Abstract
Mammalian cells primarily rejoin DNA double-strand breaks (DSBs) by the non-homologous end-joining (NHEJ) pathway. The joining of the broken DNA ends appears directly without template and accuracy is ensured by the NHEJ factors that are under ATM/ATR regulated checkpoint control. In the current study we report the engineering of a mono-specific DNA damaging agent. This was used to study the molecular requirements for the repair of the least complex DSB in vivo. Single-chain PvuII restriction enzymes fused to protein delivery sequences transduce cells efficiently and induce blunt end DSBs in vivo. We demonstrate that beside XRCC4/LigaseIV and KU, the DNA-PK catalytic subunit (DNA-PKcs) is also essential for the joining of this low complex DSB in vivo. The appearance of blunt end 3′-hydroxyl and 5′-phosphate DNA DSBs induces a significantly higher frequency of anaphase bridges in cells that do not contain functional DNA-PKcs, suggesting an absolute requirement for DNA-PKcs in the control of chromosomal stability during end joining. Moreover, these minimal blunt end DSBs are sufficient to induce a p53 and ATM/ATR checkpoint function.
INTRODUCTION
The core components for a repair by NHEJ are conserved from yeast to mammals and consist of the XRCC4/DNA-LigaseIV complex and the KU70/KU80 heterodimer (1–3). Vertebrates in addition also require DNA-PKcs (4,5) and as shown more recently, the Artemis nuclease (6). Whereas the XRCC4/DNA-LigaseIV complex is required for end joining of DSBs, the KU complex contributes to DSB recognition, DSB bridging and nucleolytic processing of the ends (3). Beside its essential function during V(D)J recombination in lymphocytes, the role of DNA-PKcs is less clear, but DNA-PKcs impaired cells are hypersensitive to various DSB inducing agents (4,7). Central to the understanding of the NHEJ repair function is first whether, depending on the chemical characters of the break points, the core molecular components are recruited on default and second, if broken ends that resemble already suitable substrates for DNA-ligases in the chromosomal context still require all components for the joining step. The ligation of the 5′-phosphate containing blunt ends in signal-joint formation during immunoglobulin V(D)J recombination requires both DNA-LigaseIV and KU complexes. DNA-PKcs is dispensable, but has an essential function in hairpin opening together with the Artemis nuclease during coding joint formation (6). This might suggest that DNA-PKcs is only recruited to resolve more complex DSB products. ATM is normally not required during V(D)J recombination, but has its role in protecting cells from tumors caused by aberrant V(D)J recombination (8). Cells from patients with AT support normal levels of V(D)J recombination, and mature antigen receptor-bearing lymphocytes are readily observed in ATM-deficient mice (9). Understanding the role of a minimal DSB in sensing DNA damage is an essential prerequisite for the analysis of the induction thresholds in DNA repair checkpoint signaling.
Commonly used DSB causing agents such as ionizing radiation or radiomimetic drugs introduce simultaneously a broad spectrum of chemical modifications. This limits the analysis of substrate requirements for repair in vivo. These genotoxins potentially induce double-strand as well as single-strand breaks simultaneously and concomitantly several distinct mutagenic modifications at the breakpoint nucleotides (for a recent review, see 10). Homing endonucleases, transposases and restriction enzymes were successfully introduced into different cell types. Inducible vector systems (11), or native enzymes were introduced by electroporation, lipofection or calcium phosphate precipitation (reviewed in 12–14). These types of experiments employing specific nucleases are limited by the accessibility of the cells and the side effects from transient transfections, by constitutive although low basic expression levels and the slow on-rates. Moreover, many of the nucleases used (11,15,16) (i.e. I-SceI, HO, RAG1,2) exhibit low turnover rates, additional unspecific DNA binding activities and often are single-strand or hairpin overhang producing enzymes (17; reviewed in 18). It is this complex feature that might influence the sensors for a sensible checkpoint and repair induction. Due to the lack of reagents that cause exclusively 5′ phosphate blunt end DSBs of well-defined character in vivo, it is not clear how a minimal, potentially non-mutagenic DSB is repaired by the NHEJ pathway and if ATM and p53-dependent checkpoint controls are activated.
In this study, we investigated the substrate/sensor function of the least complex DSB lesion, blunt end cleavage caused exclusively by phosphodiester hydrolysis. We used the single-chain variant of the restriction endonuclease PvuII (SCPVU) to elaborate a new procedure for an unequivocal generation of DSBs in cells. As was shown before, single-chain PvuII exhibits only specific, cognate site DNA binding activity with high catalytic turnover (19). In addition, effects resulting from nuclease activity can be distinguished from possible side effects by employing a nuclease-deficient but DNA-binding proficient SCPVU mutant.
