Abstract
Purpose
Endometrial cancer (EC) is the most common gynecologic malignancy. Type I EC has a favorable prognosis, while type II ECs account for half of all treatment failures. Little knowledge of the biological differences is available to predict EC outcomes besides their pathological distinctions. MicroRNAs (miRNA) are a family of non-translated RNAs important in regulating oncogenic pathways. Mis-expression patterns of miRNAs in EC, as well as differences in miRNA expression patterns between the subtypes of EC has not been previously evaluated. Our purpose was to identify miRNA profiles of EC subtypes, and to identify miRNAs associated with these subtypes to ultimately understand the different biological behavior between these subtypes.
Methods
95 fresh/frozen and paraffin embedded samples of endometrial type I and II cancer, carcinosarcomas and benign endometrial samples were collected. MiRNA expression profiles were evaluated by microarray analysis. Statistical analysis was performed.
Results
Distinct miRNA signatures in tumor versus normal samples and in endometrioid vs. uterine papillary serous carcinomas exist. Additionally, carcinosarcomas have a unique miRNA signature from either the type I or II epithelial tumors.
Conclusions
We hypothesize that further understanding the miRNAs that separate these subtypes of EC will lead to biological insights into the different behavior of these tumors.
Keywords: microRNAs, endometrial cancer, carcinosarcoma, uterine papillary serous carcinoma
Introduction
Cancer of the uterine corpus is the most common gynecologic malignancy and the fourth most common cancer in women. (1) The American Cancer Society estimates that 40,100 women will be diagnosed with endometrial cancer in the United States in 2009 – and 7,470 of these women will die from their disease. (2) However, endometrial carcinoma is a varied disease with five-year survival rates for localized, regional, and metastatic disease reported to be 95, 67, and 23 percent, respectively. (3) The disparity in overall patient survival is clarified by classification of endometrial carcinomas into two types of tumors carrying distinctly different characterization and prognosis. (4) Type I cancers, which are estrogen related, occur mainly in perimenopausal and obese patients, are usually low stage and low grade (frequently occurring in the background of hyperplasia) and have an excellent prognosis. (4) Type II endometrial carcinomas tend to spread aggressively and have a poor prognosis. They are unrelated to estrogen stimulation and occur in older non-obese women. Women with type II endometrial cancer have adverse histologic features, including poorly differentiated Grade 3 tumors, papillary serous and clear cell tumors. The mean age of Type II tumors is 68 years and the overall 5-year survival is only 46%. (4) Uterine papillary serous carcinomas carry a particularly poor prognosis, with extrauterine spread found in up to 72% of patients at diagnosis.
Carcinomas account for 95% of uterine malignancies and arise from the epithelial layer of the uterus. The prevalence of pathological subtype of this tissue is reported to be: adenocarcinoma as 89 percent, uterine papillary serous carcinomas as 6 percent and clear cell tumors as 5 percent. (9) (10) The remaining 6 percent of uterine cancers are sarcomas (consisting of leiomyosarcomas and endometrial stromal sarcomas) and carcinosarcomas. Carcinosarcomas have historically been classified as sarcomas, however, recent nomenclature categorizes these tumors as carcinomas. Carcinosarcomas carry a very poor prognosis with the five-year survival of 25 to 35 percent. (11) In these cancers malignant epithelial and stromal components contribute to the architecture of the tumor. The carcinomatous element is usually grade 3 endometrioid, clear cell or papillary serous histology. Many investigators have attempted to determine if these tumors represent collision tumors (made of 2 genetically distinct cell populations) or combination tumors (both cell types arise from a common progenitor cell that is capable of multlineage differentiation). (12) Immunohistochemical studies support the later, that precursor (stem) cells give rise to both components during the histogenesis of the tumor. (13) Data confirms that the carcinomatous element is the predominant element and that the sarcomatous component is derived from the metaplasia or from a stem cell that undergoes divergent differentiation. (14) Based on these findings, uterine carcinosarcomas are now classified as a type of non-endometrioid endometrial cancer rather than a uterine sarcoma by most recent treatment guidelines from the National Comprehensive Cancer Network. However, treatment of these tumors is still debated, with some endorsing chemotherapy appropriate for the high-grade epithelial component while others advocating sarcoma based adjuvant treatment. (15)
