Abstract
Neurons are known to use large amounts of energy for their normal function and activity. In order to meet this demand, mitochondrial fission, fusion, and movement events (mitochondrial dynamics) control mitochondrial morphology, facilitating biogenesis and proper distribution of mitochondria within neurons. In contrast, dysfunction in mitochondrial dynamics results in reduced cell bioenergetics and thus contributes to neuronal injury and death in many neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease, and Huntington’s disease. We recently reported that amyloid-β peptide, thought to be a key mediator of AD pathogenesis, engenders S-nitrosylation and thus hyperactivation of the mitochondrial fission protein Drp1. This activation leads to excessive mitochondrial fragmentation, bioenergetic compromise, and synaptic damage in models of AD. Here, we provide an extended commentary on our findings of nitric oxide-mediated abnormal mitochondrial dynamics.
Keywords: S-Nitrosylation, Dynamin-related protein 1, Alzheimers’s disease, Mitochondrial fission
1. Introduction
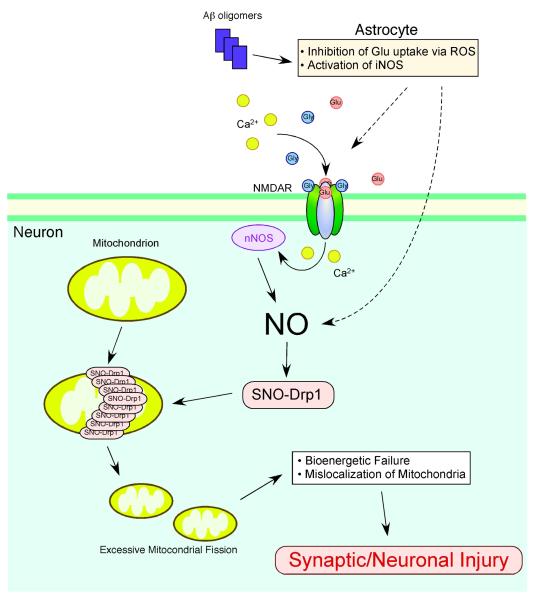
Normally, mitochondria continuously undergo fission and fusion (known as mitochondrial dynamics) to generate smaller organelles or elongated, tubular structures, respectively. This normal mitochondrial fission and fusion can facilitate formation of new mitochondria (biogenesis), repair of defective mitochondrial DNA through mixing, and redistribution of mitochondria to sites requiring high-energy production (Chen and Chan, 2006; Frederick and Shaw, 2007; Knott et al., 2008). Conversely, an imbalance in fission or fusion initiates malfunctions in mitochondrial morphology and bioenergetics, and may thus contribute to neuronal injury during neurodegeneration (Barsoum et al., 2006; Bossy-Wetzel et al., 2003; Knott et al., 2008). Dysfunction in mitochondrial dynamics can result from either (i) rare genetic mutations in fission- or fusion-related genes, as occurs in Charcot-Marie-Tooth (CMT) Disease and Autosomal Dominant Optic Atrophy (ADOA) (Delettre et al., 2000; Zuchner et al., 2004), or (ii) posttranslational changes to the fission or fusion proteins (Cho et al., 2009). In particular, a posttranslational modification engendered by nitrosative/oxidative stress may well account for the more common sporadic cases of the disease. Recently, we discovered that excessive accumulation of nitrosative stress triggers abnormal mitochondrial morphology in brains of neurodegenerative patients via S-nitrosylation of the mitochondrial fission protein dynamin-related protein 1 (Drp1) (Cho et al., 2009). S-Nitrosylated Drp1 contributes to excessive mitochondrial fission/fragmentation, synaptic injury, and neuronal apoptosis in neurodegenerative diseases such as AD (Fig. 1).
Fig. 1.
Possible mechanism whereby S-nitrosylated Drp1 contributes to excessive mitochondrial fragmentation and neuronal injury. NMDAR hyperactivation triggers generation of NO and subsequent S-nitrosylation of Drp1 (forming SNO-Drp1), contributing to synaptic injury and eventually neuronal death. Soluble oligomers of Aβ peptide, thought to be a key mediator of AD pathogenesis, can facilitate neuronal NO production, potentially in both NMDAR-dependent and -independent manners. S-Nitrosylation of Drp1 can contribute to synaptic damage and neuronal cell death by triggering excessive mitochondrial fission and bioenergetic impairment.
