Abstract
With the worldwide emergence of multi-drug resistant (MDR) and extensively-drug-resistant (XDR) strains of Mycobacterium tuberculosis (Mtb), there are serious concerns about the continued ability to contain this disease. We discuss the most promising new drugs in late stage development that might be useful in treating MDR and XDR forms of the disease. These agents have novel mechanisms of action that are not targeted by the standard drugs used presently to treat susceptible strains.
Introduction
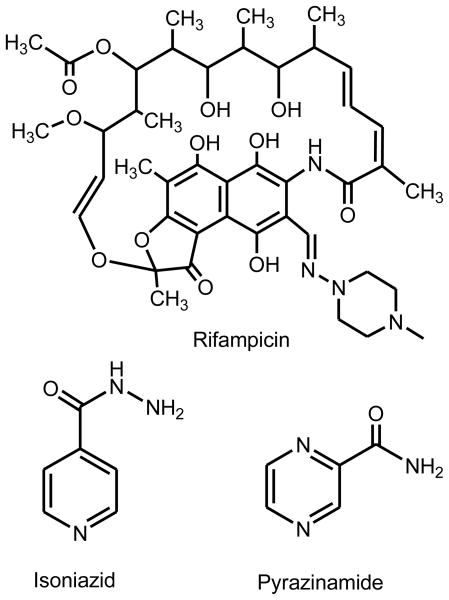
Although effective chemotherapy for tuberculosis has been in place for over 50 years, it was clear from the first clinical trials that monotherapy with any agent led to the development of resistance and clinical failure in 2–5 months [1]. Therefore, tuberculosis chemotherapy has involved the administration of multiple drugs since the late 1960’s [2]. At present, the most widely used short course therapy involves taking isoniazid, pyrazinamide and rifampicin (Figure 1) for two months, followed by an additional four months of treatment with isoniazid and rifampicin alone [3,4]. Isoniazid and rifampicin are rapidly bactericidal agents, while pyrazinamide is included to treat a presumed population of non-replicating organisms [5].
Figure 1.
In the early 1980’s, significant clinical resistance to isoniazid began to be noted and 15 years later, combined resistance to isoniazid and rifampicin similarly began to increase [6,7]. MDR is defined as resistance to these latter two rapidly bactericidal agents [8,9]. Strains that have acquired additional resistance to one of the three most potent injectables (second-generation aminoglycosides such as kanamycin and amikacin, and cyclic polypeptides such as capreomycin) and a fluoroquinolone are deemed extensively-drug-resistant (XDR) [10,11]. This combination of resistances is associated with particularly poor outcomes in patients due to the relative lack of potency of the remaining second-line agents [12,13]. In a report published in 2006, an outbreak of XDR-TB was described in KwaZulu Natal, South Africa where the strain was responsible for 100 % mortality of 54 infected individuals with an average time from diagnosis to death of 16 days [14].
In the last ten years, there has been resurgence in interest in identifying new compounds that are effective against Mtb. Much of this interest has been within academic and governmental laboratories, as opposed to the biotechnology or large pharmaceutical industries. Because of space limitations, we will be unable to discuss many promising new compounds in preclinical development (many of which have been reviewed recently [15–17], and have elected instead to discuss those compounds for which clinical trials (Phase 1 and 2) have or will begin shortly in some detail. Several of these compounds are similar to those used against other bacterial infections; however, some appear to be quite specific for Mtb.
Diarylquinolines (R207910)
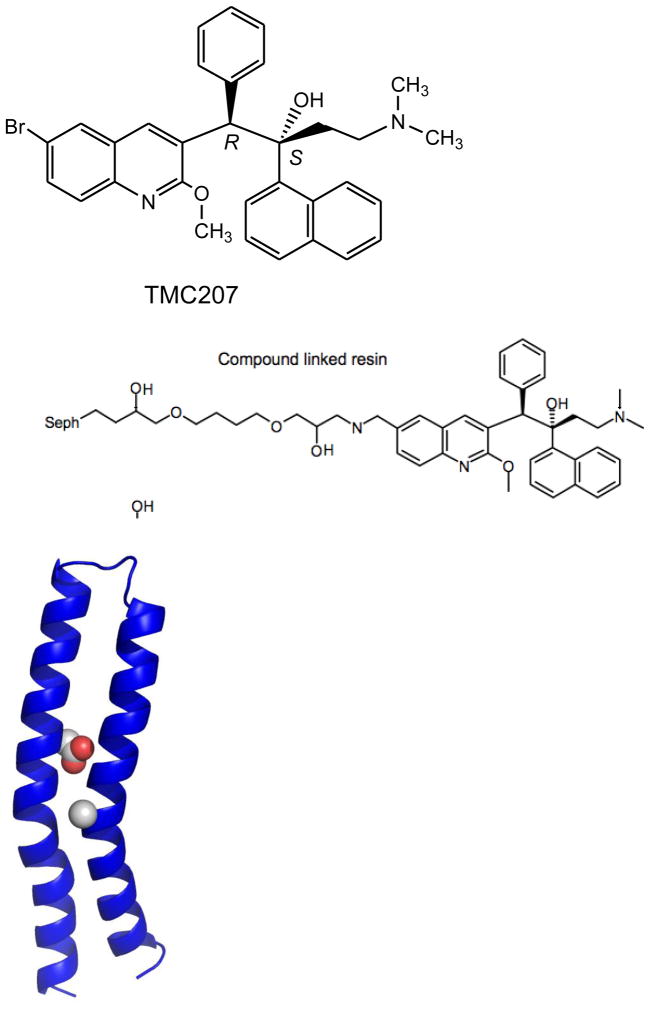
In early 2005, a report from a Johnson and Johnson group in Europe appeared describing the activity of a new class of diarylquinolines against Mtb [18]. Using a whole-cell assay against a surrogate organism (a fast-growing saprophytic cousin of Mtb called Mycobacterium smegmatis) a hit was identified that was a structurally unique, highly substituted quinoline that also showed activity against Mtb. Diarylquinolines are distinct both in structure and mechanism from fluoroquinolones (which inhibit type II topoisomerases such as DNA gyrase) and quinolines such as mefloquine. A series of analogs were prepared and assessed for enhanced activity. Unfortunately, none of this chemistry has appeared outside of the patent literature where it is sketchy and incomplete limiting the amount of available structure-activity information (US Patent 2007/0249667). A smaller series of similarly substituted quinolines have subsequently been reported by another group but these have much more limited potency [19]. R207910 was selected as the lead compound in the series after a mouse infection study showed it was the only compound in a series of three tested to have significant activity. As shown in Figure 2, R207910 is a single enantiomer of a compound with two chiral centers with the carbon bearing the phenyl substituent of the R configuration and with the carbon bearing the hydroxyl group of the S configuration.
Figure 2.
Homology model of the Mtb c subunit of ATP synthase. Position 28 (left, spheres) is the aspartate residue equivalent to D32 in M. smegmatis. Position 63 (right sphere) is the Ala residue mutated to a Pro in the Mtb strain reisitant to TMC207. This figure was generously provided by Josh Goldman and Mark Girvin, Department of Biochemistry, Albert Einstein College of Medicine.