Varying risk factors and prognosis between the different subtypes of uterine cancer suggest that they harbor distinct molecular alterations, some of which have been previously delineated through single gene analysis. Mutations of the p53 gene have been found in up to 90% of epithelial tumors that are grade 3 or papillary serous carcinoma but are absent in grade one type I tumors. (16) The presence of p53 overexpression and high S phase fraction increases the risk of recurrence seven-fold, and the risk of cancer-related death almost 10-fold when compared to tumors with neither factor. (17) In a multivariate analysis p53 was identified as the strongest predictor of survival. (18) In contrast, PTEN, a tumor suppressor gene on chromosome 10, is often mutated or deleted and is associated with endometrioid histology and a favorable prognosis. (19) Other altered oncogene/tumor suppressor gene expression patterns have been demonstrated in endometrial cancer; MDR-1 and ER/PR positivity have been reported to be favorable prognostic factors, while microsatellite instability, HER2/neu receptor positivity, Ki 67, PCNA and EGF-R over-expression have been shown to carry an unfavorable prognosis. (20-25) Expression of the Her-2/neu gene has been shown to be present in 27 percent of women with metastatic disease compared to 4 percent of patients where disease is limited to the uterus. (26)
Although the above findings reflect important molecular insights into uterine cancer, a better and more global understanding is necessary to both identify new targets for therapy and to better predict an individual's outcome. MicroRNAs (miRNAs) are a class of 22-nucleotide noncoding RNAs, which are evolutionarily conserved and function by negatively regulating gene expression at the post-transcriptional level. MiRNAs are global regulatory RNAs that each control hundreds of mRNA transcripts. Recent studies have shown that miRNAs are aberrantly expressed in virtually all human cancer types (27) and that specific miRNAs misregulated in each cancer type may act as biomarkers of outcome for that cancer type. (28) The miRNA signatures of uterine cancer or specifically uterine cancer subtypes has not been previously explored, prompting the current investigation.
By miRNA microarray we were able to identify unique miRNA signatures that could separate type I (endometrioid) from type II (papillary serous) uterine cancers. Furthermore, we found that carcinosarcomas have a distinct miRNA signature that is unique from epithelial uterine cancer miRNA signatures, adding further credence to the belief that they are biologically unique tumors.
Materials and Methods
Fresh/Frozen Tissue Collection
After approval from the Human Investigation Committee at Yale University, uterine tumor samples and normal endometrial tissues were obtained from untreated patients undergoing surgery at Yale-New Haven Hospital (New Haven, CT). All patients underwent staging surgery as initial treatment. Patient data was collected including age, race, parity and risk factors. All tumors were from primary sites. The carcinoma samples were histologically examined for the presence of tumor. Specimens were immediately snap-frozen and stored at -80 C. The fresh/frozen tissue collection used for microarray analysis included five benign endometrial tissues, eleven endometrioid adenocarcinomas, six papillary serous tumors and six carcinosarcomas. All were examined microscopically and microdissected to ensure greater then 75% tumor cellularity.
Paraffin Embedded Uterine Tumors
For addition tumor specimens paraffin embedded tumors (FFPE) were microdissected and used for microarray analysis. In all cases sections of tumor used had greater then 75% tumor cellularity. Twenty-one papillary serous tumors from Yale were identified, microdissected, analyzed by microarray and included in the analysis. Forty-six endometrioid adenocarcinomas from RTOG (Radiation Therapy Oncology Group) trials 9708 and 9905 were microdissected, analyzed by microarray and used in the analysis. There was no difference in miRNA signatures identified between fresh/frozen and FFPE tissues in these analyses (data not shown).
RNA Extraction
Total RNA isolation, including small RNAs, was performed with the mirVana RNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions for all fresh frozen tissue. Each sample was derived from a single specimen. Integrity of the RNA was assessed using Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). RNA was extracted from paraffin-embedded slides using Trizol, per protocol.
MiRNA Profiling
cDNA was synthesized from 160ng-800 ng of total RNA using TaqMan MiRNA primers and the TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems). Expression of 384 mature miRNAs was then analyzed with the Asuragen TLDA assay and the Applied Biosystems 7900 Taqman Real-Time PCR machine in accordance with manufacturer's instructions. Fold changes in miRNA expression between benign and malignant samples as well as between different malignant subtypes were determined by delta-delta CT values. Normalization was done to two internal small RNA controls RNU44 and RNU48. In the majority of samples 102 miRNAs were detected from the 384 measured, and a CT cutoff of 34 was used in all of the samples. To confirm data the first 12 samples were run in duplicate, and all were statistically similar in results.