2. Dysfunctional mitochondrial fission and fusion in neurodegeneration
Neurons are particularly vulnerable to mitochondrial defects because they require high levels of energy for their survival and specialized function. In particular, mitochondrial biogenesis is required at synapses that demand high concentrations of ATP. The distribution of mitochondria at the nerve terminal can indeed facilitate synaptic transmission and maintain synaptic structure (Chen and Chan, 2006; Li et al., 2008; Li et al., 2004).
In healthy neurons, the fission/fusion machinery proteins maintain mitochondrial integrity and insure their presence at critical locations. These proteins includes Drp1 and Fis1, acting as fission proteins, and Mitofusins (Mfn1/2) and Opa1, operating as fusion proteins (Youle and Karbowski, 2005). In both familial and sporadic neurodegenerative conditions, abnormal mitochondria regularly appear in the brain as a result of dysfunction in the fission/fusion machinery. Genetic mutations in Mfn2 can cause CMT disease, a hereditary peripheral neuropathy that affects both motor and sensory neurons (Kijima et al., 2005; Zuchner et al., 2004). Additionally, mutations in Opa1 cause ADOA, characterized by the loss of retinal ganglion cells and the optic nerve, representing their axons (Delettre et al., 2000). Recently, Waterham and colleagues described a heterozygous, dominant-negative mutation of Drp1 in a patient whose symptoms were broadly similar to those of CMT neuropathy and ADOA (Waterham et al., 2007). Taken together, it is apparent that the balance between fission and fusion is critical for normal function of mitochondria and determination of phenotype in neurological disease. Additionally, these fission/fusion proteins are widely expressed in human tissues, clearly supporting the notion that neurons are particularly sensitive to mitochondrial dysfunction.
Mitochondrial dysfunction also represents a hallmark of sporadic neurodegenerative diseases. For example, patients with early stage AD regularly exhibit declining mitochondrial energy metabolism and ATP production, which may subsequently cause synaptic loss and neuronal damage (Liang et al., 2008; Parker et al., 1994; Reddy, 2007; Wang et al., 2009c). Neurons in AD and other neurodegenerative brains often display abnormal mitochondrial morphology (Baloyannis, 2006; Hirai et al., 2001; Wang et al., 2009b). In cell-based experiments, β-amyloid (Aβ) production resulted in the appearance of fragmented and abnormally distributed mitochondria (Barsoum et al., 2006; Wang et al., 2008), suggesting that Aβ (possibly in the form of soluble oligomers) may trigger excessive mitochondrial fission in AD patients. Pathological forms of tau may also contribute to mitochondrial fragmentation in AD brains since expression of caspase-cleaved tau induced mitochondrial fission in a calcineurin-dependent manner (Quintanilla et al., 2009).
3. S-Nitrosylation and neurodegenerative diseases
Brains with neurodegenerative diseases often manifest excessive generation of reactive nitrogen species (RNS) and reactive oxygen species (ROS), which can contribute to neuronal cell injury and death via a series of redox reactions (Barnham et al., 2004; Beal, 2001; Emerit et al., 2004; Lin and Beal, 2006; Muchowski, 2002). While many intra- and extracellular molecules may participate in neuronal injury, accumulation of nitrosative stress due to excessive generation of nitric oxide (NO) appears to be a potential factor contributing to neuronal cell damage and death (Lipton, 2006; Lipton and Rosenberg, 1994). A well-established model for NO production entails a central role of N-methyl-D-aspartate (NMDA)-type glutamate receptors in the nervous system. Excessive activation of NMDA receptors drives Ca2+ influx, which in turn activates neuronal NO synthase (nNOS) as well as the generation of ROS (Bredt et al., 1991; Garthwaite et al., 1988; Lafon-Cazal et al., 1993) (Fig. 1). Accumulating evidence suggests that NO can mediate both protective and neurotoxic effects by redox reactions with cysteine residues of target proteins to form S-nitrosothiols (SNOs), a process termed S-nitrosylation because of its effects on the chemical biology of protein function. Importantly, normal mitochondrial respiration may also generate free radicals, principally ROS, and one such molecule, superoxide anion (O2.−), reacts rapidly with free radical NO. to form the very toxic product peroxynitrite (ONOO−) (Beckman et al., 1990; Lipton et al., 1993).