R207910 has extraordinary activity against both drug-susceptible and drug-resistant strains of M. tuberculosis, exhibiting MIC (Minimum Inhibitory Concentration) values of 30–120 ng/ml, equal to or lower than isoniazid and rifampicin. The compound exhibits a rapid bactericidal activity in vitro, causing losses of 3 log orders of CFU (Colony Forming Units)/ml in 12 days. Surprisingly, the compound exhibits a very narrow spectrum of activity and loses activity against even closely related actinomycetes such as Corynebacteria, and it is also inactive against other Gram-positive and Gram-negative pathogens. Amongst the mycobacteria, R207910 shows consistent activity against many species, including many medically important opportunistic pathogens [20]. Recently it has even been shown to have activity against Mycobacterium leprae, the causative agent of leprosy, in a mouse model of the disease [21].
The target of R207910 was identified by selecting resistant mutants in both Mtb and M. smegmatis and subsequently applying whole genome resequencing to identify candidate polymorphisms associated with resistance. Mutations common to the organisms that had acquired resistance were found in the atpE gene, encoding the membrane-bound subunit of the F0 ATP synthase complex. In an Mtb mutant, a single point mutation that generated an A63P substitution was observed, while in one M. smegmatis mutant, a point mutation generated a D32V substitution. Subsequent investigations of large numbers of in vitro generated mutants have confirmed a role for AtpE in resistance to R207910, and the few atypical mycobacteria that show intrinsic resistance to R207910 have naturally occurring polymorphisms at A63 [20,22]. Similarly, the eukaryotic mitochondrial ATP synthase has a methionine at position 63, a fact that may explain the remarkable selectivity (>20,000 fold) of R207910 for Mtb versus mammals [23]. Based on a homology model built from the NMR-derived structure of the c-subunit from E. coli, both A63 and D32 are within the membrane-spanning portion of the antiparallel α-helices (Figure 2). Computational docking studies based on this model structure suggest that important contacts are possible with R186, E61 and F65 but do not convincingly explain the observed mutations at A63 and D32, nor do they produce a convincing ranking of the stereoisomeric versions of this compound [24]. The intrinsically low resolution of NMR structures amplified by uncertainties in deriving homology models from such structures limit the confidence in the predicted binding site and contacts of R207910 with this protein.
A recent study looking at the development of resistance to R207910 in nearly 100 spontaneously resistant mutants obtained from seven different clinical isolates found mutation of atpE only partially accounted for the resistance observed. In 38 of these isolates no mutations were found anywhere in the ATP synthase operon, suggesting the possibility that R207910 has additional, as yet unknown, targets [22]. However, inhibition of ATP synthesis clearly plays a major role in the mechanism of R207910. Treatment of whole cells with the drug reduces ATP concentrations and the drug inhibits ATP synthesis even in isolated vesicles. Finally, coupling of a derivative of the drug (Figure 2) to an affinity column effectively bound the α and β subunits of ATP synthase from solubilized membrane proteins of M. smegmatis (but unfortunately not the c subunit which is highly hydrophobic) [25]. These results are not overly surprising given the nuances of drug-mediated bacterial cell death that are being revealed for even “simple” antibiotics like aminoglycoside inhibitors of protein synthesis [26,27].
One of the major challenges for tuberculosis drug development is the ability of a subpopulation of the organism to exist in a non-replicating, but viable form. Several models of this physiological state have been developed, but the most commonly used is the Wayne model [28]. Although assumed to be an obligate aerobe, when deprived of oxygen, M. tuberculosis can enter into a metabolic state termed “persistence”. In this state, the organisms are insensitive to, or substantially less sensitive to common drugs used to treat the disease, including isoniazid and rifampicin. In this state, intracellular ATP concentrations decrease to approximately 20% of those in actively growing cells. Administration of R207910 to cultures of anaerobic cells causes a dose-dependent reduction of ATP levels and a corresponding decrease in CFU’s recoverable after readmission of oxygen [29]. These results have led to the conclusion that ATP homeostasis is critical in non-replicating cells and validate the ATP synthase as a valuable target in this important subpopulation [30].
R207910 has performed exceptionally well in various mouse models of TB [31–35] and initial Phase I safety and Phase II efficacy studies continue to look very promising [18,36,37]. The most compelling results so far are from a Phase 2, randomized, controlled clinical trail in South Africa of patients diagnosed with MDR-TB. In this study, patients were treated with either R207910 or a placebo in combination with five other second-line antituberculosis agents. In the placebo control group fewer than 10 % of patients had converted their sputum from culture-positive to culture-negative after two months, while 50% of the patient group treated with R207910 converted in the same period.
R207910 represents one of the most promising of the new drug candidates for the treatment of tuberculosis but there remains a critical issue. The drug is rapidly metabolized by cytochrome P-450 isoform 3A4, an isoform that is strongly upregulated with the use of rifampicin, perhaps the single most important agent used for first line chemotherapy [33]. In healthy volunteers receiving both drugs the level of R207910 was reduced by 50%. Although additional drug-drug interaction studies are certainly necessary it remains to be seen whether R207910 will be able to be used as a component of first line therapy for drug-susceptible disease. In retrospect it is surprising that a compound with such a liability was selected as a candidate since screening for metabolic instability typically occurs very early in drug development. Unfortunately, the complexity of an integral membrane target with little structural information, added to the chemical complexity of the scaffold that requires chiral HPLC resolution at the last step, make it very difficult to address this liability from an informed medicinal chemistry perspective.
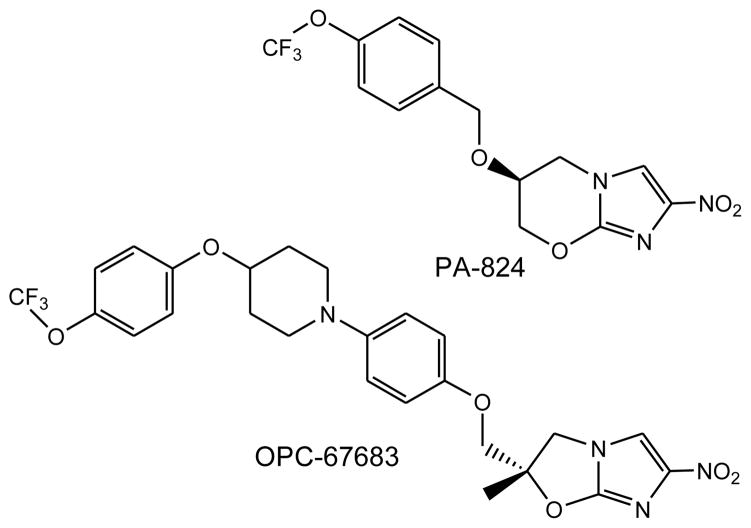
Nitroimidazoles (PA-824 and OPC67683)
Metronidazole is perhaps the most well known antibiotic of the nitroimidazole class. It is effective against anaerobic bacteria, protozoa and hypoxically-adapted, non-replicating Mtb [38]. In 1989, a group from Ciba-Geigy India reported the powerful antitubercular activity of a series of bicyclic nitroimidazo[2,1-b]oxazoles. The most potent of these, CGI 17341, inhibited both drug-susceptible and multi-drug resistant strains of Mtb and was specifically active against Mtb with significantly less active against M. avium, M. intracellulare and M. fortuitum [39]. In preliminary tests in a mouse model of infection, the compound exhibited a dose-dependent increase in mean survival time. However, further development of the compound was halted, presumably due to concerns relating to mutagenicity.