Statistical Analysis
All normalization and data analyses were performed in the statistical programming environment R (29) using customized functions and functions available from Bioconductor (30) and the limma software package. We normalized the sample input CT values for each miRNA by quantitating small nuclear RNAs using the TaqMan(R) MiRNA Assay Controls (Applied Biosystems). Each of the 8 pools are normalized separately by the associated small nuclear RNAs. The intensities are scaled to have similar distributions across the entire series of samples to have the same median absolute deviation across samples. The miRNA expression data for different tumor types was analyzed together by using linear modeling methods. (31) The linear models allowed for general changes in gene expression between different conditions and across different biological replicates. Assessment of differential expression was assessed using a moderated t-statistic. P values were adjusted for multiple testing based on all the miRNAs which were expressed in samples (excluding control and unexpressed miRNAs) according to the method of Benjamini and Hochberg (32) to control the false discovery rate. Hierarchical clustering was performed with Pearson correlation and average linkage, based on miRNAs selected for differential expression between any of the groups of interest.
Patient Characteristics
Table 1 describes the clinicopathologic parameters of the study population. Pathologic examination confirmed malignancies in 90 patients while 5 patients had no malignancy and represent the benign cases.
Table 1. Patient characteristics.
| Clinicopathologic Parameters (n=95) | |
| Pathology: | |
| Malignant | 90 |
| Endometrioid | 57 |
| Uterine Papillary Serous Carcinoma | 27 |
| Carcinosarcoma (MMMT) | 6 |
| Benign | 5 |
| Age: | |
| Malignant | |
| Endometrioid | 60 (36-82) |
| Uterine Papillary Serous Carcinoma | 70 (55-90) |
| Carcinosarcoma (MMMT) | 62 (48-75) |
| Benign | 53 (45-63) |
| Ethnicity: | |
| Malignant | |
| Endometrioid | |
| Caucasian | 46 |
| African American | 8 |
| Hispanic | 3 |
| Uterine Papillary Serous Carcinoma | |
| Caucasian | 18 |
| African American | 8 |
| Unknown | 1 |
| Carcinosarcoma (MMMT) | 6 |
| Caucasian | 5 |
| African American | 1 |
| Benign | |
| Caucasian | 2 |
| African American | 1 |
| Hispanic | 2 |
| FIGO stage: | |
| Endometrioid carcinoma | |
| Stage I | 27 |
| Stage II | 12 |
| Stage III | 18 |
| Uterine Papillary Serous carcinoma | |
| Stage I | 8 |
| Stage II | 5 |
| Stage III | 7 |
| Stage IV | 7 |
| Carcinosarcomas | |
| Stage I | 4 |
| Stage III | 2 |
Results
A miRNA expression signature discriminates type I endometrial cancer from benign endometrium
When the expression of miRNAs was compared between endometrioid endometrial cancer samples and normal endometrial benign tissues, 10 of the 384 miRNAs showed significantly differential expression. Several miRNAs were significantly up-regulated (with FDR < 0.03) in endometrial carcinoma samples, while two miRNAs were down-regulated (Table 2, Supplemental Figure 1). Among the top differentially expressed miRNAs, miR-205, miR-182 and miR-200a are most up regulated in endometrioid samples while mir-411 was most down-regulated in cancerous samples compared to benign (Table 2). There was no significant difference between Grade 1 and 3 endometrioid tumors in this analysis, so they were grouped together.
Table 2. Type 1 endometrioid uterine carcinoma miRNA signatures compared to benign endometrium miRNA signatures.
| Upregulated | Fold Change | Nominal P Value | Adjusted P. Value (FDR) |
|---|---|---|---|
| miR-650 | 4.8 | 6.3E-05 | 0.0065 |
| miR-183 | 5.3 | 1.2E-04 | 0.0065 |
| miR-572 | 4.5 | 1.5E-04 | 0.0065 |
| miR-200a | 5.4 | 1.7E-04 | 0.0065 |
| miR-182 | 6.2 | 4.7E-04 | 0.0111 |
| miR-622 | 4.8 | 5.0E-04 | 0.0111 |
| miR-34a | 3.7 | 1.7E-03 | 0.0301 |
| miR-205 | 6.7 | 1.9E-03 | 0.0301 |
| Downregulated | |||
| miR-411 | -3.8 | 4.8E-03 | 0.0111 |
| miR-487b | -2.7 | 1.9E-03 | 0.0301 |
A miRNA expression signature discriminates between Type I (endometrioid) from Type II (uterine papillary serous) cancers
We next compared miRNA expression patterns between endometrioid and papillary serous tumors (UPSC). MiRNA expression patterns were also distinct between these carcinomas (Supplemental Figure 2). Eight miRNAs were significantly lower in endometrioid tumors compared to UPSC tumors (with FDR < 0.025) (Table 3). The most down-regulated miRNAs in endometrioid tumors compared to UPSC included miR-19a and miR-19b.