Production of NO from inducible NOS (iNOS) can also contribute to the pathogenesis of neurodegenerative diseases, including Alzheimer’s disease (AD) (Medeiros et al., 2007; Wang et al., 2004). A classic feature of AD pathology is the generation of Aβ peptides. Recently, several lines of evidence have suggested that soluble oligomers of Aβ represent the most toxic form of the peptide (Haass and Selkoe, 2007). Consistent with this notion, Aβ oligomers, but not fibrillar Aβ, induce high expression of iNOS in astrocytes and thus generation of NO (White et al., 2005). Additionally, Aβ is known to inhibit glutamate re-uptake, at least in part via generation of ROS, producing an increase extracellular glutamate; this can lead to pathological activation of NMDA receptors, thereby disturbing synaptic function in AD (Li et al., 2009; Matos et al., 2008; Trotti et al., 1998). Excessive stimulation of NMDA receptors also leads to activation of nNOS, as discussed above, thus representing another source of NO emanating from Aβ oligomers.
Nitrosative stress can result in defects in mitochondrial function. For example, NO affects mitochondrial respiration by reversibly inhibiting complexes I and IV (Cleeter et al., 1994; Clementi et al., 1998). Mitochondria thus compromised will release ROS, and this in turn could contribute to brain aging and/or pathological conditions associated with neurodegenerative diseases. Additionally, increased nitrosative and oxidative stress can elicit dysfunction of mitochondrial dynamics (Barsoum et al., 2006; Bossy-Wetzel and Lipton, 2003; Yuan et al., 2007). However, until recently little was known regarding the molecular and pathogenic mechanisms by which NO contributes to the formation of fragmented mitochondria. Our recent findings have shed light on the molecular events underlying this relationship, particularly in AD. Specifically, we recently discovered physiological and chemical evidence that S-nitrosylation modulates the GTPase activity of Drp1, thus contributing to mitochondrial fragmentation, bioenergetic impairment, synaptic damage, and eventually frank neuronal loss in cell-based models of AD.
4. S-Nitrosylation of Drp1 results in excessive mitochondrial fission in AD
In addition to rare hereditary mutations seen in the genes encoding mitochondrial fission and fusion proteins, recent studies have demonstrated that posttranslational modification of these molecules can contribute to altered mitochondrial dynamics. For example, phosphorylation, ubiquitination, sumoylation, and proteolytic cleavage of Drp1 regulate mitochondrial fission by affecting Drp1 activity, at least in cell culture systems (Breckenridge et al., 2008; Chang and Blackstone, 2007; Cribbs and Strack, 2007; Karbowski et al., 2007; Nakamura et al., 2006; Taguchi et al., 2007; Wasiak et al., 2007; Yonashiro et al., 2006). We therefore posited that excessive activation of mitochondrial fission or fusion proteins by posttranslational modification could contribute to neurodegeneration by compromising mitochondrial function. Interestingly, along these lines, we recently reported that NO can also lead to S-nitrosylation of Drp1 at Cys644 and excessive activation of its fission activity (Cho et al., 2009). Drp1 includes four distinct structural domains: an N-terminal GTPase domain, a dynamin-like middle domain, an insert B domain, and a C-terminal GED domain. Cys644 resides within the GTPase effector domain (GED) of Drp1, which influences both GTPase activity and oligomer formation of Drp1 (Low and Lowe, 2006; Low et al., 2009; Pitts et al., 2004; Ramachandran et al., 2007; Zhu et al., 2004). S-Nitrosylation of Drp1 (forming SNO-Drp1) induces formation of Drp1 dimers, which function as building blocks for tetramers and higher order structures of Drp1, and stimulates Drp1 GTPase activity. In contrast, we found that substitution of Cys644 for an Ala [Drp1(C644A)] abrogated these effects of NO.
