SUMMARY
Cell adhesion is a key feature in the regulation of many biological processes. In the budding yeast Saccharomyces cerevisiae, Flo11p is the major adhesion molecule that controls filamentous growth [1–3] and the expansion of interconnected cells in mats or biofilms [4]. We show here that Flo11p is shed from cells. Flo11p shedding attenuated adherence and contributed to the overall balance in adherence properties that was optimal for filamentous growth and mat formation. Shed Flo11p comprised an essential component of a fluid layer surrounding yeast mats that may be functionally analogous to the mucus secretions of higher eukaryotes. Genome-wide secretion profiling of Flo11p identified new regulatory proteins, including the furin protease Kex2p, which was required for cleavage and maturation of the Flo11p protein. Secreted mucin-like proteins may play unexpected roles in the adherence properties and virulence of microbial pathogens.
Keywords: filamentous growth, microbial mats, invasive growth, pseudohyphal growth, extracellular matrix, flocculation, anti-adhesion, Flo11
RESULTS
Flo11p is Shed From Cells
The yeast adhesion molecule Flo11p/Muc1p is a typical fungal adhesion molecule that is homologous to pathogenic adhesins [5], which contains a putative N-terminal signal sequence and transmembrane domain, an internal Ser/Thr/Pro-rich repeat region, and a C-terminal glycosylphosphatidylinositol (GPI) anchor. The FLO11 gene is regulated by an unusually large promoter where multiple signal transduction pathways converge, including RAS-cAMP-PKA, TOR, and a Cdc42p-dependent MAPK pathway [6]. Changes in the FLO11 gene/promoter can have dramatic effects on cell-surface variation [7, 8] and can induce novel cellular properties, including the formation of buoyant aggregates of cells on broth surfaces [9]. We previously showed that two signaling mucins that show homology with Flo11p - Msb2p and Hkr1p [10, 11] - are shed from cells [12, 13]. This discovery provoked the questions of what other yeast mucin-like proteins are secreted and what the functional roles of shed mucins might be.
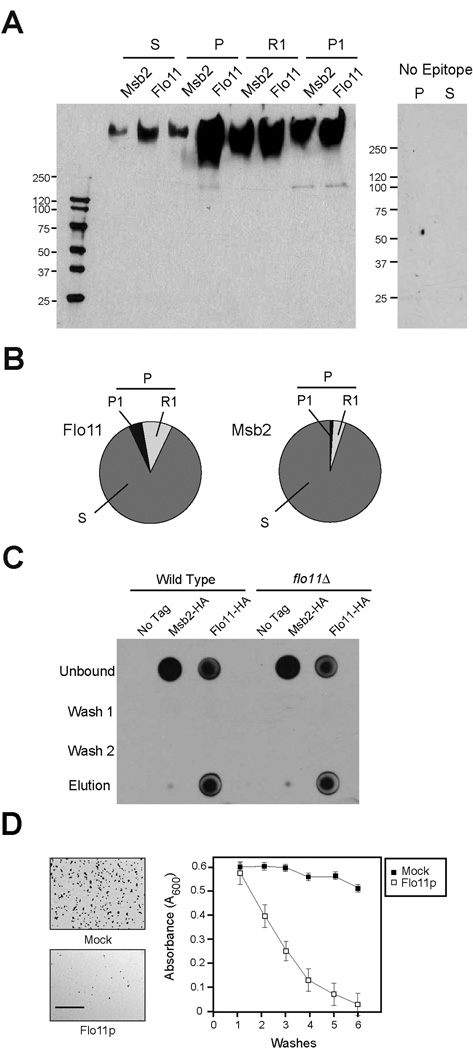
To determine whether Flo11p is shed from cells, a functional epitope fusion of Flo11p was generated (Flo11p-HA). The Flo11p-HA protein was identified in culture supernatants by immunoblot analysis (S, Fig. 1A) indicating that it is a shed protein. The properties of shed Flo11p were examined. Densitometric analysis of immunoblots and normalization to input volumes showed that most of the total protein is in the shed form (~85% S, Fig. 1B). In addition, most of the cell-associated Flo11p could be released from cells by incubation in a neutral buffer (~70% R1, Fig. 1, A and B). Immunofluorescence microscopy confirmed that Flo11p-HA was evenly released from cell surfaces by washing (Fig. S1) [14]. An in vitro binding assay with shed Flo11p showed that shed Flo11p bound weakly to cells in trans (Fig. 1C), equivalently to wild-type cells and the flo11Δ mutant (Fig. 1C), indicating that binding does not result from homotypic interactions in line with a previous report [15]. Another shed mucin-like protein had similar properties (Msb2p, Fig. 1, A and B), except that it failed to adhere to cells in trans (Fig. 1C), indicative of a specific adherence property associated with the shed Flo11p molecule.