Ten years later, a related compound, PA-824, featuring a six-membered oxazine rather than the five-membered oxazole, was reported [40]. From more than 300 6-substituted 6,7-dihydro-5H-imidazo[2,1-b][1,3]oxazines, PA-824 was selected as a lead molecule. Much of the initial excitement around the series was due to the fact that this compound, like its distant progenitor metronidazole, had activity against hypoxic bacilli, an observation interpreted as indicating potential for shortening of the overall duration of treatment. PA-824, though was selected based on one characteristic – activity in the mouse model of TB, activity that has subsequently been confirmed many times and in many models [41–45]. From the very beginning bioreductive activation of the molecule was suspected as being a key feature of its potent activity, and examination of the fate of radioactive drug in the presence of sensitive cells confirmed a complex mixture of bacterial metabolites were produced. In addition, reductions in protein synthesis and a specific effect on ketomycolate synthesis were noted.
Because of the tantalizing anaerobic activity, and the strange connection with an important component of the bacterial cell wall PA-824 became a powerful tool to unravel some of the biochemistry surrounding anaerobic metabolism. Because the primary mechanism of resistance to this agent involved the loss of reduction capacity in resistant organisms, the major cofactor involved in this reductive process, the deazaflavin F420 was quickly identified, as was F420-dependent mycobacterial glucose-6-phosphate dehydrogenase (G6PDH), its major redox partner. Careful examination of PA-824 resistant mutants also allowed the identification of the major biosynthetic genes involved in F420 biosynthesis [46,47]. What became clear ultimately was that F420 was essential for reduction, and Fdg (the G6PDH) was essential for reducing F420 at the expense of G6P, but that there had to be a missing enzyme that catalyzed the actual reduction.
Because the major class of mutants that were obtained always mapped to the biosynthetic genes for F420 or the Fdg protein whether selected in vivo (from treated mice) or in vitro searching for the missing enzyme required screening through many mutants and quickly ruling out those with defects in either of these. Eventually, a rarer class of mutants were identified and whole genome resequencing revealed that thes mutants all had mutations in a single gene annotated as Rv3547 [48]. This gene encoded a conserved predicted 151 amino acid protein with no sequence homology to any protein of known function. Three close homologs of Rv3547 were identified in the TB genome, and one of these appeared to be structurally related to FMN-binding proteins. Homologs of this enzyme were also found throughout the Actinobacteria suggesting an important, and as yet unidentified, physiological role for this enzyme family. Based on these results, Rv3547 was proposed to be the nitroreductase that bound reduced F420 and catalyzed the reduction of PA-824.
Several years later this was finally confirmed by reconstitution of the enzyme, cofactor and PA-824 and both the enzymatic function of Rv3574 (renamed Ddn: deazaflavin-dependent nitroreductase) and the mechanism of anaerobic killing became somewhat clearer [49]. In the presence of reduced F420 and Ddn, PA-824 was converted to three products, the major one being the des-nitro form of PA-824. As reduced F420 is an obligate hydride ion donor, the reaction proceeds via hydride ion transfer to the C3 position of the imidazole ring, followed by protonation at the C2 position. This generates an intermediate that can decompose with the generation of nitrous acid that will rapidly decompose to generate nitric oxide. Indeed using both the Greiss reagent to directly detect nitrous acid in vitro and diaminofluorescein, that specifically fluoresces in the presence of NO, in mycobacterial cells, this chemistry was confirmed. Finally, the correlation between the anaerobic activity of a series of nitroimidazopyrans and the amount of denitrated product produced supports the role of Ddn as the nitroreductase responsible for the activity of PA-824 in the Wayne model.
PA-824 and its derivatives were initially optimized for activity against organisms during mouse infection. Even with a rapid assay for assessing bacterial burden in mice [50] this was an inefficient and labor-intensive way to screen analogs. The reason this was necessary was because there was a poor correlation between in vivo and in vitro activities. Likewise there is a poor correlation between aerobic and anaerobic activities across series of analogs of these compounds [51]. Both activities appear to be dependent upon unique structural features of the molecule suggesting that there are two different modes of action for the drug following reduction. In fact a reexamination of the transcriptional response of the organism to drug treatment revealed two discrete responses, one that appeared to be based on inhibition of respiration and the other based on inhibition of cell wall synthesis [52].
Simultaneously with the PA-824 development program another program to optimize bicyclic nitroimidazoles based upon the inhibition of cell wall mycolic acid synthesis was undertaken in Japan at Otsuka Pharmaceuticals. The resulting compound, dubbed OPC-67683, has now reached Phase II clinical trials and is highly likely to be activated by the Ddn/F420 reductase [53,54]. It is clear that the two activities are both dependent upon reduction but are nonetheless distinct. The distribution of products formed by partitioning between products formed may play a role in this dual activity and the availability of a known enzyme that catalyzes this reaction may allow a more informed structure-based interpretation of the mechanism and may facilitate design of a new generation molecule with improved properties.
Diamines (SQ109)
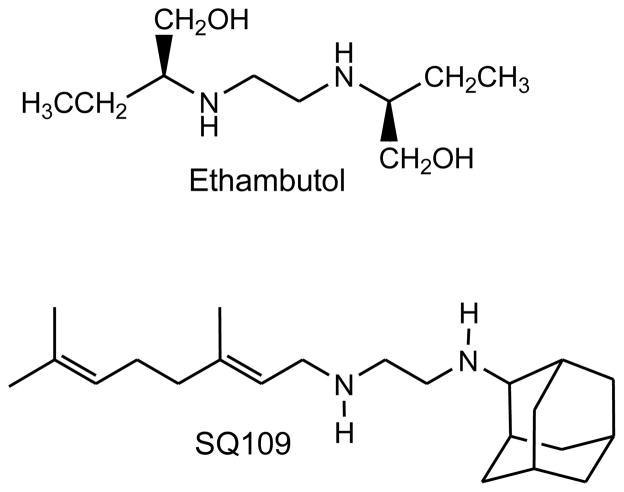
SQ109 was first synthesized in an attempt to generate more effective analogs of ethambutol (Figure 4). A report in 2003 used a combinatorial approach to generate a library of 1,2-disubstitued ethylenediamine derivatives. Some 288 amines were tested and using a split and pool strategy four libraries were generated containing >60,000 asymmetrically disubstituted ethylenediamines. Compounds were screened for in vitro activity against Mtb in pools of ten to thirty compounds. More than 2000 active wells were then deconvoluted and of these, 69 of the most active compounds were resynthesized, purified and evaluated in detail. Of these, 25 exhibited MIC values equal to or less than ethambutol. In particular, compound 109, N-geranyl-N′-(2-adamanthyl)ethane-1,2-diamine, exhibited submicromolar MIC values in broth microdilution methods (MIC = 0.2 uM) [55]. This compound, dubbed SQ109 has been shown to have good selectivity and efficacy in mouse models of disease [56]. It also has acceptable pharmacokinetics and pharmacodynamics and concentrates more than 100 fold in the lung [57,58]. SQ109 has synergistic effects both in vitro and in vivo with most of the major front-line agents [59,60]. SQ109 is currently in late-stage Phase 1 (safety) trials.
Figure 4.
So why hasn’t SQ109 generated as much enthusiasm as the other novel classes of agents that have arisen recently? One factor is that the actual target of SQ109 remains elusive. SQ109 was derived intellectually from ethambutol, a 1,2-diamine of similar structure. Unfortunately it was discovered early on that SQ109 is active against isoniazid and rifampicin resistant strains of TB, but surprisingly was also active against ethambutol resistant strains [56]. The target of ethambutol is an arabinosyltransferase involved in construction of the polymeric cell wall unique to this group of organisms [61,62]. Somewhere in the process of analoging around the diamine structure the series may have acquired other, related cellular targets or might have acquired a different binding mode to the target of ethambutol. The strategy used for screening these analogs was to incorporate a green fluorescent protein driven by a promoter known to be highly sensitive to cell wall inhibitors [63]. Although it was thought at the time that the combination of keeping a minimal pharmacophore (the 1,2-diamine) and utilizing this genetic marker of cell wall inhibition would be sufficient to stay on target the suspicion arose from the sensitivity of the ethambutol resistant strains to SQ109 that the target had drifted.