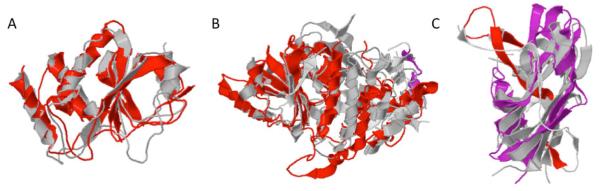
Recently, another group of scientists has raised some concerns regarding this work, in particular, over our homology modeling of Drp1, the formation of dimers or higher order structures of Drp1, the ability of S-nitrosylation to increase Drp1 GTPase activity, and the effect of SNO-Drp1 on mitochondrial fragmentation (Bossy et al., 2010). In response to these questions, in our report we constructed an atomic model of Drp1 to attempt to offer a structural interpretation for our empirical findings. Our homology modeling rests upon the assumption that the three dimensional structure of human Drp1 is similar to the structure of bacterial dynamin-like protein (PDB ID: 2J68-A). Although the overall homology is distant, the reliability of the model was supported by statistically significant sequence similarities. We would like to emphasize that low primary sequence similarity by itself does not prove that the structures must be different. In fact, there are ample examples showing that at this level of homology, three-dimensional protein structures can look remarkably similar (see Fig. 2) (Li, Z., Bakolitsa, C., Jaroszewski. L. and Godzik, A., unpublished data), although examples to the contrary can also be found. Therefore, while homology between human Drp1 and the bacterial dynamin-like protein is strongly supported and is essentially unquestionable (E-value of 10−15), accuracy of specific features of the model is difficult to access. Hence, specific predictions have to be verified by experiments. For example, in this case we found that one of the predictions of our model, namely that the solvent exposure of Cys644 is located on the surface of the molecule, fits well with our experimental data. Another prediction is that the C-terminal GED domain of Drp1, recognized and defined solely on the basis of sequence conservation, does not form a proper domain but instead is intertwined with the middle domain, and therefore probably regulates GTPase activity only indirectly. This prediction is also consistent with our experimental data, and in fact resembles the role that the C-terminal domain plays in other, even more distant members of the dynamin family, such as human guanylate-binding protein 1 (Prakash et al., 2000). This dynamin homologue was previously proposed to model the relationship between the GTPase and GED-like domains (Praefcke and McMahon, 2004).
Fig. 2.
Examples of proteins showing conserved three-dimensional structures despite extreme sequence divergence and evolutionary distance. (A) Crystal structures of putative S. Putrefaciens phosphatase (3GXG; grey) aligned with H. sapiens dual specificity protein phosphatase 23 (2IMG; red). The two proteins show 2.8 Å root mean square deviation (RMSD) over a region of 138 amino-acids residues with 10% sequence identity. (B) Crystal structures of E. coli lactaldehyde reductase (1RRM; grey) aligned with A. Nidulans dehydroquinate synthase (1SG6; red). The two proteins show 2.9 Å RMSD over a region of 295 amino acids with 9% sequence identity. (C) Crystal structures of B. thetaiotamicron BT_1233 (3GF6; grey) aligned with H. sapiens Ectodysplasin A (1RJ7; red and magenta). The two proteins show 3.1 Å RMSD over a region of 123 amino acids with 4% sequence identity. All structural alignments were prepared with the flexible structural alignment program FATCAT (http://fatcat.burnham.org/fatcat/).
Dynamin and Drp1 have both been shown to dimerize and form higher-order structures upon activation (Ingerman et al., 2005; Ramachandran et al., 2007; Zhang and Hinshaw, 2001). For instance, extensive published work has demonstrated that dimeric yeast Drp1 functions as a building block for higher order structures (Ingerman et al., 2005). Further along these lines, Schmid and colleagues proposed that the related molecule, dynamin, forms a tetramer that actually represents a ‘dimer of dimers’ (Ramachandran et al., 2007). Thus, the observation presented in our original report is consistent with previously published structural models. In fact, using computer graphics manipulations, we could fit our predicted atomic structure of Drp1 by superimposition onto the published low-resolution cryo-electron micrographs of the dynamin dimer (Zhang and Hinshaw, 2001).