Figure 1. Properties of shed Flo11p.
A) Immunoblot analysis of supernatant (S) and pellet (P) fractions derived from cells expressing Flo11p-HA (PC2043) or Msb2p-HA (PC999). For panels A to C, strains were grown for 16 h in YEPD medium. Cells were concentrated by centrifugation and supernatants were used in immunoblotting with anti-HA antibodies or to test the properties of shed Flo11p. For panel A, the pellet fraction was further separated into R1 and P1 fractions by washing cell pellets in a neutral buffer (50 mM Tris pH 7) at 4° C for 30 min. At right, control S and P fractions from a non-tagged strain (PC538). B) Quantitation of shed (S), released (R1), and cell-associated (P, P1) Flo11p-HA and Msb2p-HA proteins. Band intensities in Fig. 1A were compared by densitometry and normalized to total input volumes (S, 10 mls, P, 200 µl, S1, 400 µl, P1, 100 µl). C) In vitro binding assay. Equal volumes of cells of untagged strains of wild type (PC538) and the flo11Δ (PC1029) mutant were incubated with equal volumes of conditioned media from wild type strains (non-tagged, PC538; Flo11p-HA, PC2043 and Msb2p-HA, PC999). After 2 h incubation at 30°C, cells were centrifuged and the supernatant fraction was used as the ‘Unbound’ fraction. Cells were washed twice with water (Wash 1 and 2) and treated with buffer (50mM Tris pH 9.5, 10mM DTT) to remove bound proteins. Eluates from this treatment along with washed and unbound fractions were spotted (50 µl) onto a nitrocellulose filter and examined by immunoblot analysis. D) Polystyrene surfaces were pre-coated with purified Flo11p (open squares) or water (Mock, filled squares). Cells were applied to pre-coated surfaces, which were sequentially washed with water (washes). Released cells were measured by absorbance at OD (A600) after each wash. The experiment was performed in duplicate; error bars represent the standard deviation between trials. At left, low magnification microscopic images showing cells adhering to surfaces pre-coated with water (upper panel) or purified shed Flo11p (lower panel). Bar = 500 microns.
We hypothesized that shed Flo11p, which resembles mucin glycoproteins of higher organisms [16], may coat cells and surfaces and play a role in cellular lubrication. Yeast cells adhere to plastic surfaces in a Flo11p-dependent manner [4]. Pretreatment of plastic surfaces with purified shed Flo11p (Fig. S2) reduced the adherence of cells to plastic (Fig. 1D). Because binding of cells to surfaces requires Flo11p [4], shed Flo11p may compete with cell-associated Flo11p to prevent cellular adherence.
The Impact of Flo11p Shedding on Mat Expansion and Invasive Growth
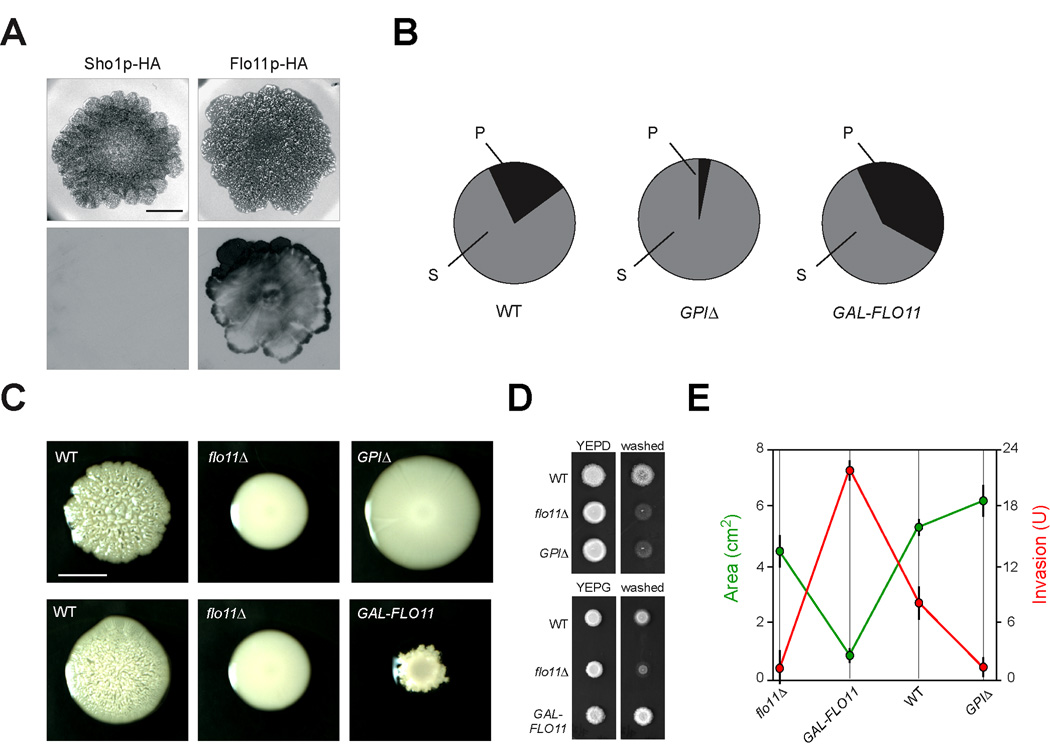