Subsequent studies of the transcriptional response to various inhibitors confirmed that SQ109 had distinct effects from ethambutol, suggesting that the compound was inhibiting a different target (albeit still a target involved in cell wall biosynthesis since the locus used to generate the reporter used in the screen was still prominent in the array profiles) [64]. This result was confirmed by an analysis of whole cells of Mtb treated with both inhibitors that showed that incorporation of arabinose into cell wall arabinogalactan, well known to be inhibited by ethambutol, was not inhibited at all with SQ109.
In an attempt to define the target of SQ109 and its possible mechanism of action, a proteomic study of the effects of SQ109, ethambutol and isoniazid was performed. When tested at their respective MIC values for 24 hours, relatively few and small changes were observed for any of the compounds with the exception of the increased levels of the secreted antigens ESAT-6/CFP-10 and AhpC, a small disulfide-containing protein that catalyzes the reduction or organic hydroperoxides [64]. The effects on ESAT-6/CFP-10 expression were more pronounced for isoniazid and ethambutol but equal for all three compounds in the case of AhpC. This again points out that the intracellular targets of ethambutol and SQ109 are likely different.
Two other groups have reported more recently additional analogs of SQ109 with either alterations in the geranylamine side chain or substitutions in the adamantine ring and although some slight changes in overall activity were noted in general the SAR of both of the amine substituents is quite flat [65,66]. In an effort to improve upon the bioavailability of orally administered SQ109, a series of carbamate derivatives was prepared that would be chemically stable but hydrolyzed once inside the host [67]. One of these compounds, the N2-isopropyl carbamate of SQ109 exhibited quite superior pharmacokinetic properties when administered orally compared to SQ109. The tolerance of this series to significant changes in the two amine substituents is very high and suggests the possibility that the binding site of the drug must show little specificity except for requiring the presence of large hydrophobic groups. It has proven very difficult to select resistant mutants (a good thing for the patients if this drug makes it into efficacy trials, but a bad thing for attempting to use this methodology for identifying the target). The mechanism of SQ109 remains a very important outstanding issue in the field of tuberculosis drug discovery.
β-lactams (Clavulanate/Carbapenems)
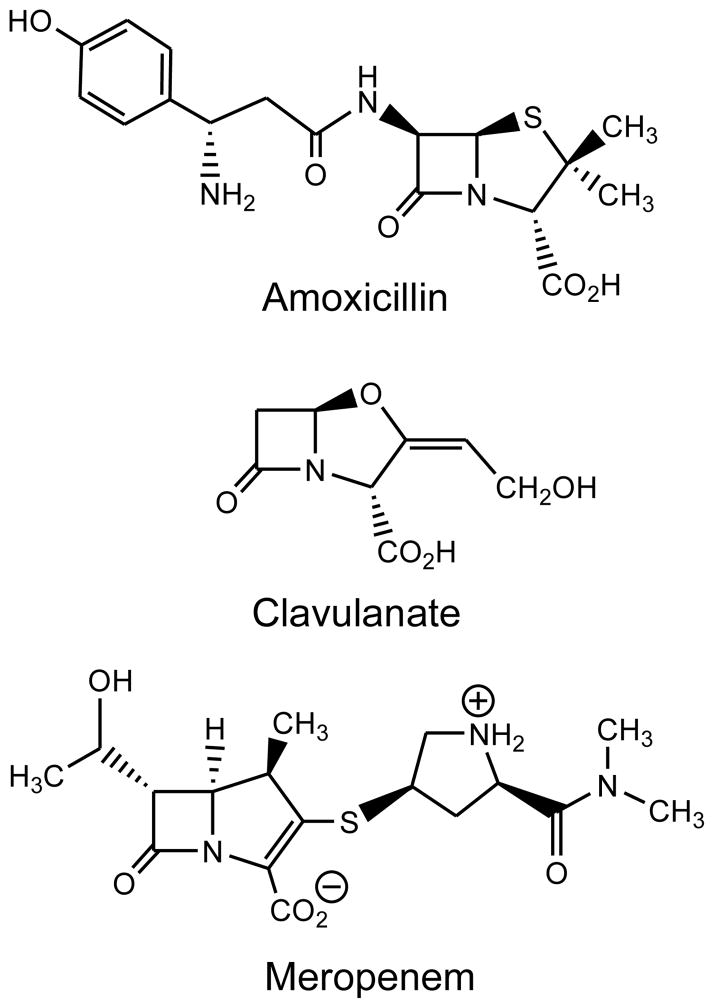
The β-lactam class of antibacterials is one of the most important and successful classes of antibiotics ever developed. However, tuberculosis has only episodically been evaluated for susceptibility to this class. This likely stems from the report in 1949 [68] that Mycobacterium tuberculosis produces a “penicillinase” that prevented the penicillin class of β-lactams from inhibiting mycobacterial growth. In 1989, the MIC value of amoxicillin was shown to drop precipitously when clavulanate, an FDA-approved β-lactamase inhibitor, was added to amoxicillin (Figure 5) [69]. This combination (trademark name: Augmentin, Glaxo Smith Kline) was used ten years later to successfully treat a single MDR-TB-infected individual [70]. Given this success, it is curious that no additional reports of using Augmentin to treat drug-susceptible or drug resistant TB infected individuals. When Cole and colleagues published the genome of Mycobacterium tuberculosis in 1999, a single chromosomally-encoded β-lactamase homologue of Class A (Ambler) β-lactamases was annotated [71]. This gene, Rv3587, included an N-terminal secretory signal sequence as well as a sequence that likely allowed for the anchoring of the protein in the outer aspect of the cell membrane and its periplasmic location [72,73]. Removal of these first 40 residues allowed the production of a soluble and active form of the enzyme that was crystallized and whose three-dimensional structure was determined [74]. The activity for a series of β-lactams, including penicillins, cephalosporins and imipenem, a carbapenem, were reported, and the structure exhibited a fold similar to previously reported Class A β-lactamases.
Figure 5.
This same construct was used to express and purify the TB BlaC β-lactamase for a more complete kinetic description. The enzyme catalyzes hydrolysis of all penicillin-class β-lactams at nearly the diffusion-limited rate, While cephalosporins are less rapidly hydrolyzed, all compounds tested were substrates for the enzyme [75]. Finally, we tested two carbapenems, compounds that are derivatives of thienamycin, a natural product: imipenem and meropenem. Imipenem, the first carbapenem approved for human use, was a good substrate for the enzyme with a turnover number of 10 min−1. Meropenem, which contains a methyl-substituted pyrroline ring, was an extremely poor substrate, which a low micromolar Km value and a turnover number of 0.08 min−1, some 120 times slower than imipenem. In an attempt to define the nature and extent of inhibition of BlaC by FDA-approved β-lactamase inhibitors, sulbactam, tazobactam and clavulanate were tested as inhibitors of the enzyme. Neither of the penicillanic acid sulfone inhibitors, sulbactam and tazobactam, irreversibly inhibited BlaC, with activity being regained within 20–30 minutes of incubation with these compounds. However, addition of clavulanate at near stoichiometric concentrations, resulted in rapid and irreversible inhibition, suggesting a mechanistic rationale for its in vitro efficacy in combination with amoxicillin against H37Rv, and its in vivo activity against MDR-TB.