Based on this homology model, we investigated whether S-nitrosylation of Drp1 at Cys644 alters its GTPase activity. In line with the model, we showed empirically that SNO-Drp1 triggers increased GTPase activity, thus causing excessive mitochondrial fragmentation, bioenergetic compromise, and synaptic damage in AD (Cho et al., 2009). Concerning the GTP hydrolysis activity assay of Drp1, it is critical when performing these experiments to use chemically reduced Drp1 (the physiological form of the protein) and not to mistakenly use Drp1 that has already been oxidized, and is thus already maximally dimerized. Artifactual oxidation can occur because of the experimental conditions, for example, if recombinant protein is prepared in ambient air, representing an oxidizing condition, and this artifactual oxidation is apparently why the Bossy-Wetzel group had difficulty observing the effect of NO on Drp1 activity. Claims by Bossy et al. (2010) that Drp1 is not predominantly oxidized based on the fact that the protein can form SNO-Drp1 by biotin switch assay are fallacious. Chemically- speaking, redox reactions rarely go to completion, and the standard biotin-switch assay is not quantitative. Hence, even if the vast majority of Drp1 were already oxidized (as evidenced in Bossy et al.’s blots showing Drp1 oligomer formation), a small fraction of remaining reduced Drp1 could still be S-nitrosylated. Importantly, if the majority of cysteine residue(s) of Drp1 are artifactually oxidized in this manner, it would prevent S-nitrosylation of these thiol groups, which itself represents an oxidation reaction. Since the activity of Drp1 GTPase is physiologically regulated by redox state, artifactual oxidation by preparing the protein in ambient air or other oxidizing conditions could artificially increase enzyme activity to maximal levels, thereby preventing an S-nitrosylation—mediated increase in Drp1 dimer formation and activation. In fact, as we previously showed (Cho et al., 2009), Drp1 normally exists as a monomer under basal physiological conditions, and S-nitrosylation stimulates Drp1 dimerization and increased GTPase activity.
Additionally, in experiments analyzing the effect of S-nitrosylation on Drp1, it is critical to employ an assay that directly monitors GTPase activity. If, for instance, a Drp1 GTPase assay is utilized in which GTP substrate is continuously regenerated from GDP, as the Bossy-Wetzel group used, potential problems can arise. One problem with this approach is that the other enzymes present in the assay system, which are used to regenerate GTP, i.e., pyruvate kinase and lactate dehydrogenase, can also be S-nitrosylated (Gao et al., 2005; Hao et al., 2006; Paige et al., 2008). This fact can totally obfuscate any attempt to determine the specific effect of S-nitrosylation on Drp1 activity. For example, one cannot tell if S-nitrosylation of multiple enzymes offsets the effects of SNO-Drp1. To avoid such confusion, we measured the release of inorganic phosphate (Pi) catalyzed by Drp1 from a physiologically-relevant concentration of GTP (0.5 mM) (Traut, 1994); in this manner we were able to directly monitor the GTPase activity of Drp1. Recently, in corroboration of our findings, two other laboratories independently found that S-nitrosylation increases GTPase activity of both dynamin 1 and 2, close homologues of Drp1 (Kang-Decker et al., 2007; Wang et al., 2006). Notably, similar to our approach, these two other groups utilized unoxidized enzyme and chose a direct GTPase activity assay in order to eliminate confounding factors. Under these conditions, both groups replicated our results, whereas others using oxidized Drp1 or regenerating GTPase assays have failed to do so.
In our original report, we further demonstrated that exposure to oligomeric Aβ peptide results in formation of SNO-Drp1 in cell culture models. Moreover, we and others have observed that Drp1 is S-nitrosylated in the brains of virtually all cases of sporadic AD (Cho et al., 2009; Wang et al., 2009b). In order to determine the consequences of S-nitrosylated Drp1 in neurons, we exposed cultured cerebrocortical neurons to the physiological NO donor, SNOC, or to Aβ oligomers and found that both induced SNO-Drp1 formation and led to the accumulation of fragmented mitochondria. Moreover, mutation of a specific cysteine residue in Drp1 (C644A) prevented these effects of SNOC or Aβ on mitochondrial fragmentation, consistent with the notion that SNO-Drp1 triggered excessive mitochondria fission or fragmentation. Finally, in response to Aβ, we found that SNO-Drp1—induced mitochondrial fragmentation caused synaptic damage, an early characteristic feature of AD, and eventually apoptotic neuronal cell death (Fig. 1). Importantly, blockade of Drp1 nitrosylation (using the Drp1(C644A) mutant) prevented Aβ-mediated synaptic loss and neuronal cell death, suggesting that SNO-Drp1 may represent a potential therapeutic target to protect neurons and their synapses in AD.
