009). In the adult mosquito, HR3 is involved in 20E action during vitellogenesis as indicated by several findings such as kinetics and dose response of HR3 to 20E in in vitro tissue culture experiments and expression in vitellogenic tissues, the fat body and the ovary of the female mosquito, Ae. aegypti (Kapitskaya et al., 2000).
FTZ-F1 is also a 20E induced gene, and was shown to play key roles in regulation of vitellogenesis in the adult mosquito (Zhu et al., 2003). DHR3 binding sites were identified in the promoter of FTZ-F1 gene (Kageyama et al., 1997). Also, Drosophila, DHR4 coordinates growth and maturation by controlling the attainment of critical weight during larval development (King-Jones et al., 2005). As identified by genome-wide mapping of binding sites, HR4 is EcR and USP direct target and is required for cellular differentiation in response to 20E (Gauhar et al., 2009). In vertebrates, GCNF (germ cell nuclear factor, a HR4 homologue in the human) is required for the repression of pluripotency genes during embryonic stem cell differentiation (Gu et al., 2005), during gamete regulation of female fertility (Lan et al., 2003). However, the precise function of HR3 and HR4 in regulation of female reproduction still remains unknown.
Seven-up (homolog of chicken ovalbumin upstream promoter transcription factor, COUP-TF) is highly expressed in the fat body during vitellogenesis in T. castaneum (this study) as well as in the mosquito (Miura et al., 2002). Moreover, the Drosophila SVP is important for development of photoreceptor cells of the ommatidium (Kramer et al., 1995; Mlodzik et al., 1990), fat body (Hoshizaki et al., 1994), Malpighian tubules (Kerber et al., 1998) and neuronal tissues (Mlodzik et al., 1990).
Eight NRs play essential roles in the embryogenesis of T. castaneum
The second major contribution of this study is the identification of eight NRs (Knirps-like, HR96, Tll, HR51, Dsf, HR38, HR39 and E78) that are required for embryogenesis in T. castaneum. To date, five of these eight NRs, Tll, Dsf, HR51, HR38 and Knirps-like have been shown to have some function in reproduction and embryogenesis. The current study identified three additional NRs (HR38, HR39 and E78) as those required for successful completion of embryogenesis.
Tll is required for formation of the terminal domains of Drosophila embryo (Steingrimsson et al., 1991). In the previous study, we showed that eggs laid by T. castaneum females developed from final instar larvae injected with Tll dsRNA fail to hatch (Tan and Palli, 2008). In this study, we found that the eggs laid by Tll dsRNA injected insects have problems completing embryogenesis and stop at the germ band stage. DHR51 binds NO and CO and may be either a gas or a heme sensor (de Rosny et al., 2008) and DHR51 mutant flies show wing expansion failure and compromised fertility (Sung et al., 2009). Our studies showed that the embryos developed from eggs laid by beetles injected with the HR51 dsRNA completed embryonic development but first instar larvae were not able to come out of the egg shells. HR38 was shown to function as a negative regulator in the EcR-USP mediated 20E pathway by competing with EcR for USP binding (Sutherland et al., 1995). HR38 has no ligand binding pocket and coactivator binding site as shown by X-ray crystallographic analysis (Baker et al., 2003). Fisk and Thummel (1995) reported that both ~4.0–kb and ~5.0 kb HR38 mRNA are highly abundant during the late embryonic stage (19–23 hr post-egg laying) in D. melanogaster. Our results from nuclear staining of HR38 RNAi embryos showed that the development in these embryos was blocked at the stage of germ band growth completion suggesting that expression of HR38 during the late embryonic stage of T. castaneum is required for successful embryogenesis.
In D. melanogaster Knirps-like NR is involved in the wing development during early embryogenesis (Lunde et al., 1998), initially Knirps-like is expressed in three identical regions of the blastoderm: in an anterior cap domain, in an anterior stripe and in a posterior broad band linked to the kni gap gene function (Rothe, 1994). In a recent study, TcKnirps-like was found to play a major role in head segmentation and a minor role in abdominal patterning (Cerny et al., 2008). Taken together, these data suggest that expression of Knirps-like NR is required during early stages of embryogenesis when blastoderm formation and segmentation occur. DHR39 is related to fly FTZ-F1 and human liver receptor homologue 1 (LRH1) and steroidogenic factor 1. Horner et al.(1995) showed that DHR39 is induced by 20E and suggested that it may function together with the early regulatory gene to coordinate 20E action. DHR96 is also 20E inducible, and known to regulate xenobiotic responses in Drosophila (Fisk and Thummel, 1995; King-Jones et al., 2006).