MATERIALS AND METHODS
Constructs
The PvuII endonuclease gene was cloned into the pRIZ′ vector (20) as described, to obtain pRIZ′-PvR NBt (19). In this vector, the endonuclease gene is followed by a polycloning region containing BamHI and HindIII terminal sites. This polycloning region was replaced by a synthetic dsDNA to obtain a fusion gene coding for PvuII (amino acids 1–157) followed by a C-terminal GSYGRKKRRQRRRGGS HHHHHH extension. The PvuII gene of this vector (pRIZ′-wtPvR-Tat-His6) was duplicated by inserting the XbaI–XbaI fragment of the single-chain (sc) PvuII gene [the region coding for residues (121–157)-GSGG-(2–120)] as described (19) to obtain the vector pRIZ′-scPvR-Tat-His6. With this vector, the fusion protein (1–157)-GSGG-(2–157)-GSYGRKKRRQRRRGGSHHHHHH is expressed (we call this SCPVU-TAT). SC34 (D34G substitution in both subunits) was constructed in a similar way using the mutated endonuclease version as a template for fusion to Tat-His6 (19). Vectors for the dominant negative ATM constructs kdATM (as described in 21) were a kind gift from M.B. Kastan, St Jude Children’s Research Hospital, USA. The EGFP-histone2B expressing vector was obtained by PCR amplification of a H2B fragment from human genomic DNA and subsequent cloning into the vector pEGFP-N1 (Clontech).
Protein expression and purification
Protein expression was performed by using Escherichia coli XL1 MRF′ host carrying the pLGM methylase expressing plasmid as described (19). Cell pellets from 1.6 l of culture of a SCPVU-TAT expression experiment were suspended in 40 ml of 50 mM sodium phosphate (pH 8.0) containing protease inhibitors (Complete EDTA-free, four tablets from Roche Diagnostic) and 1 mg/ml of egg white lysozyme. After incubation on ice for 1 h, suspensions were sonicated and centrifuged (16 000 r.p.m., 20 min). The supernatants were adjusted to 500 mM NaCl and 10 mM imidazole and loaded on a HiTrap Chelating affinity column (5 ml, Amersham Pharmacia Biotech). Columns were washed with 100 mM imidazole and proteins were then eluted with 300 mM imidazole. The eluted fractions were subsequently purified on a SP Sepharose column (5 ml HiTrap SP HP, Amersham Pharmacia Biotech) using a linear NaCl gradient (0.3–1 M NaCl in 20 mM sodium phosphate and 1 mM EDTA at pH 7.0). The SCPVU-TAT proteins eluted between 0.63 and 0.67 M NaCl. Preparations yielded about 10 mg of protein. Concentrations of the recombinant proteins were estimated based on the calculated molar extinction coefficient 71 120 M–1 cm–1 at 280 nm as described before (19). Purification of wtPvuII variants was performed similarly, except these proteins eluted between 0.81 and 0.85 M NaCl from the SP Sepharose column. All proteins were homogeneous on 15% SDS–PAGE and analysis by electron spray mass spectrometry using an API 150 EX apparatus (Perkin Elmer) revealed the theoretical molecular masses. The purified, C-terminally Tat-His6 tagged and untagged versions exhibited the same specific nuclease activities in vitro, as assayed with λ phage DNA as a substrate.
SCPVU-TAT transduction assay
For the protein transduction of various PvuII enzyme derivatives, recombinant proteins were added to the complete growth medium including FCS and antibiotics. Protein stock solutions for transductions contained enzyme concentrations between 20 and 50 µM in SP-Sepharose elution buffer.
Cell culture and transfections
The rodent cell lines AA8, V3 (DNA-PKcs(–/–)), xrss5 (KU80(–/–)) and XR-1 (XRCC4(–/–)) [a kind gift from P. Jeggo, University of Sussex, UK (22)]; AT-5, MRC-5 (a kind gift from L. Zentilin; ICGEB, Trieste, Italy), and U2OS, IMR90 (early passages; a kind gift from G. del Sal, CIB, University of Trieste, Italy) cell lines were maintained in DMEM supplemented with 10% fetal calf serum (FCS). M059J and M059K cell lines [a kind gift from Susan Lees Miller, University of Calgary, Canada; (23)] were maintained in DMEM/Hams12 (1:1) supplemented with 10% FCS. The various HCT116 derivates [a kind gift from B. Vogelstein, Johns Hopkins University, USA (24)] were cultivated in McCoy’s medium supplemented with 10% FCS. For transient DNA transfections of U2OS cells, the SuperFect Transfection Reagent (Qiagen GmbH) was used as directed by the manufacturer.