Table 3. Endometrioid carcinoma miRNA signatures compared to UPSC carcinoma miRNA signatures.
| Downregulated | Fold Change | Nominal P Value | Adjusted P. Value (FDR) |
|---|---|---|---|
| miR-19a | -5.1 | 7.6E-08 | 1.2E-05 |
| miR-19b | -4.2 | 2.0E-06 | 1.5E-04 |
| miR-30e-5p | -3.8 | 7.2E-06 | 3.7E-04 |
| miR-101 | -3.8 | 1.6E-05 | 6.1E-04 |
| miR-452 | -3.9 | 2.5E-04 | 6.5E-03 |
| miR-15a | -3.3 | 7.2E-04 | 1.3E-02 |
| miR-29c | -3.7 | 1.1E-03 | 1.6E-02 |
| miR-382 | -3.8 | 1.2E-03 | 1.6E-02 |
A miRNA expression signature discriminates between uterine carcinomas and uterine carcinosarcomas
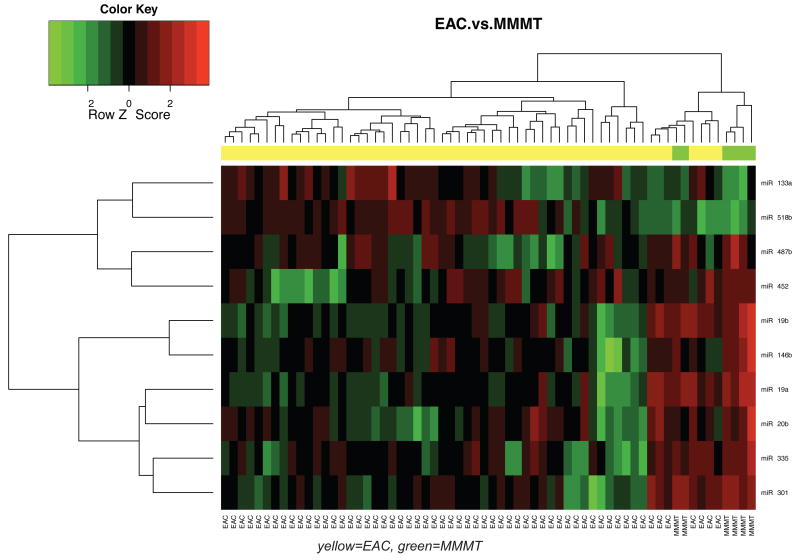
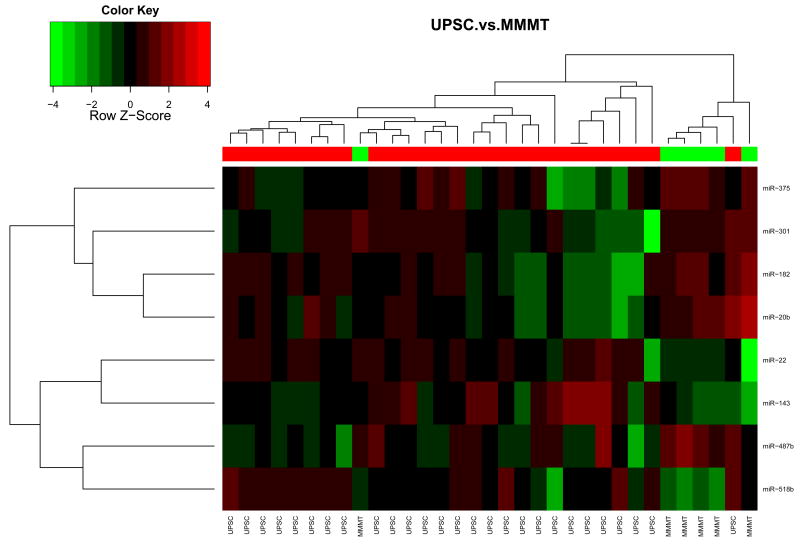
We compared miRNA signatures between carcinosarcomas and the epithelial type uterine tumors. We found that carcinosarcomas have a unique miRNA signature that is unlike either endometrioid (Figure 1A) or uterine papillary serous tumors (Figure 1B). Some unique miRNAs differentiate carcinosarcomas from type I endometrioid tumors and others from UPSC tumors. In endometrioid tumors compared to carcinosarcomas specifically, miR-133a is upregulated in endometrioid tumors, while miR-19a and miR-19b are down-regulated (Table 4). When comparing UPSC to carcinosarcomas, miR-22 is upregulated in UPSCs while miR-182 is down-regulated with a FDR < 0.05. (Table 5). Interestingly, there were also miRNAs that were similarly misregulated in endometrioid and UPSC tumors compared to carcinosarcomas: miR-518b was upregulated and miR-301, miR-20b and miR-487b were down-regulated. These miRNAs may have a specific role in carcinosarcomas compared to carcinomas of the uterus, and may warrant further investigation.