The determination of the structure of the complex gave hints as to why the inhibitory activity of clavulanate was unique [76]. Once clavulanate binds, and the active site serine attacks the β-lactam ring, the oxazol ring subsequently opens, generating a C6 carbonyl. The C5 carboxyl group is beta to the carbonyl and rapidly decarboxylates to a product that accumulates as observed by mass spectrometry. The final step is the tautomerization of the C3-N4 imine to the C2-C3 eneamine. This final step reorients the clavulante skeleton to increase the distance between the ester carbonyl and the catalytic water molecule, and change the orientation of the catalytic water molecule.
When clavulanate is added to any beta-lactam, the MIC values are dramatically reduced at concentrations as low as 0.5 μg/ml. However, meropenem was selected for detailed analysis due to its extremely low turnover number, and its excellent safety profile. The combination of meropenem (2–4 μg/ml) and clavulanate (2 μg/ml) was rapidly bactericidal against aerobically growing organisms and sterilized cultures in ~ 2 weeks [77]. In similar types of experiments performed in the Wayne model of anaerobic noon-replicative growth, we were surprised to find that the combination of clavulanate-meropenem was bactericidal in a dose-dependent manner. This suggests that while organisms in this physiological state are not producing new peptidoglycan, that there must be dynamic remodeling occurring in this state, and that inhibiting the re-cross-linking of peptidoglycan has cidal consequences for the organism. Finally, this combination was tested for activity against 13 clinical strains of TB that are phenotypically XDR. Many of these strains exhibited MIC values for amoxiciilin or imipenem that were greater than 5 μg/ml, even in the presence of 2.5 μg/ml of clavulanate. However, all 13 strains exhibited MIC values for meropenem less than 1.25 μg/ml in the presence of 2.5 μg/ml of clavulanate. Given that these FDA-approved compounds have been shown to be individually (meropenem) bactericidal, or mycobactericidal in combination with a second beta-lactam (Clavulanate/Amoxicillin), the rapid in vivo testing of clavulanate/carbapenem combinations should be a high priority.
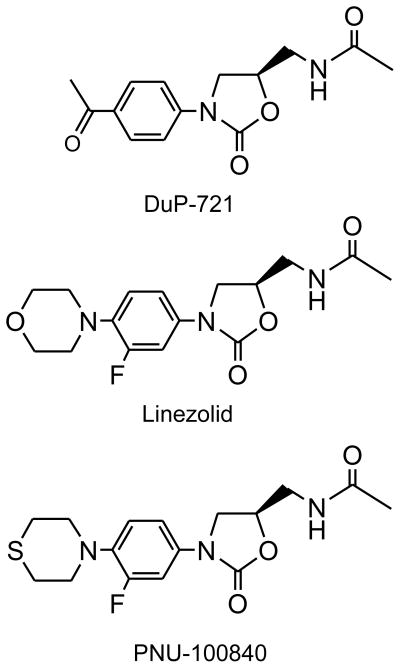
Oxazolidinones (Linezolid, PNU-100480)
N-aryl substituted 1,3-oxazolidin-2-ones were first identified in the 1950s as antidepressant monoamine oxidase inhibitors. Their antibacterial activity was uncovered in a screen against drug-resistant Staphylococci by researchers at E. I. Dupont de Nemours and Co in 1978 [78]. Chemical optimization was based upon both whole cell activity and in vitro protein synthesis. Improvements in potency were realized by numerous groups working in this class of molecules (summarized in [79]) but the breakthrough that propelled Linezolid to the status of a marketed drug was at Pfizer’s laboratories in Kalamazoo, Michigan. The primary target of these compounds was known to be protein synthesis, therefore toxicity problems arising from inhibiting mammalian protein synthesis were both predictable and frequent [78]. Early evidence from key compounds suggested strongly that the SAR (Structure Activity Relationships) and STR (Structure Toxicity Relationship) were distinct and that such toxicity could be minimized to produce a drug with acceptable toxicity. Although it was known that these compounds inhibited prokaryotic protein synthesis, their precise mechanism of action was only described in 2001. As a result, this discovery program was achieved by classical medicinal chemistry since the highly complex structure of the bacterial ribosome was not known at the time.
Linezolid received approval by the US FDA in April of 2000 for the treatment of severe Gram-positive skin infections (e.g., MRSA, methicillin-resistant Staphylcoccus aureus) and hospital-acquired pneumonia. Linezolid, and other oxazolidinones, bind to the 23S rRNA in the 50S subunit and prevent the binding of the initiating formyl-Met-tRNA [80]. By inhibiting this binding, oxazolidinones also prevent the formation of the intact 70S ribosome. Mutations in the 23S RNA that confer linezolid resistance (G2032A and G2447A) allow the site of binding to be mapped quite precisely to the region between the A (Acceptor) and P (Peptidyl transfer) sites on the 50S ribosomal subunit [81]. Linezolid has been successfully used off-label to treat MDR-infected tuberculosis patients (for example [82–84]), although the effects of extended linezolid therapy are still unclear.
PNU-100480 (originally U-100480) is a close structural analog of Linezolid developed by Upjohn, where the morpholino ring has been replaced by the thiolmorpholino group [85]. The sulfur atom of the thiomorpholine ring is rapidly oxidized first to the sulfoxide and eventually to the sulfone. The antimycobacterial activity of both the parent and the sulfoxide are nearly equal with reported MIC values of ~0.12 ug/ml, values comparable to isoniazid. Likewise in murine models PNU-10048 was more effective than Linezolid and approached INH in potency [86]. Curiously, a related oxazolidinone, eperezolid, showed almost no activity.
The identification of the oxazolidinone class of antibacterials represents a significant new addition to our antibacterial armementum, and the identification of PNU-100480 similarly bodes well in the development of new treatments for tuberculosis. TB has, of course, been an incidental bystander in the story of the development of oxazolidinones. Fortunately the ultimate broad-spectrum molecule (Linezolid) retains considerable activity against TB and mounting evidence of efficacy of this drug in highly drug-resistant patients, combined with two on-going prospective clinical trials (ClinicalTrials. gov Identifiers: NCT00727844 and NCT00664313) have dramatically increased enthusiasm for this class of molecules and the therapeutic potential of oxazolidinones for TB treatment. The extended courses of chemotherapy with Linezolid required for TB have already been shown to cause serious side effects [87]. Whether the toxicity associated with long-term administration of linezolid [88] can be avoided with PNU-100480 is unknown, but increased study is certainly warranted.
There are many other candidate oxazolidinones that could be developed for TB but these need to be reconsidered in light of the significant toxicology of these to date and they were not specifically developed with an extended dosing problem in mind. The recent solution of the co-crystal structure of Linezolid bound to the 50S subunit of the ribosome has enabled the possibility that further analogs could maximize differences between activity on the TB ribosome and activity on the mamallian ribosome to develop a much more highly selective agent with reduced toxicity specifically for the treatment of TB [89,90].
Conclusions and perspectives
The pipeline of promising agents for the treatment of TB has certainly advanced from where it was ten years ago but the glass is far from half-full. Statistically there is about a 10% chance that any given agent will advance from Phase II to larger Phase III studies and there are only two classes of new agents currently in Phase II. There is therefore a critical need for dramatically expanding efforts in TB drug discovery. The existing new agents also stand a negligible chance of slowing the rising epidemic of MDR, XDR and now TDR (totally drug-resistant) TB strains since, unless multiple agents can be introduced simultaneously, further development of resistance is inevitable.