This initial study identified 17 NRs play key roles in regulation of oogenesis, vitellogenesis and embryogenesis. These studies could serve as solid foundation on future studies on mechanism of action of these NRs in regulation of female reproduction and embryogenesis in T. castaneum. Knock-down in the expression of genes coding for four NRs did not show much effect on reproduction or embryogenesis. Comparison of knock-down efficiency of the NRs with RNAi phenotypes (e.g. hatch rate, Fig. 2B) showed no correlation between RNAi phenotype and knock-down efficiency. For example, the gene coding for ERR was knocked-down by 81% but no detectable effect on reproduction or embryogenesis was observed in the RNAi beetles. In contrast, the genes coding for Eagle and HNF4 were knock-down by 44 and 54% respectively in dsRNA injected insects but only 50% of the eggs laid by these RNAi beetles hatched successfully. These data suggest that knock-down in the expression of genes coding for these NRs was achieved but these NRs did not show any detectable effects on reproduction or embryogenesis therefore, they may not have a major role in these processes. Since, we were not able to achieve 100% knock-down in the expression some of these NRs, it is possible that the amount of these NRs required for regulation of reproduction and embryogenesis may be present in the RNAi beetles. Therefore, the function in female reproduction and embryogenesis for four NRs that did not show any effect could not be excluded completely. Further studies are required to determine whether or not these four NRs are required for female reproduction and embryogenesis.
Acknowledgments
We would like to dedicate this paper to Professor Judy Willis for her encouragement and insightful discussions during the past several years. This work was supported by USDA-CSREES (2007-04636). This is contribution number 06-08-075 from the Kentucky Agricultural Experimental Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AK, Spradling AC. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development. 2008;135:311–321. doi: 10.1242/dev.015156. [DOI] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annual Review of Physiology. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, Wisely B, Caravella JA, Lambert MH, Reinking JL, Krause H, Thummel CS, Willson TM, Mangelsdorf DJ. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- Beeman RW, Stuart JJ. A gene for lindane 1 cyclodiene resistance in the red flour beetle (Coleoptera: Tenebrionidae) Journal of Economic Entomology. 1990;83:1745–1751. [Google Scholar]
- Bitra K, Palli SR. The members of bHLH transcription factor superfamily are required for female reproduction in the red flour beetleTribolium castaneum. Journal of Insect Physiol. 2010 doi: 10.1016/j.jinsphys.2010.03.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Freeman MR, Carlson JR, Bender M, Cooley L, Segraves WA. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 1999;126:4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- Carney GE, Bender M. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics. 2000;154:1203–1211. doi: 10.1093/genetics/154.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny AC, Grossmann D, Bucher G, Klingle M. The Tribolium ortholog of knirps and knirps-related is crucial for head segmentation but plays a minor role during abdominal patterning. Developmental biology. 2008;321:284–294. doi: 10.1016/j.ydbio.2008.05.527. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- de Rosny E, de Groot A, Jullian-Binard C, Borel F, Suarez C, Le Pape L, Fontecilla-Camps JC, Jouve HM. DHR51, the Drosophila melanogaster homologue of the human photoreceptor cell-specific nuclear receptor, is a thiolate heme-binding protein. Biochemistry. 2008;47:13252–13260. doi: 10.1021/bi801691b. [DOI] [PubMed] [Google Scholar]