Immunodetection analysis
Extracts for Figure 1B, were prepared in cell lysis buffer (20 mM Tris–HCl pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.2% Triton X-100, 20 µM TPCK, 20 µM TLCK, 60 mM 4-nitrophenyl phosphate (all Roche Diagnostics), sonicated and cleared by centrifugation. Extracts for Figure 4B and C, and Figure 5C, D and E were prepared by direct collection of the cells in SDS loading buffer and sonication. Mono-specific α-PvuII antibody were raised in New Zealand white rabbits with native, highly purified PvuII fractions. The anti-serum was affinity-purified against PvuII immobilized to BrCN-Sepharose (Amersham-Pharmacia Biotech) as described (25). Immunofluorescence analysis was done after fixation in 3% paraformaldehyde (26), except for the phospho-H2AX-(Ser139) detection where the cells were fixed for 5 min on ice in acetone and methanol (1:1). Analysis was done by confocal microscopy with a Zeiss Axiovert 100M microscope attached to a LSM 510 confocal unit. Antibodies used were the rabbit α-phospho-H2AX-(Ser139) (Upstate Biotechnology), α-phospho-Ser15 p53 (Cell Signaling Technology, MA), α-actin (Sigma), the sheep α-ADPRT (26), and the mouse monoclonal α-p53 (DO-1, SC-126) (Santa Cruz Biotech).
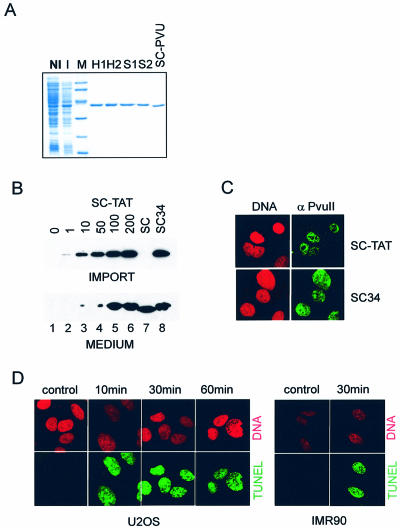
Figure 1.
Introduction of functional SCPVU-TAT protein into vertebrate cells. (A) Expression and purification of SCPVU-TAT proteins. E.coli containing the SCPVU-TAT expression plasmid were induced for protein expression and purification was performed as described in Materials and Methods. NI and I are total cell extracts of non-induced and induced E.coli cells, respectively. H1 and H2 are peak fractions from the HiTrap Chelating affinity column purification step; S1 and S2 are peak fractions from the SP Sepharose column purification step; M, molecular mass markers (from top to bottom in kDa: 94, 67, 43, 33, 20, 14); SC-PVU, protein lacking the TAT-tag purified in a similar way. (B) Immunoblot analysis of extracts after protein transduction into U2OS cells. Proteins assayed were: SCPVU-TAT, increasing protein concentrations were applied (1, 10, 50, 100, 200 nM; numbers indicated on the top represent nM protein in the cell culture supernatants; SC-TAT; lanes 2–6), SCPVU (200 nM; without TAT, SC; lane 7) or SC34 (200 nM; SCPVU-TAT nuclease impaired version; SC34; lane 8) and no protein addition (lane 1). Ten minutes after the addition of the proteins, cell extracts were prepared, separated on SDS–PAGE and analyzed by immunoblotting using PvuII specific antibodies (upper blot; IMPORT). Aliquots of the cell culture supernatants were also included in the analysis (lower blot; MEDIUM). (C) Thirty minutes after protein transduction with SCPVU-TAT or SC34, cells were trypsinized and reseeded on poly-lysine coated glass slides and stained with mono-specific antibodies to PvuII and FITC-conjugated secondary antibodies for confocal immunofluorescence analysis (green, SC-TAT or SC34 as indicated). Propidium iodide DNA co-staining was applied (red; DNA). (D) TUNEL analysis after SCPVU-TAT protein transduction. Cells were treated with SCPVU-TAT and DNA DSBs were analyzed by confocal immunofluorescence microscopy after TdT-labeling in the presence of dUTP-FITC (TUNEL) of U2OS and IMR90 cells. Treatment was done with 100 nM SCPVU-TAT and for the time periods indicated. A ‘non import competent’ SCPVU (100 nM) (containing no TAT-tag) was added for 10 min and served as a background control (control); cells positive for ‘TUNEL-labeling’ (green); propidium iodide DNA stain (red). Assays done with SC34 did not result in TUNEL positive cells (data not shown).