Many species of microorganisms congregate in multicellular communities called biofilms or microbial mats, in which cells adhere to surfaces and each other to form interconnected, multi-dimensional specialized structures [17, 18]. Budding yeast forms mats on semisolid surfaces, a foraging response in which cells expand colonially in a Flo11p-dependent manner through an unknown mechanism [4]. We tested whether Flo11p shedding influenced mat expansion. Immunoblots of mats grown on filters atop semisolid agar media showed that Flo11p-HA is shed from mats (Fig. 2A), in comparison to a control protein (Fig. 2A, Sho1p-HA). Flo11p was shed in a non-uniform pattern, providing a unique view into the complexity of these communities (Fig. S3). Mats that shed Flo11p abundantly (GPIΔ, Fig. 2B and Fig. 4E, see below) showed enhanced expansion compared to wild-type and flo11Δ mutant mats (Fig. 2C). This effect was subtle and might indicate that a combination of shed and cell-associated Flo11p is required for optimal expansion. Mats that had reduced Flo11p shedding (GAL-FLO11, Fig. 2B) expanded poorly (Fig. 2C), more so than mats lacking Flo11p entirely (flo11Δ, Fig. 2C). The expansion defect was specific to this condition and did not result from a general growth defect of the GAL-FLO11 strain (e.g see Fig. 2D).
Figure 2. Altering Flo11p shedding affects mat expansion and invasive growth.
A) Mats expressing Flo11p-HA (PC2043) or Sho1p-HA (PC1702), a transmembrane protein with an epitope tag in the cytoplasmic domain, were grown on nitrocellulose filters on low-agar YEPD medium (0.3% agar) for 14 d. Cells were rinsed off the filters, which were immunoblotted using anti-HA antibodies. Bar, 1 cm. B) Quantitation of shed (S) and cell-associated (P) fractions of Flo11p-HA in wild type (PC2043), GAL-FLO11 (PC2712), and FLO11GPIΔ (GPIΔ, PC3422) strains. Band intensities from immunoblot analysis using anti-HA antibodies were compared by densitometry and normalized to total input volumes (S, 10 mls, P, 200 µl). C) Example of the differences in mat expansion for the indicated strains in panel 3B. Top panel, YEPD bottom panel, YEP-GAL. The differences in mat size in terms of area (cm2) of strains exhibiting different Flo11p levels and S/P ratios. Strains were grown for 4d on YEP-GAL medium (0.3% agar) to induce GAL-FLO11 expression in strain PC2712; or 4d in YEPD medium (0.3% agar), Bar, 0.5 cm. D) The effect of overexpression of FLO11 or loss of the GPI anchor on agar invasion by the plate-washing assay. Strains in Fig 2C were grown for 3 d in YEPD or YEP-GAL (YEPG) media and washed to reveal invaded cells. E) Graph of mat expansion and invasive growth of strains carrying different versions of Flo11p. For mat expansion, mat areas (in cm2) was determined for the strains described in panel 2C in triplicate (n = 3). The flo11Δ strain grown in YEPD was used. Error bars represent the standard deviation between experiments. For invasive growth, densitometry of the invasive patch was determined by ImageJ after background subtraction and correction for colony size (prewash). The analysis was performed from two independent replicates (n=2) and expressed as units (U) of invasion. Strains were plotted in order of increasing S/P ratios. A two-sample paired t-Test showed the statistical difference for wild type (P=0.08), GPIΔ (P=0.002) and GAL-FLO11 (P=0.007) mat areas compared to the flo11Δ mutant.