5. Potential implication of S-nitrosylated Drp1 in other neurodegenerative diseases
In addition to AD, mitochondrial dysfunction and nitrosative/oxidative stress have long been implicated in the pathogenesis of Parkinson’s disease (PD) and Huntington’s disease (HD). For instance, mitochondrial respiratory electron transport chain NADPH dehydrogenase (Complex I) activity is reduced in the substantia nigra of PD patients, and complex I inhibitors, such as rotenone, MPP+ and pesticides, result in the production of ROS/RNS and subsequent neuropathological changes similar to PD (Schapira et al., 2006). Not only are levels of multiple mitochondrial proteins altered in postmortem samples of PD brains, but also PD-linked genetic mutations in PINK1, Parkin and DJ-1, have been identified, suggesting that mitochondrial dynamics may be altered (Abou-Sleiman et al., 2006; Jin et al., 2007). Recent evidence indeed suggests that abnormal mitochondrial dynamics may contribute to neuronal injury and death in animal models of PD. For example, both rotenone and 6-hydroxydopamine, have been shown to induce Drp1-dependent mitochondrial fragmentation as well as oxidative stress (Barsoum et al., 2006; Gomez-Lazaro et al., 2008). Additionally, loss of function of PINK1 or Parkin leads to mitochondrial fragmentation, which is associated with enhanced mitophagy (Dagda et al., 2009; Exner et al., 2007; Lutz et al., 2009). Moreover, fibroblasts carrying PINK1 mutations from PD patients (Q456X nonsense or V170G missense) also exhibit more fragmented mitochondrial networks (Grunewald et al., 2009). In contrast, the normal PINK1/Parkin pathway appears to promote mitochondrial fission and/or inhibits mitochondrial fusion in Drosophila (Deng et al., 2008; Poole et al., 2008; Yang et al., 2006). While further studies will be needed to fully understand the implications of these findings, it is clear that several PD-associated gene products are related to mitochondrial dynamics and that abnormal mitochondrial morphology is associated with PD pathology. Nonetheless, we were unsuccessful in observing increased SNO-Drp1 formation in both cortex and substantia nigra of PD patients (Cho et al., 2009 and unpublished data). This finding may suggest that formation of SNO-Drp1 plays a role in PD pathogenesis only in the early stages of the disease, if at all. Additional studies with cell-based and animal models of PD will be needed to address these questions.
Unlike AD and PD, HD is a purely genetic disease caused by mutations that result in CAG expansion in the first exon of the huntingtin gene (Htt), with more than ~35 CAGs being pathogenic and resulting in polyglutamine expression. Mitochondrial dysfunction is also associated with pathogenesis of HD (Reddy et al., 2009). Respiratory electron transport chain activity and ATP levels are decreased in mitochondria from HD patients and Htt transgenic mice (Pandey et al., 2008; Seong et al., 2005; Tabrizi et al., 1999). Additionally, the complex II inhibitor, 3-nitropropionic acid (3-NP), causes a movement disorder similar in many respects to HD (Brouillet et al., 1995). Moreover, mutant Htt directly impairs mitochondrial membrane potential, calcium homeostasis, and mitochondrial axonal trafficking (Chang et al., 2006; Panov et al., 2002). Recently, Wang et al. demonstrated that expression of mutant Htt sensitizes cells to oxidative stress-induced mitochondrial fission and reduces ATP levels by inhibiting mitochondrial fusion (Wang et al., 2009a). Overexpression of the Drp1 dominant negative K38A or the fusion protein Mfn2 reduces mutant Htt-induced mitochondrial fragmentation as well ATP loss and cell death. Importantly, RNAi against Drp1 also reduces the motility defect in a worm model of HD. In addition, 3-NP exposure induces increased mitochondrial fragmentation in an NMDA receptor-dependent manner in cerebrocortical neurons (Liot et al., 2009). Remarkably, we have also observed S-nitrosylation of Drp1 in HD brains similar to that seen in AD brains, raising the possibility that this redox event may play a pathogenic role in HD in addition to AD. Collectively, these studies suggest that alterations in mitochondrial dynamics may be involved in the pathogenesis of HD.