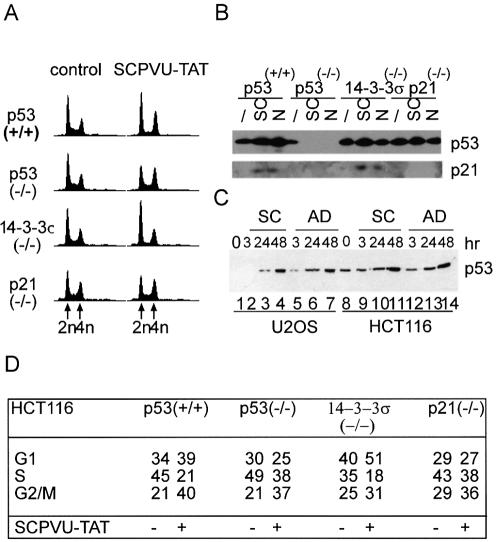
Figure 4.
p53 induction by blunt end DSBs. (A) FACS analysis of HCT116 cell lines and derivatives with homozygous knockout mutations for p53, p21WAF1/CIP1 and 14-3-3σ. Cells were either untreated (control) or treated with SCPVU-TAT (100 nM) for 36 h. Analysis was done as in Figure 2; All histogram plots represent a measure of DNA content versus cell number. DNA content equivalents indicated at the bottom lane: 2n (G1), 4n (G2/M); positions valid for all histogram plots in a row. (B) Immunoblot analysis from extracts prepared from the HCT116 cell lines as described in (A) that were treated with SCPVU-TAT (100 nM; SC) or nocodazol (0.2 µg/ml; N) for 24 h, extracts from non-treated cells were used for comparison (/). Extracts were resolved on SDS–PAGE and immunoblotted with p53 or p21WAF1/CIP1 specific antibodies as indicated. (C) U2OS (lanes 1–7) and HCT116 (lanes 8–14) cells were treated either with SCPVU-TAT (100 nM; SC) or with adriamycin (0.02 µg/ml; AD) for the time periods indicated, non-treated cells were used for comparison (0). Equal amounts of protein extracts were resolved on SDS–PAGE and immunoblotted with p53 specific antibodies. (D) Analysis of the histograms in (A). The percentage of cells in the various cell cycle phases is summarized.
Figure 5.
ATM is essential for checkpoint control of SCPVU-TAT induced repair. (A) Colony forming assays with ATM mutant cell lines. Different dilutions of AT-5 or the ATM positive cells MRC-5 were either ‘not treated’ (white bars) or treated with SCPVU-TAT (25 nM, gray bars; 100 nM, black bars) for 60 min; cells were then washed and analyzed after 7–10 days with GIEMSA stain as described in Figure 3. Standard deviations in the range of 10–15% as indicated. ‘No treatment’ was set equal to 100%, black bars in AT-5 cells nearly 0% and thus not indicated. (B) Example of a colony-forming assay with AT-5 cells at high density; 50% confluent AT-5 cells were either untreated (control) or treated with SCPVU-TAT (10, 50, 100 nM) or with the nuclease impaired SCPVU-TAT version SC34 (100 nM; SC34) or adriamycin (0.02 µg/ml; AD) for 24 h; cells were then washed, grown for an additional 7 days and stained as described in (A). (C) SCPVU-TAT dependent Ser15 phosphorylation of p53. ATM positive MRC-5 cells were treated either with SCPVU-TAT (100 nM; SCTAT) or with hydroxyurea (1 mM; HU) for the time periods as indicated, ‘not-treated’ cells were used for comparison (0). Extracts were resolved on SDS–PAGE and immunoblotted with p53-specific antibodies (upper panel; p53), p53 Ser15 phospho-specific antibodies (middle panel; S15P) and PARP-specific antibodies (lower panel; PARP). (D) ATM negative AT-5 cells were treated and assayed as in (C). (E) SCPVU-TAT dependent Ser15 phosphorylation of p53 is caffeine-sensitive. ATM negative AT-5 cells were treated with SCPVU-TAT (100 nM; SCTAT) or with hydroxyurea (1 mM; HU) in the presence and absence of caffeine (2 mM) for 4 h, ‘not-treated’ cells and caffeine ‘only’ treated cells were used for comparison. Extracts were resolved on SDS–PAGE and immunoblotted with p53-specific antibodies (upper panel; p53), p53 Ser15 phospho-specific antibodies (middle panel; S15P) and actin-specific antibodies (lower panel; actin). (F) H2AX-(Ser139) phosphorylation in U2OS cells after SCPVU-TAT treatment. U2OS cells grown on microscopic slides were transiently transfected with pcDNA3 and pEGFP-H2B (a, b, d), or pcDNA3kdATM and pEGFP-H2B (c) expression constructs. Twenty hours after transfections, samples were treated with SC-34 (100 nM; a) as a control, SCPVU-TAT (100 nM; b, c), or Wortmanin (50 µM) in combination with SCPVU-TAT (100 nM) (d), for 40 min; objects were stained with H2AX-(Ser139) phospho-specific antibodies (red), co-transfection of pEGFP-H2B (green). (G) Summary of H2AX-(Ser139) phosphorylation in U2OS cells after SCPVU-TAT treatment. The means of three experiments are shown as histograms. For each experiment, 50 transfected cells [EGFP-H2B positive cells (green bars)] were scored for H2AX-(Ser139) positive cells (red bar). Results are expressed as percentage of cells. Assay conditions marked by letters on the top and indicated on the bottom as in (F).