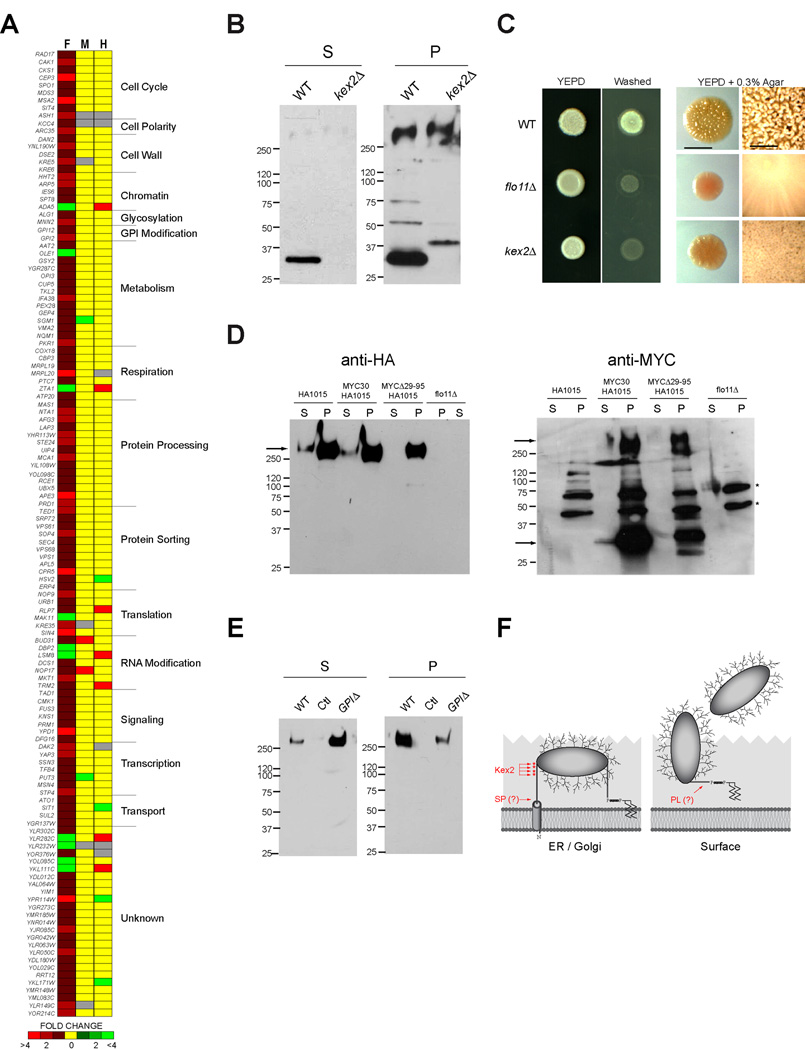
Figure 4. Kex2p and other genes influence Flo11p shedding.
A) The secretion profile of Flo11p-HA (F) compared to the profiles of Msb2p-HA (M) and Hkr1p-HA (H). Key refers to fold secretion of mucins from strains overexpressing the indicated genes. Functional categories were determined from Saccharomyces Genome Database. B) Immunoblots with anti-Myc antibodies of Flo11p-Myc@30aa showing the absence of a processing intermediate (~33 kDa) in the kex2Δ (PC3700) mutant compared to wild-type cells (PC3425). S, supernatant; P, pellet. The ~33 kDa band is indicative of processing at position ~120 aa residues. C) Left, plate-washing assay. Wild type (PC2043), flo11Δ (PC1029), and kex2Δ (PC3420) strains were spotted onto YEPD medium for 3 d. The plate was washed in a stream of water (Washed) to reveal invaded cells. Right, mat formation. Strains were spotted onto YEPD medium (0.3% agar) atop a filter for 7 d and photographed with transmitted light. Bar, 0.2 cm. At right, close up of colony morphologies of mat interiors. Bar, 0.05 cm. D) Wild type (PC2043, Flo11p-HA), flo11Δ (PC1029), Myc-Flo11p-HA (HA@1015aa and Myc@30aa PC3425) and MycΔ29–95-Flo11p-HA (HA@1015aa and Myc@Δ29–95aa PC3426) were analyzed for Flo11p shedding by immunoblot analysis. At left, immunoblot with anti-HA antibodies. At right, immunoblot with anti-Myc antibodies, which shows processing intermediates. Arrows indicate the expected low and high molecular weight bands. Asterisks represent background bands. E) The effect of loss of the GPI anchor on Flo11p-HA shedding. Equal amounts of protein were loaded in each lane. Immunoblots with anti-HA antibodies of wild type (PC2043), non-tagged (PC538, Ctl), and a strain lacking the GPI anchor (PC3422). F) Model for posttranslational processing events in Flo11p. Diagram of the Flo11p protein with N-terminal transmembrane (TM) domain and C-terminal GPI attachment site is shown. The protein is shown as heavily glycosylated. Two processing sites are shown: site 1; 27 to 30 residues; site 2; 80–320 residues. Dibasic potential Kex2p cleavage sites (289, 297, 298 and 308aa) are marked by red asterisks. Processing by Kex2p likely occurs in the Golgi, and processing by a phospholipase (PL) or other enzyme is likely also required for release of the protein from the cell.