Figure 1. Comparison of miRNA expression patterns in endometrioid carcinoma, UPSC and carcinosarcoma.
A. A heat map of 6 Carcinosarcomas and 57 endometrioid endometrial carcinoma samples indicates that there is a difference in miRNA expression between the two groups. B. Comparison of 6 Carcinosarcomas and 27 UPSC show that there is a difference in miRNA expression between these groups. MicroRNA expression is displayed as higher (red) or lower (green).
Table 4. Endometrioid miRNA signatures compared to carcinosarcoma miRNA signatures.
| Upregulated | Fold Change | Nominal P Value | Adjusted P. Value (FDR) |
|---|---|---|---|
| miR-518b | 2.4 | 2.6E-05 | 1.4E-03 |
| miR-133a | 2.5 | 3.6E-04 | 8.3E-03 |
| Downregulated | |||
| miR-19a | -7.0 | 1.4E-07 | 2.2E-05 |
| miR-19b | -5.7 | 2.8E-06 | 2.2E-04 |
| miR-301 | -6.2 | 4.9E-05 | 1.9E-03 |
| miR-146b | -4.6 | 1.4E-04 | 4.3E-03 |
| miR-335 | -5.1 | 3.8E-04 | 8.3E-03 |
| miR-487b | -4.8 | 8.0E-04 | 1.6E-02 |
| miR-20b | -5.2 | 1.2E-03 | 2.0E-02 |
| miR-452 | -4.8 | 2.7E-03 | 4.0E-02 |
Table 5. UPSC miRNA signatures compared to carcinosarcoma miRNA signatures.
| Upregulated | Fold Change | Nominal P Value | Adjusted P. Value (FDR) |
|---|---|---|---|
| miR-22 | 2.4 | 2.6E-05 | 1.4E-03 |
| miR-518b | 2.5 | 3.6E-04 | 8.3E-03 |
| miR-143 | |||
| Downregulated | |||
| miR-182 | -7.0 | 1.4E-07 | 2.2E-05 |
| miR-301 | -6.2 | 4.9E-05 | 1.9E-03 |
| miR-20b | -4.6 | 1.4E-04 | 4.3E-03 |
| miR-375 | -5.1 | 3.8E-04 | 8.3E-03 |
| miR-487b | -4.8 | 8.0E-04 | 1.6E-02 |
MiRNA signatures slightly differ by age and ethnicity
MiRNA profiles were compared between patients of different ages and ethnicities, including Caucasian and AA, to determine if miRNA expression patterns would vary depending on these factors. We found only one miRNA, miR-486, that was significantly higher in younger patients with endometrioid uterine cancer (p<0.03, Supplemental Figure 3). This was primarily driven by elderly AA EAC patients where the expression was virtually absent. While these results are based on small sample sized they suggest that ethnicity and age should be considered in miRNA signatures.
Discussion
We report unique miRNA signatures for endometrial type I endometrioid carcinomas, type II papillary serous carcinomas and uterine carcinosarcomas. While multiple human cancer miRNA signatures have been described, only breast cancer has been previously profiled by subtype. (33-38) Perhaps because our numbers were small, there was no significant miRNA subset classifying different stages of disease, and only one that could separate patients by age, miR-486. However, our findings support the unique biology of these tumor types, and may represent future means to distinguish these tumors in difficult cases as well as to identify novel targets for therapy.