Acknowledgments
We thank Rongsheng Jin for critical discussions on the structure of Drp1. This work was supported in part by NIH grants P01 HD29587, R01 EY05477, R01 EY09024, P01 ES016738, and P30 NS057096 (S.A.L.).
Abbreviations
- Aβ
β-amyloid
- AD
Alzheimer’s disease
- ADOA
Autosomal Dominant Optic Atrophy
- CMT
Charcot-Marie-Tooth
- Drp1
dynamin-related protein 1
- GED
GTPase effector domain
- HD
Huntington’s disease
- Mfn
Mitofusin
- NMDA
N-methyl-D-aspartate
- NO
nitric oxide
- NOS
nitric oxide synthase
- PD
Parkinson’s disease
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Experimental models of Parkinson’s disease. Nat Rev Neurosci. 2001;2:325–334. doi: 10.1038/35072550. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy B, Petrilli A, Klinglmayr E, Chen J, Lütz-Meindl U, Knott AB, Masliah E, Schwarzenbacher R, Bossy-Wetzel E. S-Nitrosylation of DRP1 Does Not Affect Enzymatic Activity and is Not Specific to Alzheimer’s Disease. J Alzheimers Dis. 2010 doi: 10.3233/JAD-2010-100552. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr Opin Cell Biol. 2003;15:706–716. doi: 10.1016/j.ceb.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Lipton SA. Nitric oxide signaling regulates mitochondrial number and function. Cell Death Differ. 2003;10:757–760. doi: 10.1038/sj.cdd.4401244. [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Kang BH, Kokel D, Mitani S, Staehelin LA, Xue D. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Brouillet E, Hantraye P, Ferrante RJ, Dolan R, Leroy-Willig A, Kowall NW, Beal MF. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc Natl Acad Sci USA. 1995;92:7105–7109. doi: 10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006;18:453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerit J, Edeas M, Bricaire F. Neurodegenerative diseases and oxidative stress. Biomed Pharmacother. 2004;58:39–46. doi: 10.1016/j.biopha.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Guo H, Wei J, Mi Z, Wai PY, Kuo PC. Identification of S-nitrosylated proteins in endotoxin-stimulated RAW264.7 murine macrophages. Nitric Oxide. 2005;12:121–126. doi: 10.1016/j.niox.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Garthwaite J, Charles SL, Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Lazaro M, Bonekamp NA, Galindo MF, Jordan J, Schrader M. 6-Hydroxydopamine (6-OHDA) induces Drp1-dependent mitochondrial fragmentation in SH-SY5Y cells. Free Radic Biol Med. 2008;44:1960–1969. doi: 10.1016/j.freeradbiomed.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Grunewald A, Gegg ME, Taanman JW, King RH, Kock N, Klein C, Schapira AH. Differential effects of PINK1 nonsense and missense mutations on mitochondrial function and morphology. Exp Neurol. 2009;219:266–273. doi: 10.1016/j.expneurol.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amylo id beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J. Identification of novel proteins associated with both alpha-synuclein and DJ-1. Mol Cell Proteomics. 2007;6:845–859. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- Kang-Decker N, Cao S, Chatterjee S, Yao J, Egan LJ, Semela D, Mukhopadhyay D, Shah V. Nitric oxide promotes endothelial cell survival signaling through S-nitrosylation and activation of dynamin-2. J Cell Sci. 2007;120:492–501. doi: 10.1242/jcs.03361. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, Ogawa M, Ishizaki Y, Kitamura T, Shozawa Y, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Cazal M, Culcasi M, Gaven F, Pietri S, Bockaert J. NMDA-dependent superoxide production and neurotoxicity. Nature. 1993;364:535–537. doi: 10.1038/364535a0. [DOI] [PubMed] [Google Scholar]