Flo11p is also required for a distinct foraging response called filamentous/invasive/pseudohyphal growth, in which cells remain connected to each other in branched filaments that can invade into substrates [1–3]. Flo11p is thought to mediate the adhesive contacts between cells to promote filament formation. As expected, the reduction in cell-associated Flo11p (GPIΔ) resulted in an agar invasion defect that was comparable to the flo11Δ mutant (Fig. 2D). Increasing the levels of cell-associated Flo11p maximized agar invasion (GAL-FLO11, Fig 2D). Mat expansion and invasive growth were quantified in strains with altered Flo11p shedding. Direct comparison showed that reducing Flo11p shedding maximized invasive growth at the cost of expansion (Fig. 2E, GAL-FLO11 red and green data points). Increasing Flo11p shedding promoted expansion at the expense of invasive growth (Fig. 2E, GPIΔ red and green data points). Wild-type cells with intermediate levels of shedding had intermediate expansion and invasive growth (Fig. 2E). These data suggest that wild-type cells maintain an overall balance in Flo11p adherence, such that different foraging responses are optimized but not maximized. Recalibration of Flo11p-mediated adherence through changes in shedding, transcriptional regulation, and epigenetic mechanisms may alter that balance to differentially promote specific responses in different environments that cells may encounter.
Shed Flo11p Is An Essential Component of a Secreted Fluid in Mats
To further explore the role of Flo11p shedding in microbial communities, mat behaviors were examined in more detail. We found that mats produce an encapsulating fluid that extended beyond the perimeter (Fig. 3A). The fluid contained abundant quantities of Flo11p and other shed mucins (Fig. 3B). The amount of fluid was dependent on Flo11p and was reduced >10-fold in the flo11Δ mutant (Fig. 3C). The fluid might result from water absorption by shed Flo11p molecules, which are hydrophilic (Fig. 3D), similar to the gel-forming properties of some mammalian mucins [19] or by exclusion of fluid from mats as a result of Flo11p-dependent cell-cell adhesion, as cells expressing Flo11p possess hydrophobic properties [4]. An alternative possibility is that the fluid is produced by degradation/solubilization of the agar matrix by secreted enzymes. Fluid derived from mats did not deform the agar surface, and secreted enzymes that degrade polysaccharides, like Pgu1p [20], were not required for mat expansion or to produce the fluid boundary (Fig. 3C). As expected, fluid production was dependent on the MAPK pathway that regulates FLO11 expression (ste12Δ; Fig. 3C). The fluid may promote hydration and the diffusion of materials such as nutrients and small molecules throughout the biofilm, including molecules that function in cellular communication and quorum sensing [21].
Figure 3. Flo11p comprises an essential component of a fluid layer that surrounds yeast mats.