We have demonstrated both up-regulation and down-regulation of miRNAs in uterine endometrioid cancer compared to benign specimens. There were several miRNAs of interest that were different between benign endometrium and endometrioid cancers. Up regulation of the mir-200 family has been recently demonstrated in well-differentiated cancers, and is seen in our endometrioid samples. (39) Likewise, expression of miR-183 is inversely correlated with the metastatic potential of lung cancer cells. A 2–3 fold decrease of miR-183 was demonstrated in highly metastatic lung cancer cells versus non-metastatic counterparts derived from same parental cell lines. (40) Finding that mir-183 is relatively high in endometrioid cancer, which metastasizes infrequently, is thus not surprising. We find miR-205 and miR-182 to be up-regulated in endometrioid carcinomas. MiR-205 has previously been described to be up-regulated in ovarian cancer as well as bladder and kidney cancers. (41-42) MiR-205 is down-regulated in prostate cancer and esophageal cancer compared to normal tissue. (43-44) MiR-205 along with the mir-200 family has been demonstrated to cooperatively regulate expression of the E-cadherin transcriptional repressors ZEB1 (also known as δEF1) and SIP1 (also known as ZEB2), factors previously implicated in epithelial to mesenchymal transition and tumor metastasis. (45) MiR-182, member of a miRNA cluster in a chromosomal locus (7q31–34), up-regulated in endometrioid cancer is also up-regulated in melanoma cell lines and tumor samples. MiR-182 expression stimulates migration of melanoma cells in vitro and their metastatic potential in vivo, whereas miR-182 down-regulation inhibits tumor invasion and triggers apoptosis. (46)
Compared to the endometrioid subtypes (Type I), UPSC (Type II) had unique miRNA signatures, and showed higher miRNA expression of some specific miRNAs. MiR-19a & b, the key oncogenic component of mir-17-92, is up-regulated in UPSC tumors. These miRNAs have been shown to be altered in hematologic cancers and to promote lymphomagenesis in vivo. (47) The oncogenic activity of miR-19 is has been shown to be at least partly due to its repression of the tumor suppressor Pten. (48) MiR-101, another miRNA altered in UPSC, is down-regulated in hepatocellular carcinoma and was further reported to promote apoptosis and affect tumorigenicity. (49) MiR-30e-5p is also up-regulated in UPSC tumors. Interestingly, this miRNA has been reported to be aberrant in ovarian and peritoneal endometriosis. Its up-regulation was described to be specific to endometriosis independently from the site of the lesion. (50) Furthermore, up-regulation of miR-452 seen in UPSC, has been shown to be associated with lymph node positivity and serve as a prognostic marker for death in urothelial cancers. (51) This described finding in consistent with UPSC tumors having overall poor prognosis and widespread metastasis to the lymph nodes. MiR-29c, also misregulated in UPSC, is up-regulated in epithelial mesotheliomas. Increased expression of hsa-miR-29c predicted a more favorable prognosis in these tumors, and it's overexpression of resulted in significantly decreased proliferation, migration, invasion, and colony formation in these tumor cell lines. (52)
We have further shown that uterine carcinosarcomas demonstrate a unique miRNA signature, easily distinguishing these tumors from endometrial epithelial cancers. This is interesting as carcinosarcomas consist of both epithelial and sarcomatous components, and many advocate treating the epithelial component. However, our studies suggest that based on the miRNA signature of these tumors, they are biologically unique, and further support the hypothesis that these tumors likely require therapy unique from other epithelial tumors. Certain miRNAs appear to be consistently altered in the carcinosarcomas compared to epithelial tumors. MiR-518b is down-regulated in carcinosarcomas compared to both endometrioid and UPSC tumors, while miR-20b, miR-301 and miR-487 are up-regulated. MiR-20b has been described to accumulate in tumor cells and to play an oncogenic role. (53) MiR-20b is a regulator of VEGF, the critical angiogenic factor in response to hypoxia. (54) Low expression of miR-20b inhibits tumor cell growth but gives tumor cell more resistance to apoptosis in hypoxia. (55)
While limitations of our study were the lack of clinical follow up for these patients to correlate miRNA signatures with outcome, this is the first report in our knowledge of different miRNA signatures across these subtypes of uterine cancer. Due to the large number of patient samples the differences in our miRNA signatures are strongly significant and represent real differences between these tumor subtypes. Because these subtypes have such different biological behavior, their baseline differences in miRNA signatures are certainly likely to represent meaningful insight into their behavior. Our findings thus represent insight into the basic biological differences between these types of uterine cancers, and when further validated may represent the first steps towards identifying important biomarkers of patient outcome and targets for therapy for these patients.
Supplementary Material
A. A heat map of 5 benign endometrial and 57 endometrioid endometrial carcinoma samples indicates that there is a difference in miRNA expression between the two groups that clusters them separately. MicroRNA expression is displayed as higher (red) or lower (green).
The heat map of 57 EAC and 27 UPSC displays grouping of these two types of uterine cancers by miRNA signatures. The subset that most clearly separates EAC and UPSC are higher in UPSC. MicroRNA expression is displayed as higher (red) or lower (green).
Acknowledgments
This work was supported by the RTOG Translational Research Program, funded through grant U10CA21661 by the National Cancer Institute. This research was further supported in part by the Yale Center of Excellence in Molecular Hematology, NIH P30 DK072442. JW was supported by K08 (CA124484).