- Li H, Chen Y, Jones AF, Sanger RH, Collis LP, Flannery R, McNay EC, Yu T, Schwarzenbacher R, Bossy B, et al. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2008;105:2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liot G, Bossy B, Lubitz S, Kushnareva Y, Sejbuk N, Bossy-Wetzel E. Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell Death Differ. 2009;16:899–909. doi: 10.1038/cdd.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Laemmermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, et al. Loss of parkin or PINK1 function increases DRP1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos M, Augusto E, Oliveira CR, Agostinho P. Amyloid-beta peptide decreases glutamate uptake in cultured astrocytes: involvement of oxidative stress and mitogen-activated protein kinase cascades. Neuroscience. 2008;156:898–910. doi: 10.1016/j.neuroscience.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Medeiros R, Prediger RD, Passos GF, Pandolfo P, Duarte FS, Franco JL, Dafre AL, Di Giunta G, Figueiredo CP, Takahashi RN, et al. Connecting TNF-alpha signaling pathways to iNOS expression in a mouse model of Alzheimer’s disease: relevance for the behavioral and synaptic deficits induced by amyloid beta protein. J Neurosci. 2007;27:5394–5404. doi: 10.1523/JNEUROSCI.5047-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron. 2002;35:9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M, Varghese M, Sindhu KM, Sreetama S, Navneet AK, Mohanakumar KP, Usha R. Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3-nitropropionic acid-induced rat model of Huntington’s disease. J Neurochem. 2008;104:420–434. doi: 10.1111/j.1471-4159.2007.04996.x. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Parker WD, Jr., Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer’s disease brain. Neu rology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- Pitts KR, McNiven MA, Yoon Y. Mitochondria-specific function of the dynamin family protein DLP1 is mediated by its C-terminal domains. J Biol Chem. 2004;279:50286–50294. doi: 10.1074/jbc.M405531200. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- Quintanilla RA, Matthews-Roberson TA, Dolan PJ, Johnson GV. Caspase-cleaved tau expression results in mitochondrial dysfunction in cortical neurons. Implications for the pathogenesis of Alzheimer disease. J Biol Chem. 2009;284:18754–18766. doi: 10.1074/jbc.M808908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH. Mitochondrial dysfunction in aging and Alzheimer’s disease: strategies to protect neurons. Antioxid Redox Signal. 2007;9:1647–1658. doi: 10.1089/ars.2007.1754. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Mao P, Manczak M. Mitochondrial structural and functional dynamics in Huntington’s disease. Brain Res Rev. 2009;61:33–48. doi: 10.1016/j.brainresrev.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Bezard E, Brotchie J, Calon F, Collingridge GL, Ferger B, Hengerer B, Hirsch E, Jenner P, Le Novere N, et al. Novel pharmacological targets for the treatment of Parkinson’s disease. Nat Rev Drug Discov. 2006;5:845–854. doi: 10.1038/nrd2087. [DOI] [PubMed] [Google Scholar]
- Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- Tabrizi SJ, Cleeter MW, Xuereb J, Taanman JW, Cooper JM, Schapira AH. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Trotti D, Danbolt NC, Volterra A. Glutamate transporters are oxidant-vulnerable: a molecular link between oxidative and excitotoxic neurodegeneration? Trends Pharmacol Sci. 1998;19:328–334. doi: 10.1016/s0165-6147(98)01230-9. [DOI] [PubMed] [Google Scholar]
- Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci USA. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009a;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Rowan MJ, Anwyl R. Beta-amyloid-mediated inhibition of NMDA receptor-dependent long-term potentiation induction involves activation of microglia and stimulation of inducible nitric oxide synthase and superoxide. J Neurosci. 2004;24:6049–6056. doi: 10.1523/JNEUROSCI.0233-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci. 2009b;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Neurochem. 2009c;109(Suppl 1):153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- White JA, Manelli AM, Holmberg KH, Van Eldik LJ, Ladu MJ. Differential effects of oligomeric and fibrillar amyloid-beta 1-42 on astrocyte-mediated inflammation. Neurobiol Dis. 2005;18:459–465. doi: 10.1016/j.nbd.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- Yuan H, Gerencser AA, Liot G, Lipton SA, Ellisman M, Perkins GA, Bossy-Wetzel E. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–471. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]