A) Wild-type mats (PC538) were grown for 7d on YEPD (0.3% agar) and examined by light microscopy. The arrow points to the extracellular fluid around the mat. Bar, 200 µM. B) Flo11p and Msb2p are abundantly shed in mat exudates. Mats expressing Flo11p-HA (PC2043) and Msb2p-HA (PC999) were grown on YEPD medium (0.3% agar) for 12 d. Plates were slightly tilted to collect the fluid produced by mats, without otherwise disturbing the mats. The fluid was spun down to remove cells and 10 µl of exudate (EX) was spotted onto nitrocellulose membranes. As controls, the above strains were grown for 16 h in liquid YEPD medium. Supernatants (S) were collected and 50 µl was spotted onto the same nitrocellulose filter. Spots were allowed to dry and the filter was probed with anti-HA antibodies. Darker exposures showed Msb2p-HA and Flo11p-HA in S fractions (not shown). C) Fluid was collected from wild-type (PC538), flo11Δ (PC1029), ste12Δ (which encodes a transcription factor for the filamentation MAPK pathway, PC2382), and pgu1Δ (PC1519) mats grown for 14 d on YEPD (0.3% agar). Fluid levels were adjusted to total biofilm mass (in mg) and plotted. The experiment was performed in two independent trials (n = 2); error bars represent the standard deviation between trials. D) Strains PC2043 (Flo11p-HA), PC999 (Msb2p-HA), and PC538 (Control) were grown for 16 h in YEPD medium. Conditioned medium was separated from cells by centrifugation, and 100 µl of medium was mixed with 1 ml octane by vortexing. Hydrophilic (A) and octane (O) layers were separated by centrifugation at 13,000 rpm for 5 min, and mucins were visualized by spotting on nitrocellulose membranes that were probed by anti-HA antibodies and quantitated by ImageJ analysis and spot intensities (Intensity) were plotted. The experiment was performed in duplicate (n = 2); error bars represent the standard deviation between experiments.
Flo11p Shedding Requires Cleavage at Multiple Sites
We investigated the molecular basis of Flo11p secretion. A high-throughput screening approach called secretion profiling [22] was used to identify new regulators of Flo11p secretion. The secretion profile of Flo11p was determined using a genomic overexpression collection [23] and compared to the secretion profiles of two other shed proteins, Msb2p and Hkr1p (Fig. 4A). Comparative genomic secretion profiling identified few common genes (Fig. 4A), which indicates that mucin shedding in this organism is highly protein specific. Comparative secretion profiling identified known transcriptional regulators of FLO11 expression and more than one hundred new Flo11p regulatory proteins (Table S2). Many of these proteins function at the post-translational level (Fig. 4A) and have functions in protein modification (including GPI anchor modification), protein processing, and protein trafficking.
Several candidate proteases were identified by secretion profiling and by direct testing that influenced Flo11p secretion (Fig. 4A, Protein Processing). Genes encoding candidate proteases were deleted in a wild-type strain of the Σ1278b background, and mutants were assessed for effects on Flo11p shedding, mat expansion, and invasive growth. The furin protease Kex2p, which has an established function in the cleavage of pro-proteins in the Golgi apparatus [24, 25], was required for Flo11p cleavage (Fig. 4B). Specifically, a dual-tagged fusion of Flo11p, Myc-Flo11p-HA, in which the MYC epitope was inserted at 30 amino acids in the Flo11p-HA molecule by homologous recombination, produced a low-molecular weight product (~33 kDa with anti-MYC antibodies), indicative of cleavage at ~120 amino acids in the N-terminus of the protein (S, Fig. 4B). Kex2p was required for the production of this processing intermediate (Fig. 4B). Kex2p was also required for Flo11p-dependent invasive growth (Fig. 4C, left panels) and mat expansion (Fig. 4C, right panels), indicating that Kex2p processes Flo11p to its mature form. Kex2p may directly cleave Flo11p at one of four dibasic sites in the N-terminus or indirectly by activating a Flo11p-dependent protease.
To further examine the posttranslational processing of Flo11p, an N-terminal deletion (from 29–95 amino acids) was constructed, which showed reduced levels of the protein and decreased shedding (Fig 4D, Flo11pMYCΔ29–95-HA). Deletion of the GPI anchor attachment site caused increased shedding (Fig 4E, GPIΔ), confirming that Flo11p is anchored to cells by its GPI anchor. A fraction of Flo11pGPIΔ remained associated with cells (P, GPIΔ, Fig 4E), consistent with the idea that some form of posttranslational processing occurs in the N-terminus. Together, the data support a model for Flo11p processing, in which Flo11p is processed by a signal peptidase in the endoplasmic reticulum and by Kex2p in the Golgi to produce the mature form of the protein (Fig. 4F). At the cell surface, Flo11p is attached to the cell wall by its GPI anchor (Fig. 4F). Presumably other proteases and/or cell wall glucanases contribute to the release of Flo11p from the cell wall (Fig. 4F). Two other proteases (Rbd2p, Ecm14p) and a phospholipase (Spo1p) were identified that contributed in a minor way to Flo11p shedding (Fig. S4).