It is not enough to simply continue to develop drugs against whole cells and optimize them based upon activity in mice (a notably poor surrogate for human TB that fails to predict the activity observed in clinical trials in humans [91]). Until we begin to unravel, in detail, the mechanisms involved in cell death and survival it will be difficult to prospectively select targets for structure-guided drug design. It is noteworthy that chemical biology has played an important role in this respect in understanding the mechanisms of killing of various bacteria [92] and chemical biologists are uniquely positioned to consider the mechanisms of cell death.
Figure 3.
Figure 6.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Reference number and short description, * important, **most important
- 1.Tsukamura M. A comparison of the time of in vivo resistance development of tubercle bacilli to rifampicin, kanamycin, ethionamide, lividomycin, and enviomycin (tuberactinomycin-N) in patients with chronic cavitary tuberculosis (author’s transl) Kekkaku. 1978;53:495–498. [PubMed] [Google Scholar]
- 2.Mitchison DA. The diagnosis and therapy of tuberculosis during the past 100 years. Am J Respir Crit Care Med. 2005;171:699–706. doi: 10.1164/rccm.200411-1603OE. [DOI] [PubMed] [Google Scholar]
- *3.Fox W, Mitchison DA. Short-course chemotherapy for tuberculosis. Lancet. 1976;2:1349–1350. doi: 10.1016/s0140-6736(76)91989-9. An authoritative history and discussion of the development of the multi-drug chemotherapy currently used in the treatment of Tuberculosis. [DOI] [PubMed] [Google Scholar]
- 4.Neff M. ATS, CDC, and IDSA update recommendations on the treatment of tuberculosis. Am Fam Physician. 2003;68:1854, 1857–1858, 1861–1852. [PubMed] [Google Scholar]
- 5.Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis. 2003;7:6–21. [PubMed] [Google Scholar]
- 6.Fox W. The chemotherapy of pulmonary tuberculosis: a review. Chest. 1979;76:785–796. doi: 10.1378/chest.76.6_supplement.785. [DOI] [PubMed] [Google Scholar]
- 7.Outbreak of multidrug-resistant tuberculosis--Texas, California, and Pennsylvania. MMWR Morb Mortal Wkly Rep. 1990;39:369–372. [PubMed] [Google Scholar]
- 8.Edlin BR, Tokars JI, Grieco MH, Crawford JT, Williams J, Sordillo EM, Ong KR, Kilburn JO, Dooley SW, Castro KG, et al. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 9.Dooley SW, Jarvis WR, Martone WJ, Snider DE., Jr Multidrug-resistant tuberculosis. Ann Intern Med. 1992;117:257–259. doi: 10.7326/0003-4819-117-3-257. [DOI] [PubMed] [Google Scholar]
- 10.Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs--worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 11.Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, Martin-Casabona N, Drobniewski F, Gilpin C, Havelkova M, Lepe R, et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007;13:380–387. doi: 10.3201/eid1303.061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DH, Kim HJ, Park SK, Kong SJ, Kim YS, Kim TH, Kim EK, Lee KM, Lee SS, Park JS, et al. Treatment outcomes and long-term survival in patients with extensively drug-resistant tuberculosis. Am J Respir Crit Care Med. 2008;178:1075–1082. doi: 10.1164/rccm.200801-132OC. [DOI] [PubMed] [Google Scholar]
- 13.Jeon CY, Hwang SH, Min JH, Prevots DR, Goldfeder LC, Lee H, Eum SY, Jeon DS, Kang HS, Kim JH, et al. Extensively drug-resistant tuberculosis in South Korea: risk factors and treatment outcomes among patients at a tertiary referral hospital. Clin Infect Dis. 2008;46:42–49. doi: 10.1086/524017. [DOI] [PubMed] [Google Scholar]
- **14.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. A detailed report of an outbreak of extensively drug-resistant Tuberculosis in the Kwa-Zulu Natal province of South Africa. [DOI] [PubMed] [Google Scholar]
- 15.Ma Z, Lienhardt C. Toward an optimized therapy for tuberculosis? Drugs in clinical trials and in preclinical development. Clin Chest Med. 2009;30:755–768. ix. doi: 10.1016/j.ccm.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Spigelman MK. New tuberculosis therapeutics: a growing pipeline. J Infect Dis. 2007;196 (Suppl 1):S28–34. doi: 10.1086/518663. [DOI] [PubMed] [Google Scholar]
- 17.Barry CE., 3rd Preclinical candidates and targets for tuberculosis therapy. Curr Opin Investig Drugs. 2001;2:198–201. [PubMed] [Google Scholar]
- **18.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. The first report on the discovery, activity, specificity and target of R207910, a novel diarylquinoline with exquisite activity against Tuberculosis. [DOI] [PubMed] [Google Scholar]
- 19.Upadhayaya RS, Vandavasi JK, Vasireddy NR, Sharma V, Dixit SS, Chattopadhyaya J. Design, synthesis, biological evaluation and molecular modelling studies of novel quinoline derivatives against Mycobacterium tuberculosis. Bioorg Med Chem. 2009;17:2830–2841. doi: 10.1016/j.bmc.2009.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Huitric E, Verhasselt P, Andries K, Hoffner SE. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2007;51:4202–4204. doi: 10.1128/AAC.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelber R, Andries K, Paredes RM, Andaya CE, Burgos J. The diarylquinoline R207910 is bactericidal against Mycobacterium leprae in mice at low dose and administered intermittently. Antimicrob Agents Chemother. 2009;53:3989–3991. doi: 10.1128/AAC.00722-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, Andersson DI. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 54:1022–1028. doi: 10.1128/AAC.01611-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, Krab K, Vergauwen K, Guillemont J, Andries K, Lill H, Koul A, Bald D. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob Agents Chemother. 2009;53:1290–1292. doi: 10.1128/AAC.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jonge MR, Koymans LH, Guillemont JE, Koul A, Andries K. A computational model of the inhibition of Mycobacterium tuberculosis ATPase by a new drug candidate R207910. Proteins. 2007;67:971–980. doi: 10.1002/prot.21376. [DOI] [PubMed] [Google Scholar]
- 25.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol. 2007;3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 26.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Stelten J, Silva F, Belin D, Silhavy TJ. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. Description of a reliable and reproducible anaerobic model of persistence. [DOI] [PubMed] [Google Scholar]
- 29.Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, Gohlmann HW, Willebrords R, Poncelet A, Guillemont J, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- 30.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibrahim M, Truffot-Pernot C, Andries K, Jarlier V, Veziris N. Sterilizing activity of R207910 (TMC207)-containing regimens in the murine model of tuberculosis. Am J Respir Crit Care Med. 2009;180:553–557. doi: 10.1164/rccm.200807-1152OC. [DOI] [PubMed] [Google Scholar]
- 32.Veziris N, Ibrahim M, Lounis N, Chauffour A, Truffot-Pernot C, Andries K, Jarlier V. A once-weekly R207910-containing regimen exceeds activity of the standard daily regimen in murine tuberculosis. Am J Respir Crit Care Med. 2009;179:75–79. doi: 10.1164/rccm.200711-1736OC. [DOI] [PubMed] [Google Scholar]