TUNEL assay
For in vivo detection of dsDNA breaks, the dUTP-FITC, TUNEL labeling technique was used in the presence of 1 mM CoCl2 as described by the suppliers (in situ Cell Death Detection Kit, Roche Diagnostic, Mannheim, Germany). Reactions were performed on adhesion slides in the dark. Prior to the TUNEL reactions, the cell lines were fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100/PBS for 5 min (U2OS) or 20 min (IMR90). Following TUNEL reactions, the cells were extensively washed with PBS and then counterstained with propidium iodide as described for immunofluorescence analysis (26).
FACS analysis
The cells from the supernatants and the cells detached by trypsinization were pooled and washed with PBS containing 1% FCS and fixed with 70% ethanol on ice for 1 h. The fixed cells were then washed twice with PBS containing 1% FCS and incubated for 1 h at room temperature with 200 µg/ml of RNAseA and propidium iodide (20 µg/ml) was added before measurements. DNA content analysis was done with a FACSCalibur™ (Becton Dickinson) using CellQuest™ software and the MODFIT LT™ statistics program.
Colony forming assays
The various cell lines were incubated with different concentrations of SCPVU-TAT for 1 h (AT-5, MRC-5 cells) or 24 h (NHEJ-cells), the medium was changed by two washes and cells were grown for a further 7–10 days. For the estimation of colony formation, cells were then fixed with 10% formaldehyde and stained with 10% GIEMSA (Merck) solution. Assays were done in triplicates.
RESULTS
Introduction of functional single-chain PvuII into vertebrate cells
We engineered a new tool for the induction of DSBs in vivo. We used the HIV-TAT protein transduction domain (11 amino acids, residues 47–57) (27) for protein import (reviewed in 28) of the class II restriction endonuclease PvuII into vertebrate cells. PvuII cleaves phosphodiesters in the palindromic sequence CAG/CTG to produce blunt ends. This sequence does not contain any known consensus site for DNA methylation in vertebrate cells. A single-chain variant of PvuII enzyme (SCPVU) was used, consisting of covalently joined subunits of PvuII (19). Proteins were purified using a two-step column purification scheme under non-denaturing conditions (Fig. 1A). This purification (see Materials and Methods) results in homogeneous, highly active and transport-competent fusion enzymes. We chose to call these proteins SCPVU-TAT in the following.
Uptake of the SCPVU-TAT protein into U2OS cells was analyzed with a polyclonal antibody against PvuII. Extracts prepared from cells that were protein transduced with either SCPVU-TAT or SCPVU (without TAT) for 10 min were analyzed in immunoblots. Enrichment for SCPVU-TAT versus proteins with no TAT was obtained (Fig. 1B). Moreover, microscopic analysis detected uptake into virtually all the nuclei of the SCPVU-TAT or SC34 (see below) treated cells (Fig. 1C). These results demonstrate that significant amounts of SCPVU-TAT fusion proteins are targeted to the nucleus of a cell. The uptake was not dependent on the denatured state of the protein as reported for other TAT-tag containing proteins (29).
Terminal deoxynucleotidyl transferase labeling in the presence of CoCl2 (TUNEL) was used after SCPVU-TAT treatment to detect the nuclease activity of the imported restriction enzyme in vivo (Fig. 1D). For U2OS and IMR90 cells, TUNEL-positive cells were readily detected as soon as 10 min after the addition of the SCPVU-TAT enzyme. The TUNEL positive staining was cell cycle state independent and no signs of apoptosis could be detected (Fig. 1D, and data not shown). Protein uptake and TUNEL positive staining could be detected in response to an SCPVU-TAT treatment, contrary to treatments with either SCPVU lacking the TAT-tag fusion (Fig. 1D, control), or the nuclease-deficient and TAT-tag containing version SC34 (not shown). TUNEL staining was observed with all human and rodent cell lines analyzed so far, including epithelial cells (HEK293, MCF7, HCT116) and primary fibroblasts (WI83, IMR90, MEFs).