DISCUSSION
In summary, we have shown that the yeast flocculin Flo11p, one of the most intensively studied microbial adhesion molecules, is shed from cells. This discovery challenges the prevailing dogma for the regulation of microbial adhesion glycoproteins and their adhesive properties, including their roles in cell-surface variability and interactions with the cell wall/cell surface. Shed Flo11p may prevent cell adhesion in two ways: 1) the release of Flo11p from cells directly attenuates adherence, and 2) shed Flo11p binds to surfaces and may compete with cell-associated Flo11p for surface sites. As a result of the calibration of Flo11p’s adherence properties, cells maintain a balance that is optimal for the different foraging behaviors in which Flo11p is required.
We also show that multicellular communities of yeast secrete a fluid rich in shed mucins that may be functionally analogous to the mucus secretions produced by gastropods and other metazoans. Mucus production in microbes has implications in mucin evolution, fungal pathogenesis, and social evolution, in that production of a secreted material by individual cells can benefit the entire community [26]. Glycoprotein shedding may contribute to the formation or regulation of an extracellular matrix-like material that along with other proteins [27, 28] may regulate biofilm expansion and architecture.
Mucin-like glycoproteins may be shed from pathogens and have unappreciated roles in virulence. Shed mucins would be among the first molecules encountered by the host, and their anti-adhesion properties may prevent host cells from attaching to the fungal surface. Detection of shed fungal glycoproteins may allow early diagnosis of pathogenic infections. Shedding of mucin glycoproteins adds to the repertoire of surface variability [7, 8] by contributing to cell-surface variation and by generating complex, non-uniform collections of cells with varying adherence properties. Mucin shedding in microbes may not be limited to fungi: the protozoal parasite Trypanosoma cruzi express as many as 850 different mucin-coding genes [29], and it is tempting to speculate that mucin shedding in eukaryotic pathogens is tailored to optimize virulence.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Microbiological Techniques
Yeast strains are listed in Table S1. Yeast and bacterial strains were manipulated by standard methods [30, 31]. Epitope fusions were at 500 aa residues for Msb2p, 1015 aa residues for Flo11p, 298 aa residues for Hkr1p, and 367 aa residues for Sho1p. All fusion proteins were created at their genomic loci under the control of their endogenous promoters and were functional with respect to agar invasion, MAPK signaling, and mat expansion. Details of strain construction and manipulation, immunoblot analysis, and microscopy can be found in the supplemental materials.
Secretion Profile Analysis
An ordered collection of ~5,400 ORFs under the control of the GAL1 inducible promoter was used [23] (Open Biosystems). The details of the protocol can be found in the supplemental materials. Genes that showed altered Flo11p-HA secretion were confirmed by retesting and examined by standard immunoblot analysis to determine S/P ratios. False positives were uncovered at a frequency of ~35%. To enrich for candidate proteases that process Flo11p, a genetic miniarray containing known proteases was pinned onto a 96-well plate and examined in strains containing Flo11p-HA1015 (PC2043).
Purification of Shed Flo11p-HA
Procedures were adapted from [32]. Pilot experiments showed a maximum yield of Flo11p-HA at 8% of PEG 8000 pH 5.45, estimated by immunoblots and silver staining. 300 mls of cells (PC2714, GAL-FLO11-HA) were grown in SC-GAL medium for 48 h. Cells were harvested by centrifugation and discarded, and supernatants were collected. Supernatant volumes were adjusted to pH 5.45, and MgCl2 was added to a final concentration 10mM. PEG 8000 (initial concentration 50%) was added drop wise to 8% saturation. After 30 min stirring at 4°C, proteins were precipitated by centrifugation at 15,000 RPM for 15 min. The pellets were resuspended in 20 mM NaPO4 and loaded on a Sepharose CL-4B column. Flo11p-HA was collected in the void volume. For some experiments, Flo11p-HA was overexpressed and purified in cells lacking MSB2 (PC2714). Msb2p-HA was similarly purified using strains PC1083 (GAL-MSB2-HA) or PC2716 (GAL-MSB2-HA flo11Δ).
HIGHLIGHTS
The major yeast adhesion molecule Flo11p is abundantly shed from cells.
Flo11p shedding attenuates adherence and optimizes filamentous growth and mat expansion.