Footnotes
Conflict of Interest Statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Cancer Society. Cancer facts and figures. Society AGAC. 2008 [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. Cancer J Clin. 2008 Mar-Apr;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Rose P. Endometrial Carcinoma. NEJM. 1996 Aug;:640–649. doi: 10.1056/NEJM199608293350907. [DOI] [PubMed] [Google Scholar]
- 4.Bokhman J. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson M, Ross J, Eifel P, Martinez A, Kempson R. Uterine papillary serous carcinoma: a highly malignant form of endometrial adenocarcinoma. Am J Surg Pathol. 1982;6:93–108. doi: 10.1097/00000478-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Carcangiu ML, Chambers JT. Uterine papillary serous carcinoma: a study on 108 cases with emphasis on the prognostic significance of associated endometrioid carcinoma, absence of invasion, and concomitant ovarian carcinoma. Gynecol Oncol. 1992;47:298–305. doi: 10.1016/0090-8258(92)90130-b. [DOI] [PubMed] [Google Scholar]
- 7.Goff BA, Kato D, Schmidt RE, et al. Uterine papillary serous carcinoma: patterns of metastatic spread. Gynecol Oncol. 1994;54:264–268. doi: 10.1006/gyno.1994.1208. [DOI] [PubMed] [Google Scholar]
- 8.Abeler VM, Kjorstad KE. Endometrial adenocarcinoma in Norway: a study of a total population. Cancer. 1991;67:3093–3103. doi: 10.1002/1097-0142(19910615)67:12<3093::aid-cncr2820671226>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson WM, Alberhasky RC, Connelly PJ. Carcinoma of the endometrium. Papillary adenocarcinoma: a clinical pathological study, 46 patients. Am J Clin Pathol. 1982;77:534–40. doi: 10.1093/ajcp/77.5.534. [DOI] [PubMed] [Google Scholar]
- 10.Christopherson WM, Connelly PF, Alberhasky RC. Carcinoma of the endometrium. An analysis of prognosticators inpatients with favorable subtypes and stage I disease. Cancer. 1983;51:1705–9. doi: 10.1002/1097-0142(19830501)51:9<1705::aid-cncr2820510924>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 11.Sartori E, Bazzurini L, Gadducci A, Landoni F, Lissoni A, Maggino T, Zola P, La Face B. Carcinosarcoma of the uterus: a clinicopathological multicenter CTF study. Gynecol Oncol. 1997 Oct;67(1):70–5. doi: 10.1006/gyno.1997.4827. [DOI] [PubMed] [Google Scholar]
- 12.Zelmanowicz A, Hildesheim A, Sherman M, et al. Evidence for a common etiology for endometrial carcinomas and malignant mixed mullerian tumors. Gynecol Oncol. 1998;69:253–7. doi: 10.1006/gyno.1998.4941. [DOI] [PubMed] [Google Scholar]
- 13.Gorai I, Yanagibashi T, Taki A, et al. Uterine carcinosarcoma is derived from a single stem cell: an in vitro study. Int J Cancer. 1997;72:821–7. doi: 10.1002/(sici)1097-0215(19970904)72:5<821::aid-ijc19>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 14.McCluggage W. Uterine carcinosarcomas (malignant mixed Mullerian tumors) are metaplastic carcinomas. Int J Gynecol Cancer. 2002;12:687–90. doi: 10.1136/ijgc-00009577-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 15.McCluggage W. Malignant biphasic uterine tumours: carcinosarcomas or metaplastic carcinomas? J Clin Pathol. 2002;55:321–5. doi: 10.1136/jcp.55.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K, Watanabe K, Nasim S, et al. Prognostic significance of p53 overexpression in endometrial cancer. Cancer Res. 1994;54:4667–70. [PubMed] [Google Scholar]
- 17.Silverman M, Roche P, Kho R, Keeney G, Li H, Podratz K. Molecular and cytokinetic pretreatment risk assessment in endometrial carcinoma. Gynecol Oncol. 2000;77:1–7. doi: 10.1006/gyno.2000.5751. [DOI] [PubMed] [Google Scholar]
- 18.Pisani AL, Barbuto DA, Chen D, Ramos L, Lagasse LD, Karlan BY. HER-2/neu, p53, and DNA analyses as prognosticators for survival in endometrial carcinoma. Obstet Gynecol. 1995;85:729–734. doi: 10.1016/0029-7844(95)00037-r. [DOI] [PubMed] [Google Scholar]
- 19.Tashiro H, Blazes M, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–40. [PubMed] [Google Scholar]
- 20.Todo Y. Analysis of p53, MDR-1, and GST-pi expression in endometrial carcinoma. Hokkaido Igaku Zasshi. 2003;78:117–27. [PubMed] [Google Scholar]
- 21.Hirasawa A, Aoki D, Inoue J, et al. Unfavorable prognostic factors associated with high frequency of microsatellite instability and comparative genomic hybridization analysis in endometrial cancer. Clin Cancer Res. 2003;9:5675–82. [PubMed] [Google Scholar]
- 22.Cirisano F, Karlan B. The role of the HER-2/neu oncogene in gynecologic cancers. J Soc Gynecol Investig. 1996;3:99–105. [PubMed] [Google Scholar]
- 23.Razorenova T, Samsonova E, Pozharisskiĭ K, Razorenov G. Mathematical evaluation of prognostic significance of clinico-morphological and immunohistochemical features of endometrioid adenocarcinoma. Vopr Onkol. 2007;53:682–9. [PubMed] [Google Scholar]