- 33.Lounis N, Gevers T, Van Den Berg J, Andries K. Impact of the interaction of R207910 with rifampin on the treatment of tuberculosis studied in the mouse model. Antimicrob Agents Chemother. 2008;52:3568–3572. doi: 10.1128/AAC.00566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim M, Andries K, Lounis N, Chauffour A, Truffot-Pernot C, Jarlier V, Veziris N. Synergistic activity of R207910 combined with pyrazinamide against murine tuberculosis. Antimicrob Agents Chemother. 2007;51:1011–1015. doi: 10.1128/AAC.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji B, Chauffour A, Andries K, Jarlier V. Bactericidal activities of R207910 and other newer antimicrobial agents against Mycobacterium leprae in mice. Antimicrob Agents Chemother. 2006;50:1558–1560. doi: 10.1128/AAC.50.4.1558-1560.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diacon AH, Pym A, Grobusch M, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, et al. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med. 2009;360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 37.Barry CE., 3rd Unorthodox approach to the development of a new antituberculosis therapy. N Engl J Med. 2009;360:2466–2467. doi: 10.1056/NEJMe0903012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barry CE, 3rd, Boshoff HI, Dowd CS. Prospects for clinical introduction of nitroimidazole antibiotics for the treatment of tuberculosis. Curr Pharm Des. 2004;10:3239–3262. doi: 10.2174/1381612043383214. [DOI] [PubMed] [Google Scholar]
- 39.Ashtekar DR, Costa-Perira R, Nagrajan K, Vishvanathan N, Bhatt AD, Rittel W. In vitro and in vivo activities of the nitroimidazole CGI 17341 against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1993;37:183–186. doi: 10.1128/aac.37.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, Anderson SW, Towell JA, Yuan Y, McMurray DN, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. Discovery and description of the antimycobacterial properties of the nitroimidazole PA-824. [DOI] [PubMed] [Google Scholar]
- 41.Tyagi S, Nuermberger E, Yoshimatsu T, Williams K, Rosenthal I, Lounis N, Bishai W, Grosset J. Bactericidal activity of the nitroimidazopyran PA-824 in a murine model of tuberculosis. Antimicrob Agents Chemother. 2005;49:2289–2293. doi: 10.1128/AAC.49.6.2289-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenaerts AJ, Gruppo V, Marietta KS, Johnson CM, Driscoll DK, Tompkins NM, Rose JD, Reynolds RC, Orme IM. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother. 2005;49:2294–2301. doi: 10.1128/AAC.49.6.2294-2301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nuermberger E, Tyagi S, Tasneen R, Williams KN, Almeida D, Rosenthal I, Grosset JH. Powerful bactericidal and sterilizing activity of a regimen containing PA-824, moxifloxacin, and pyrazinamide in a murine model of tuberculosis. Antimicrob Agents Chemother. 2008;52:1522–1524. doi: 10.1128/AAC.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji B, Lefrancois S, Robert J, Chauffour A, Truffot C, Jarlier V. In vitro and in vivo activities of rifampin, streptomycin, amikacin, moxifloxacin, R207910, linezolid, and PA-824 against Mycobacterium ulcerans. Antimicrob Agents Chemother. 2006;50:1921–1926. doi: 10.1128/AAC.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tasneen R, Tyagi S, Williams K, Grosset J, Nuermberger E. Enhanced bactericidal activity of rifampin and/or pyrazinamide when combined with PA-824 in a murine model of tuberculosis. Antimicrob Agents Chemother. 2008;52:3664–3668. doi: 10.1128/AAC.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi KP, Bair TB, Bae YM, Daniels L. Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG. J Bacteriol. 2001;183:7058–7066. doi: 10.1128/JB.183.24.7058-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi KP, Kendrick N, Daniels L. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F(420) and FO biosynthesis. J Bacteriol. 2002;184:2420–2428. doi: 10.1128/JB.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manjunatha UH, Boshoff H, Dowd CS, Zhang L, Albert TJ, Norton JE, Daniels L, Dick T, Pang SS, Barry CE., 3rd Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:431–436. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. Description of the mechanism of activation and likely mechanism of action of PA-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hickey MJ, Arain TM, Shawar RM, Humble DJ, Langhorne MH, Morgenroth JN, Stover CK. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob Agents Chemother. 1996;40:400–407. doi: 10.1128/aac.40.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim P, Zhang L, Manjunatha UH, Singh R, Patel S, Jiricek J, Keller TH, Boshoff HI, Barry CE, Dowd CS. Structure-activity relationships of antitubercular nitroimidazoles. 1. Structural features associated with aerobic and anaerobic activities of 4- and 5-nitroimidazoles. J Med Chem. 2009;52:1317–1328. doi: 10.1021/jm801246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manjunatha U, Boshoff HI, Barry CE. The mechanism of action of PA-824: Novel insights from transcriptional profiling. Commun Integr Biol. 2009;2:215–218. doi: 10.4161/cib.2.3.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki H, Haraguchi Y, Itotani M, Kuroda H, Hashizume H, Tomishige T, Kawasaki M, Matsumoto M, Komatsu M, Tsubouchi H. Synthesis and antituberculosis activity of a novel series of optically active 6-nitro-2,3-dihydroimidazo[2,1-b]oxazoles. J Med Chem. 2006;49:7854–7860. doi: 10.1021/jm060957y. [DOI] [PubMed] [Google Scholar]
- *55.Lee RE, Protopopova M, Crooks E, Slayden RA, Terrot M, Barry CE., 3rd Combinatorial lead optimization of [1,2]-diamines based on ethambutol as potential antituberculosis preclinical candidates. J Comb Chem. 2003;5:172–187. doi: 10.1021/cc020071p. The combinatorial synthesis of a library of di-substituted diamines and the analysis of their in vitro activity against Tuberculosis. [DOI] [PubMed] [Google Scholar]
- 56.Protopopova M, Hanrahan C, Nikonenko B, Samala R, Chen P, Gearhart J, Einck L, Nacy CA. Identification of a new antitubercular drug candidate, SQ109, from a combinatorial library of 1,2-ethylenediamines. J Antimicrob Chemother. 2005;56:968–974. doi: 10.1093/jac/dki319. [DOI] [PubMed] [Google Scholar]
- 57.Jia L, Tomaszewski JE, Hanrahan C, Coward L, Noker P, Gorman G, Nikonenko B, Protopopova M. Pharmacodynamics and pharmacokinetics of SQ109, a new diamine-based antitubercular drug. Br J Pharmacol. 2005;144:80–87. doi: 10.1038/sj.bjp.0705984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia L, Noker PE, Coward L, Gorman GS, Protopopova M, Tomaszewski JE. Interspecies pharmacokinetics and in vitro metabolism of SQ109. Br J Pharmacol. 2006;147:476–485. doi: 10.1038/sj.bjp.0706650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen P, Gearhart J, Protopopova M, Einck L, Nacy CA. Synergistic interactions of SQ109, a new ethylene diamine, with front-line antitubercular drugs in vitro. J Antimicrob Chemother. 2006;58:332–337. doi: 10.1093/jac/dkl227. [DOI] [PubMed] [Google Scholar]