Mucin glycoprotein shedding may be analogous to mucus secretion in higher organisms.
The furin protease Kex2p is required for Flo11p maturation.
Supplementary Material
ACKNOWLEDGEMENTS
We are deeply indebted to Drs. Gerry Fink (Whitehead Institute, MIT, Cambridge MA), Aaron Mitchell (Carnegie Mellon University, Pittsburg PN), and Todd Reynolds (University of Tennessee, Knoxville, TN), who provided strains, guidance, and suggestions throughout the work. We thank Drs. Hans-Ulrich Möesch (Philipps Universtät, Marburg, Germany), Anne Dranginis (St. John’s University, Queens NY), Kevin Verstrepen (University of Leuven, Belgium), Isak Pretorius (The Australian Wine Research Institute, Australia), and Victor Albert (University at Buffalo, Buffalo, NY) for reading the manuscript and providing helpful suggestions. Thanks also to Dr. Damian Krysan (University of Rochester Medical Center, Rochester NY) for providing plasmids and Ms. Heather Dionne for technical assistance. P.J.C. is supported from grants from the NIH (1R03DE018425-01), American Cancer Society (TBE-114083), and American Heart Association (GM 0535393T).
Sheelarani Karunanithi and Paul J. Cullen designed the research, performed the research, and wrote the paper. Nadia Vadaie, Colin Chavel, Jyoti Joshi, Laura Grell, and Barbara Birkaya performed the research.
Abbreviations
- ECM
extracellular matrix
- FG
filamentous growth
- Gal
galactose
- GPI
glycosylphosphatidylinositol
- Glu
glucose
- HA
hemaglutinin
- MAPK
mitogen activated protein kinase
- PM
plasma membrane
- SC
synthetic complete
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci U S A. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds TB, Fink GR. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 5.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 6.Vinod PK, Sengupta N, Bhat PJ, Venkatesh KV. Integration of global signaling pathways, cAMP-PKA, MAPK and TOR in the regulation of FLO11. PLoS ONE. 2008;3:e1663. doi: 10.1371/journal.pone.0001663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 8.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A. 2006;103:11228–11233. doi: 10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatebayashi K, Tanaka K, Yang HY, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. Embo J. 2007;26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF., Jr A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitoniak A, Birkaya B, Dionne HS, Vadiae N, Cullen PJ. The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation MAPK Pathway and Contribute to a Multimodal Response. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supplemental methodology figures and tables are associated with the manuscript and can be found online. Supplemental methodology figures and tables are associated with the manuscript and can be found online.
- 15.Douglas LM, Li L, Yang Y, Dranginis AM. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot Cell. 2007;6:2214–2221. doi: 10.1128/EC.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal B, Gendler SJ, Longenecker BM. The biological role of mucins in cellular interactions and immune regulation: prospects for cancer immunotherapy. Mol Med Today. 1998;4:397–403. doi: 10.1016/s1357-4310(98)01322-7. [DOI] [PubMed] [Google Scholar]
- 17.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesimer M, Sheehan JK. Analyzing the functions of large glycoconjugates through the dissipative properties of their absorbed layers using the gel-forming mucin MUC5B as an example. Glycobiology. 2008;18:463–472. doi: 10.1093/glycob/cwn024. [DOI] [PubMed] [Google Scholar]
- 20.Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci U S A. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006;20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet. 6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bussey H, Saville D, Greene D, Tipper DJ, Bostian KA. Secretion of Saccharomyces cerevisiae killer toxin: processing of the glycosylated precursor. Mol Cell Biol. 1983;3:1362–1370. doi: 10.1128/mcb.3.8.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 26.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latge JP, Fink GR, Foster KR, Verstrepen KJ. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beauvais A, Loussert C, Prevost MC, Verstrepen K, Latge JP. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 2009;9:411–419. doi: 10.1111/j.1567-1364.2009.00482.x. [DOI] [PubMed] [Google Scholar]
- 28.Straver MH, Traas VM, Smit G, Kijne JW. Isolation and partial purification of mannose-specific agglutinin from brewer's yeast involved in flocculation. Yeast. 1994;10:1183–1193. doi: 10.1002/yea.320100906. [DOI] [PubMed] [Google Scholar]
- 29.Buscaglia CA, Campo VA, Frasch AC, Di Noia JM. Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nat Rev Microbiol. 2006;4:229–236. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 32.Atha DH, Ingham KC. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J Biol Chem. 1981;256:12108–12117. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.