Nuclease-dependent cell cycle delay induced by SCPVU-TAT
For cell cycle analysis of SCPVU-TAT transduced cells, we treated the osteosarcoma cell line U2OS. These cells are p53-positive and show functional checkpoint responses to various genotoxic agents. We examined cell cycle distributions in a FACS analysis. A nuclease-deficient but high affinity DNA-binding mutant (SC34) (19) was introduced in a similar way. This allowed to test the in vivo function of the endonuclease activity and to distinguish probable nuclease-independent squelching effects, which may result from DNA-binding alone or from the TAT-tag mediated import. SCPVU-TAT at a final concentration of 10 nM readily induces an increase in the 4n DNA content within 24 h in U2OS cells (Fig. 2). This change in the steady-state cell cycle distributions was dependent on the SCPVU-TAT concentration. Initially we compared the effects on the cell cycle distribution induced by SCPVU-TAT with those of the non-covalently dimerized native PvuII containing TAT in the same arrangement. The native enzyme—although exhibiting comparable enzyme activity in vitro—was less active in our TAT-mediated in vivo assay (data not shown). Thus, only the single-chain proteins were used in the following experiments.
Figure 2.
Nuclease-dependent cell cycle delay induced by SCPVU-TAT. Cell cycle distribution of protein-transduced U2OS cells by FACS. 2n presents relative diploid, not replicated equivalent of DNA (G1), 4n presents relative tetraploid, replicated equivalent of DNA (G2/M). Ploidy indications are valid for both histogram rows. U2OS cells were grown for 24 h (upper row) or 48 h (lower row) in the presence of increasing concentrations of SCPVU-TAT (10, 50, 100 nM) as indicated and with one single addition of enzyme, or with multiple additions of enzymes (every 12 h, 10 nM*12, 100 nM*12); no treatment (control); nuclease-deficient version of SCPVU-TAT (SC34, 100 nM). All histogram plots that are shown represent a measure of the DNA content versus cell numbers as indicated only for the upper left histogram. The percentage of cells in the various cell cycle phases is given below each histogram.
The cells were released back into the cell cycle after an additional 24 h (Fig. 2; compare 24 and 48 h) suggesting that the enzyme activity was diluted and the respectively induced DSBs were repaired. No sign of immediate apoptosis was detected, as was estimated from the sub-2n DNA content populations. Addition of enzyme to the cell culture medium every 12 h (Fig. 2; *12) resulted in a stable induced cell cycle delay but no apoptosis. This was not observed for populations treated either with a preparation of a SCPVU protein without TAT-tag (not shown) or with the nuclease-deficient version (SC34). Cell cycle distributions of these populations did not change, thus proving that the catalytic-nuclease activity of the transduced protein is the main cause for the cell cycle perturbations. The cell cycle stop with a DNA content at 4n induced by SCPVU-TAT treatment in various cell lines including U2OS and HCT166 (see below) was analyzed in more detail. In the enriched 4n populations, most nuclei were of large size and no dramatic increase in mitotic α-tubulin arrays, condensed chromosomes or nuclear envelope break down was observed (data not shown). Further, cyclinB1 and Cdc25C were localized to the cytoplasma (data not shown) and histone H1 kinase activity of cyclin B1 immunoprecipitates was low compared to a 4n cell population blocked in G2/M by nocodazole (data not shown)—hallmarks for a late S/G2 population in the cell cycle.