- 60.Nikonenko BV, Protopopova M, Samala R, Einck L, Nacy CA. Drug therapy of experimental tuberculosis (TB): improved outcome by combining SQ109, a new diamine antibiotic, with existing TB drugs. Antimicrob Agents Chemother. 2007;51:1563–1565. doi: 10.1128/AAC.01326-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goude R, Amin AG, Chatterjee D, Parish T. The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2009;53:4138–4146. doi: 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belanger AE, Besra GS, Ford ME, Mikusova K, Belisle JT, Brennan PJ, Inamine JM. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci U S A. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alland D, Steyn AJ, Weisbrod T, Aldrich K, Jacobs WR., Jr Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J Bacteriol. 2000;182:1802–1811. doi: 10.1128/jb.182.7.1802-1811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 65.Meng Q, Luo H, Chen Y, Wang T, Yao Q. Synthesis of novel [1,2]-diamines with antituberculosis activity. Bioorg Med Chem Lett. 2009;19:5372–5375. doi: 10.1016/j.bmcl.2009.07.126. [DOI] [PubMed] [Google Scholar]
- 66.Onajole OK, Govender P, Helden PD, Kruger HG, Maguire GE, Wiid I, Govender T. Synthesis and evaluation of SQ109 analogues as potential anti-tuberculosis candidates. Eur J Med Chem. doi: 10.1016/j.ejmech.2010.01.046. [DOI] [PubMed] [Google Scholar]
- 67.Meng Q, Luo H, Liu Y, Li W, Zhang W, Yao Q. Synthesis and evaluation of carbamate prodrugs of SQ109 as antituberculosis agents. Bioorg Med Chem Lett. 2009;19:2808–2810. doi: 10.1016/j.bmcl.2009.03.091. [DOI] [PubMed] [Google Scholar]
- 68.Iland CN, Baines S. The Effect of Penicillin on the Tubercle Bacillus - Tubercle Penicillinase. Journal of Pathology and Bacteriology. 1949;61:329–335. [Google Scholar]
- 69.Cynamon MH, Palmer GS. In vitro activity of amoxicillin in combination with clavulanic acid against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1983;24:429–431. doi: 10.1128/aac.24.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **70.Chambers HF, Kocagoz T, Sipit T, Turner J, Hopewell PC. Activity of amoxicillin/clavulanate in patients with tuberculosis. Clin Infect Dis. 1998;26:874–877. doi: 10.1086/513945. The first demonstration of the efficacy of a β-lactam and β-lactamase inhibitor to treat an MDR-TB-infected patient. [DOI] [PubMed] [Google Scholar]
- **71.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. The determination of the genome sequence of Mycobacterium tuberculosis. [DOI] [PubMed] [Google Scholar]
- 72.Flores AR, Parsons LM, Pavelka MS., Jr Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology. 2005;151:521–532. doi: 10.1099/mic.0.27629-0. [DOI] [PubMed] [Google Scholar]
- 73.Voladri RK, Lakey DL, Hennigan SH, Menzies BE, Edwards KM, Kernodle DS. Recombinant expression and characterization of the major beta-lactamase of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1375–1381. doi: 10.1128/aac.42.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang F, Cassidy C, Sacchettini JC. Crystal structure and activity studies of the Mycobacterium tuberculosis beta-lactamase reveal its critical role in resistance to beta-lactam antibiotics. Antimicrob Agents Chemother. 2006;50:2762–2771. doi: 10.1128/AAC.00320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hugonnet JE, Blanchard JS. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry. 2007;46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *76.Tremblay LW, Hugonnet JE, Blanchard JS. Structure of the covalent adduct formed between Mycobacterium tuberculosis beta-lactamase and clavulanate. Biochemistry. 2008;47:5312–5316. doi: 10.1021/bi8001055. Mechanistic and structural rationale for the specific inhibitory effect of clavulanate on the TB β-lactamase (BlaC) [DOI] [PubMed] [Google Scholar]
- *77.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. The demonstration that the combination of clavulanate and meropenem (a carbapenem β-lactam) is rapidly bactericidal against drug-sensitive and extensively drug-resistant strains of TB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brickner SJ, Barbachyn MR, Hutchinson DK, Manninen PR. Linezolid (ZYVOX), the first member of a completely new class of antibacterial agents for treatment of serious gram-positive infections. J Med Chem. 2008;51:1981–1990. doi: 10.1021/jm800038g. [DOI] [PubMed] [Google Scholar]
- 79.Renslo AR, Luehr GW, Gordeev MF. Recent developments in the identification of novel oxazolidinone antibacterial agents. Bioorg Med Chem. 2006;14:4227–4240. doi: 10.1016/j.bmc.2006.01.068. [DOI] [PubMed] [Google Scholar]
- *80.Patel U, Yan YP, Hobbs FW, Jr, Kaczmarczyk J, Slee AM, Pompliano DL, Kurilla MG, Bobkova EV. Oxazolidinones mechanism of action: inhibition of the first peptide bond formation. J Biol Chem. 2001;276:37199–37205. doi: 10.1074/jbc.M102966200. [DOI] [PubMed] [Google Scholar]
- 81.Bobkova EV, Yan YP, Jordan DB, Kurilla MG, Pompliano DL. Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones. J Biol Chem. 2003;278:9802–9807. doi: 10.1074/jbc.m209249200. The identification of PNU-100840, a powerful analogue of the oxazolidinone, Linezolid, with potent activity against TB. [DOI] [PubMed] [Google Scholar]
- 82.von der Lippe B, Sandven P, Brubakk O. Efficacy and safety of linezolid in multidrug resistant tuberculosis (MDR-TB)--a report of ten cases. J Infect. 2006;52:92–96. doi: 10.1016/j.jinf.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 83.Fortun J, Martin-Davila P, Navas E, Perez-Elias MJ, Cobo J, Tato M, De la Pedrosa EG, Gomez-Mampaso E, Moreno S. Linezolid for the treatment of multidrug-resistant tuberculosis. J Antimicrob Chemother. 2005;56:180–185. doi: 10.1093/jac/dki148. [DOI] [PubMed] [Google Scholar]
- 84.Condos R, Hadgiangelis N, Leibert E, Jacquette G, Harkin T, Rom WN. Case series report of a linezolid-containing regimen for extensively drug-resistant tuberculosis. Chest. 2008;134:187–192. doi: 10.1378/chest.07-1988. [DOI] [PubMed] [Google Scholar]
- 85.Barbachyn MR, Hutchinson DK, Brickner SJ, Cynamon MH, Kilburn JO, Klemens SP, Glickman SE, Grega KC, Hendges SK, Toops DS, et al. Identification of a novel oxazolidinone (U-100480) with potent antimycobacterial activity. J Med Chem. 1996;39:680–685. doi: 10.1021/jm950956y. [DOI] [PubMed] [Google Scholar]
- 86.Cynamon MH, Klemens SP, Sharpe CA, Chase S. Activities of several novel oxazolidinones against Mycobacterium tuberculosis in a murine model. Antimicrob Agents Chemother. 1999;43:1189–1191. doi: 10.1128/aac.43.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vinh DC, Rubinstein E. Linezolid: a review of safety and tolerability. J Infect. 2009;59 (Suppl 1):S59–74. doi: 10.1016/S0163-4453(09)60009-8. [DOI] [PubMed] [Google Scholar]
- 88.Sood R, Bhadauriya T, Rao M, Gautam R, Malhotra S, Barman TK, Upadhyay DJ, Rattan A. Antimycobacterial activities of oxazolidinones: a review. Infect Disord Drug Targets. 2006;6:343–354. doi: 10.2174/187152606779025860. [DOI] [PubMed] [Google Scholar]
- 89.Wimberly BT. The use of ribosomal crystal structures in antibiotic drug design. Curr Opin Investig Drugs. 2009;10:750–765. [PubMed] [Google Scholar]
- 90.Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, Duffy EM. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 91.Mitchison DA, Chang KC. Experimental models of tuberculosis: can we trust the mouse? Am J Respir Crit Care Med. 2009;180:201–202. doi: 10.1164/rccm.200905-0708ED. [DOI] [PubMed] [Google Scholar]
- 92.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]