- Dhadialla TS, Le D, Palli SR, Raikhel A, Carlson GR. A photoaffinity, non-steroidal, ecdysone agonist, bisacylhydrazine compound, RH-131039: characterization of binding and functional activity. Insect Biochemistry and Molecular Biology. 2007;37:865–875. doi: 10.1016/j.ibmb.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Foss M, Gross E, Ghbeish N, Palmer RH, Taylor BJ, McKeown M. Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron. 1998;21:1363–1374. doi: 10.1016/s0896-6273(00)80655-8. [DOI] [PubMed] [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proceedings of the National Academy of Sciences U S A. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauhar Z, Sun LV, Hua S, Mason CE, Fuchs F, Li TR, Boutros M, White KP. Genomic mapping of binding regions for the Ecdysone receptor protein complex. Genome Research. 2009;19:1006–1013. doi: 10.1101/gr.081349.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, LeMenuet D, C-KChung A, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Molecular and Cellular Biology. 2005;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Horner MA, Chen TH, Thummel CS. Ecdysteroid regulation and DNA binding properties of drosophila nuclear hormone receptor superfamily members. Developmental Biology. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- Hoshizaki DK, Blackburn T, Price C, Ghosh M, Miles K, Ragucci M, Sweis R. Embryonic fat-cell lineage in Drosophila melanogaster. Development. 1994;120:2489–2499. doi: 10.1242/dev.120.9.2489. [DOI] [PubMed] [Google Scholar]
- Kageyama Y, Masuda S, Hirose S, Ueda H. Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes Cells. 1997;2:559–569. doi: 10.1046/j.1365-2443.1997.1460344.x. [DOI] [PubMed] [Google Scholar]
- Kapitskaya MZ, Li C, Miura K, Segraves M, Raikhel AS. Expression of the early-late gene encoding the nuclear receptor HR3 suggests its involvement in regulating the vitellogenic response to ecdysone in the adult mosquito. Molecular and Cellular Endocrinology. 2000;160:25–37. doi: 10.1016/s0303-7207(99)00253-1. [DOI] [PubMed] [Google Scholar]
- Kerber B, Fellert S, Hoch M. Seven-up, the Drosophila homolog of the COUP-TF orphan receptors, controls cell proliferation in the insect kidney. Genes and Development. 1998;12:1781–1786. doi: 10.1101/gad.12.12.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Charles JP, Lam G, Thummel CS. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell. 2005;121:773–784. doi: 10.1016/j.cell.2005.03.030. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metabolism. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nature Reviews Genetics. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kramer S, West SR, Hiromi Y. Cell fate control in the Drosophila retina by the orphan receptor seven-up: its role in the decisions mediated by the ras signaling pathway. Development. 1995;121:1361–1372. doi: 10.1242/dev.121.5.1361. [DOI] [PubMed] [Google Scholar]
- Lan ZJ, Gu P, Xu X, Jackson KJ, DeMayo FJ, O’Malley BW, Cooney AJ. GCNF-dependent repression of BMP-15 and GDF-9 mediates gamete regulation of female fertility. Embo J. 2003;22:4070–4081. doi: 10.1093/emboj/cdg405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde K, Biehs B, Nauber U, Bier E. The knirps and knirps-related genes organize development of the second wing vein in Drosophila. Development. 1998;125:4145–4154. doi: 10.1242/dev.125.21.4145. [DOI] [PubMed] [Google Scholar]
- Miura K, Zhu J, Dittmer NT, Chen L, Raikhel AS. A COUP-TF/Svp homolog is highly expressed during vitellogenesis in the mosquito Aedes aegypti. Journal of Molecular Endocrinology. 2002;29:223–238. doi: 10.1677/jme.0.0290223. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Moran E, Jimenez G. The tailless nuclear receptor acts as a dedicated repressor in the early Drosophila embryo. Molecular and Cellular Biology. 2006;26:3446–3454. doi: 10.1128/MCB.26.9.3446-3454.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Davis EC, Clipsham RC, Jiang M, Dehart DB, Sulik KK, McCabe ER. Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Molecular Genetics and Metabolism. 2006;88:261–271. doi: 10.1016/j.ymgme.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Eng V, Taylor J, Lubahn DB, Korach KS, Pfaff DW. Roles of estrogen receptor-alpha gene expression in reproduction-related behaviors in female mice. Endocrinology. 1998;139:5070–5081. doi: 10.1210/endo.139.12.6357. [DOI] [PubMed] [Google Scholar]