Hypersensitivity of cell lines defective in NHEJ
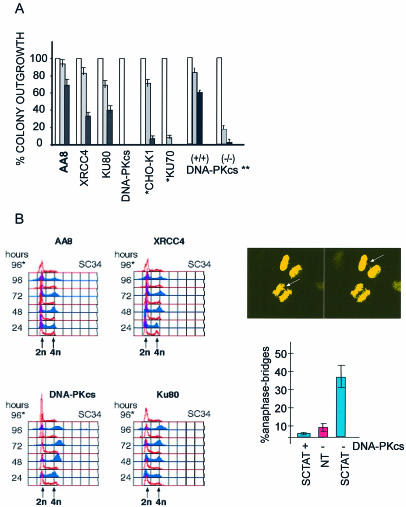
We compared rodent cells that carry homozygous loss of function mutations in one of the three essential factors for dsDNA break repair by NHEJ: the PIKK containing serine/threonine kinase DNA-PKcs, the DNA-PK regulatory subunits Ku70 and Ku80, and the DNA-Ligase IV subunit XRCC4 in the SCPVU-TAT in vivo nuclease assay. These cell lines are hypersensitive to genotoxins inducing dsDNA breaks (30), are p53-positive and do not show checkpoint defects in general (31). SCPVU-TAT treatment for 30 h resulted in distinct cell cycle stops (Fig. 3B; and data not shown). We tested SCPVU-TAT on NHEJ mutant cells in clonogenic assays and colony outgrowth was scored after a single treatment of these mutant cells. As summarized in Figure 3A, mutant cells of the DNA-Ligase IV subunit XRCC4 are hypersensitive to blunt end dsDNA breaks. Interestingly, all the DNA-PK mutant cells assayed, including mutations for the catalytic subunit also exhibit strong reduction in colony formation. A DNA-PKcs defective human glioma cell line M059J (23) was treated with SCPVU-TAT, and compared to the parental M059K cells. Similarly, colony outgrowth was scored after a single treatment. Again, these human cells showed a significant increase in sensitivity towards SCPVU-TAT treatment (Fig. 3A). This is consistent with the results obtained for the rodent DNA-PKcs mutant cell lines, which, taking into account the high specificity of SCPVU-TAT, proves an essential function for DNA-PKcs in the repair of blunt end DSBs. We analyzed cell cycle distributions of mutant cell lines treated with SCPVU-TAT up to 96 h and compared them to parental cells, non-treated cells and cells treated with the nuclease impaired SCPVU-TAT version SC34 (Fig. 3B, left panel). Wild-type and mutant cells showed an increase in 2n and 4n DNA content and a decrease in intermediate DNA content upon SCPVU-TAT treatment. But more striking after SCPVU-TAT treatment, the KU (–/–) cells and to a somehow greater extent the DNA-PKcs (–/–) cells displayed a significant increase in the DNA content beyond 4n in time (Fig. 3B). After DNA staining, microscopic examination of these cells after SCPVU-TAT treatment for 48 h displayed more than 30% of anaphase bridges (data not shown). A more detailed analysis of the human glioma DNA-PKcs defective cell lines MO59J as compared to control cells MO59K confirmed this increase in the frequency of anaphase bridges in response to SCPVU-TAT treatment. An image and summary of the results obtained from various experiments are shown in Figure 3B (right panel). The DNA-PKcs mutant cells analyzed exhibit a significant increase in anaphase bridges in response to the blunt end inducer SCPVU-TAT. This was not observed for the DNA-PKcs positive cell lines under SCPVU-TAT treatment and to a much lower extent for non-treated DNA-PKcs mutant cells. Altogether, these results strongly suggest an essential role for all the known NHEJ factors in the repair of minimal blunt end DSBs. DNA-PKcs has a possible function in protecting from transfusions of broken but ligateable chromosomes, which extends its role in preventing telomere fusions for the maintenance of chromosomal stability.
Figure 3.
Hypersensitivity of cell lines defective in NHEJ. (A) Colony forming assays of NHEJ mutated CHO cells treated with SCPVU-TAT. Cell lines used are AA8 (parental line), V3 [DNA-PKcs(–/–)], xrss5 [KU80(–/–)] and XR-1 [XRCC4(–/–)], HIS P1.13–11 [KU70(–/–)] (*) and the corresponding parental line CHO-K1 (*). Also included are assays done with human DNA-PKcs (**) mutant cells MO59J [DNA-PKcs minus (–/–)] compared to MO59K [DNA-PKcs plus (+/+)] cells. Various dilutions of the CHO and the human MO59 cells carrying the different NHEJ mutations as indicated were either ‘not treated’ (white bars) or treated with SCPVU-TAT (25 nM, gray bars; 100 nM, black bars) for 24 h; cells were then washed and grown for 7–10 days, stained with GIEMSA and colonies were counted. A summary of the results is shown as histograms and represents the mean from three experiments with standard deviations in the range of 15–20% as indicated. ‘No treatment’ was set equal to 100%. Missing gray and black bars in V3 [DNA-PKcs(–/–)] histograms indicates colony outgrowth below 1%. (B) (left panel) FACS analysis of various NHEJ mutant cell lines after the time periods indicated. Cells were treated with 100 nM SCPVU-TAT (blue histograms) or 100 nM SC34 (red histograms, SC34), or not treated (red histograms) for the time periods as indicated on the right of the histograms and analyzed in FACS analysis as in Figure 2; (B) (right panel, top) Example of an anaphase bridge observed 48 h after SCPVU-TAT treatment of MO59J [DNA-PKcs (–/–)], two different Z layers of the same object are shown; arrows indicate fused chromosomes. (B) (right panel, bottom) Analysis of anaphase bridges in MO59J [(DNA-PKcs (–/–)] and MO59K [(DNA-PKcs (+/+)] cells after 48 h of SCPVU-TAT treatment, cells and conditions as indicated; for each condition, 3 × 50 anaphases were examined by microscopy after DNA staining and results are summarized in ‘% of anaphase-bridges’ with observed standard deviations [SCTAT (blue bars), SCPVU-TAT treated; NT (red bars), not treated].