- 24.Simionescu C, Georgescu C, Mărgăritescu C, et al. P53 and PCNA immunoexpression in endometrial carcinomas. Rom J Morphol Embryol. 2006;47:137–41. [PubMed] [Google Scholar]
- 25.Wu Y, Wang J, Wang H, Yang X. Study on expression of Ki-67, early apoptotic protein M30 in endometrial carcinoma and their correlation with prognosis. Zhonghua Bing Li Xue Za Zhi. 2003;32:314–8. [PubMed] [Google Scholar]
- 26.Berchuck A, Rodriguez G, Kinney RB, et al. Overexpression of HER-2/neu in endometrial cancer is associated with advanced stage disease. Am J Obstet Gynecol. 1991;164:15–21. doi: 10.1016/0002-9378(91)90615-x. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Getz G, Miska E, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 28.Slack FJ, Weidhaas JB. MiRNAs as potential magic bullet in cancer. Future Oncology Perspective. 2006 doi: 10.2217/14796694.2.1.73. [DOI] [PubMed] [Google Scholar]
- 29.R Development Core Team: A Language and Environment for Statistical Computing. 2003 http://www.r-project.org.
- 30.Gentleman R, Carey V, Bates D, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini YaYH. Controlling the False Discovery Rate- a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 33.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jay C, Nemunaitis J, Chen P, Fulgham P, Tong A. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S, Chen H, Yang P, Chen J. Unique MicroRNA signature and clinical outcome of cancers. DNA Cell Biol. 2007;26:283–92. doi: 10.1089/dna.2006.0555. [DOI] [PubMed] [Google Scholar]
- 37.Ozen M, Creighton C, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 38.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 39.Park Sun-Mi, Gaur Arti B, Lengyel Ernst, Peter Marcus E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Guofu, Mao Weimin, Zheng Shu. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Letter. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 41.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 42.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;5:6387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino, et al. MiR-205 Exerts Tumor-Suppressive Functions in Human Prostate through Down-regulation of Protein Kinase C{varepsilon} Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 44.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory P, Bert A, Paterson E, Barry S, Tsykin A, Farshid G, Vadas M, Khew-Goodall Y, Goodall G. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 46.Segura M, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski S, Blochin E, Rose A, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. PNAS. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, Parker JS, Paddison PJ, Tam W, Ferrando A, Wendel HG. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010 Feb 28; doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olive V, Bennett M, Walker J, Ma C, Jiang I, Cordon-Cardo C, Li Q, Lowe S, Hannon G, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009 December 15;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su Hang, Yang Jian-Rong, Xu Teng, Huang Jun, Xu Li, Yuan Yunfei, Zhuang Shi-Mei. MicroRNA-101, Down-regulated in Hepatocellular Carcinoma, Promotes Apoptosis and Suppresses Tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 50.Filigheddu N, Gregnanin I, Porporato P, Surico D, Perego B, Galli L, Patrignani C, Graziani A, Surico N. Differential Expression of MicroRNAs between Eutopic and Ectopic Endometrium in Ovarian Endometriosis. Journal of Biomedicine and Biotechnology. 2010 doi: 10.1155/2010/369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, Månsson W, Rovira C, Höglund M. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009 May 1;124(9):2236–42. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 52.Pass H, Goparaju C, Ivanov S, Donington J, Carbone M, Hoshen M, Cohen D, Chajut A, Rosenwald S, Dan H, Benjamin S, Aharonov R. Hsa-miR-29c is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70:1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 54.Hua Z, Lv Q, Ye W, Wong CK, Cai G, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lei Z, Li B, Yang Z, Fang H, Zhang GM, et al. Regulation of HIF-1α and VEGF by miR-20b Tunes Tumor Cells to Adapt to the Alteration of Oxygen Concentration. PLoS ONE. 2009;4(10):e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. A heat map of 5 benign endometrial and 57 endometrioid endometrial carcinoma samples indicates that there is a difference in miRNA expression between the two groups that clusters them separately. MicroRNA expression is displayed as higher (red) or lower (green).
The heat map of 57 EAC and 27 UPSC displays grouping of these two types of uterine cancers by miRNA signatures. The subset that most clearly separates EAC and UPSC are higher in UPSC. MicroRNA expression is displayed as higher (red) or lower (green).