- Oro AE, McKeown M, Evans RM. The Drosophila retinoid X receptor homolog ultraspiracle functions in both female reproduction and eye morphogenesis. Development. 1992;115:449–462. doi: 10.1242/dev.115.2.449. [DOI] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metabolism. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palli SR, Hormann RE, Schlattner U, Lezzi M. Ecdysteroid receptors and their applications in agriculture and medicine. Vitamins and Hormones. 2005;73:59–99. doi: 10.1016/S0083-6729(05)73003-X. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Palli SR. Proliferation and differentiation of intestinal stem cells during metamorphosis of the red flour beetle, Tribolium castaneum. Devlopmental Dynamics. 2008;237:893–908. doi: 10.1002/dvdy.21475. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Sheng Z, Sun Z, Palli SR. Ecdysteroid regulation of ovarian growth and oocyte maturation in the red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2010a doi: 10.1016/j.ibmb.2010.04.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Sun ZY, Bai H, Palli SR. Juvenile hormone regulation of vitellogenin synthesis in the red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2010b doi: 10.1016/j.ibmb.2010.03.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul RK, Takeuchi H, Kubo T. Expression of two ecdysteroid-regulated genes, Broad-Complex and E75, in the brain and ovary of the honeybee (Apis mellifera L.) Zoological Science. 2006;23:1085–1092. doi: 10.2108/zsj.23.1085. [DOI] [PubMed] [Google Scholar]
- Pierceall WE, Li C, Biran A, Miura K, Raikhel AS, Segraves WA. E75 expression in mosquito ovary and fat body suggests reiterative use of ecdysone-regulated hierarchies in development and reproduction. Molecular and Cellular Endocrinology. 1999;150:73–89. doi: 10.1016/s0303-7207(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Rothe M, Wimmer EA, Pankratz MJ, Gonzaález-Gaitán M, Jäckle H. Identical transacting factor requirement for knirps and knirps-related Gene expression in the anterior but not in the posterior region of the Drosophila embryo. Mechanisms of Development. 1994;46:169–181. doi: 10.1016/0925-4773(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes and Development. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Pignoni F, Liaw GJ, Lengyel JA. Dual role of the Drosophila pattern gene tailless in embryonic termini. Science. 1991;254:418–421. doi: 10.1126/science.1925599. [DOI] [PubMed] [Google Scholar]
- Sung C, Wong LE, Chang Sen LQ, Nguyen E, Lazaga N, Ganzer G, McNabb SL, Robinow S. The unfulfilled/DHR51 gene of Drosophila melanogaster modulates wing expansion and fertility. Developmental Dynamics. 2009;238:171–182. doi: 10.1002/dvdy.21817. [DOI] [PubMed] [Google Scholar]
- Sutherland JD, Kozlova T, Tzertzinis G, Kafatos FC. Drosophila hormone receptor 38: a second partner for Drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor-induced protein B type. Proceedings of the National Academy of Sciences U S A. 1995;92:7966–7970. doi: 10.1073/pnas.92.17.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swevers L, Drevet JR, Lunke MD, Iatrou K. The silkmoth homolog of the Drosophila ecdysone receptor (B1 isoform): cloning and analysis of expression during follicular cell differentiation. Insect Biochemistry and Molecular Biology. 1995;25:857–866. doi: 10.1016/0965-1748(95)00024-p. [DOI] [PubMed] [Google Scholar]
- Tan A, Palli SR. Identification and characterization of nuclear receptors from the red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2008;38:430–439. doi: 10.1016/j.ibmb.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Trauner J, Büning J. Germ-cell cluster formation in the telotrophic meroistic ovary of Tribolium castaneum (Coleoptera, Polyphaga, Tenebrionidae) and its implication on insect phylogeny. Development Genes and Evolution. 2007;217:13–27. doi: 10.1007/s00427-006-0114-3. [DOI] [PubMed] [Google Scholar]
- Tsuchida A, Yamauchi T, Kadowaki T. Nuclear receptors as targets for drug development: molecular mechanisms for regulation of obesity and insulin resistance by peroxisome proliferator-activated receptor gamma, CREB-binding protein, and adiponectin. Journal of Pharmacological Science. 2005;97:164–170. doi: 10.1254/jphs.fmj04008x2. [DOI] [PubMed] [Google Scholar]
- Ullmann SL. Oogenesis in Tenebrio molitor: histological and autoradiographical observations on pupal and adult ovaries. Journal of Embryology and Experimental Morphology. 1973;30:179–217. [PubMed] [Google Scholar]
- Xu JJ, Shu J, Qiu XT, Wang ZP, Zhao FP, Zhang Z, Zhang Q. Effect of heat shock on ovary development and Hsp83 expression in Tribolium castaneum (Coleoptera: Tenebrionidae) Archives of Insect Biochemistry and Physiology. 2009;70:204–16. doi: 10.1002/arch.20294. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen L, Raikhel AS. Posttranscriptional control of the competence factor betaFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proceedings of the National Academy of Sciences USA. 2003;100:13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]