p53 checkpoints in response to blunt end phosphodiester hydrolysis of DNA
The tumor suppressor p53 is induced by genotoxic agents causing DSBs (reviewed in 32). Due to the poly-functional nature of all the genotoxic agents used, it is however difficult to directly assign p53-dependent checkpoint induction to DSBs and thus phosphodiester hydrolysis might not recruit sensor molecules for p53 checkpoint function (reviewed in 33). We used various derivatives of the human colorectal cancer cell lines HCT116, which contain homozygous null mutations of either p53 or of the direct p53 transcriptional targets and cell cycle inhibitors p21WAF1/CIP1 and 14-3-3δ. HCT116 parental cells are p53-positive and show normal cell cycle arrest responses following γ-irradiation (24). We assayed cell cycle distributions of these cell lines after incubation with SCPVU-TAT for 30 h (Fig. 4A and D). Regardless of the p53 status, this treatment changed steady-state distributions and the various knockout cell lines showed qualitative differences in their cell cycle stops. Parental, p53-positive cells arrested in G1 and late S/G2 with little S-phase cells, p53 minus and p21WAF1/CIP1 minus HCT116 cell lines showed similar changes, exhibiting more cells with a G2 or with an intermediate S-phase DNA content. Low G1 populations compared to p53-positive cells or to non-treated control samples were detected. For the 14-3-3δ mutants, we observed a strong enrichment of the G1 cell population, consistent with an essential function in blunt end DSB induced checkpoint signaling in G2. All these effects were nuclease-dependent, as the nuclease impaired SC34 proteins did not induce these changes (not shown). This suggests a crucial role for p53 and its downstream targets for a stress-response induced by SCPVU-TAT during cell cycle progression. We then analyzed p53 protein levels in cells treated with SCPVU-TAT and compared these to a nocodazole treatment. HCT116/p53-positive cell lines showed a moderate increase in p53 steady-state levels after 24 h in response to both agents, and this was also the case in the knockout lines for p21WAF1/CIP1 and 14-3-3δ. Moreover, p21WAF1/CIP1 was induced in a p53-dependent manner (Fig. 4B). We further compared the p53 induction in cells that were treated either with SCPVU-TAT or with adriamycin, a commonly used therapeutic DNA damaging agent. Similar rates for p53 stabilization by both agents could be detected in U2OS and HCT116 cells after 24 h (Fig. 4C). Thus, SCPVU-TAT treatment readily induces a p53 checkpoint response.
ATM is essential for SCPVU-TAT repair response
The kinases ATM and ATR resemble the central signaling elements for DNA damage response checkpoints during the cell cycle (5). We treated U2OS cells with caffeine, a methylxantine-derived drug that inhibits the kinase activities of ATM, ATR and mTOR (half-maximal inhibitor concentration: IC50 < 1 mM) (34). Cultivation for 30 h of U2OS cells in 2 mM caffeine containing medium induces a moderate G1 delay whereas growth in medium with 100 nM SCPVU-TAT leads to an increase in the 4n DNA population. In the presence of both agents, however, cell populations are distributed as seen for caffeine alone; no substantial increase in apoptosis can be detected (data not shown). In colony forming assays, the outgrowth was <0.02% after treatment with 100 nM SCPVU-TAT and only a few colonies were formed in AT cells. The ATM-positive control cell line MRC5 was less sensitive to SCPVU-TAT treatment (Fig. 5A and B). This effect was due to the endonucleolytic activity of SCPVU-TAT, as colonies were formed in comparable quality to a ‘no treatment’ in the presence of the nuclease-deficient protein (Fig. 5B, and data not shown).
ATM is an important stress induced p53-Ser15 kinase in vivo (5). p53-Ser15 phosphorylation in turn regulates p53 protein stability and transcriptional activity (reviewed in 32). We tested phosphorylation of p53 on Ser15 in response to SCPVU-TAT treatment of ATM positive and ATM impaired cell lines. For comparison, parallel experiments were done with HU treatment. We employed immunoblotting experiments with extracts obtained after treatments using phospho Ser15 specific antibodies for detection. Specific phosphorylation of p53-Ser15 was detected after treatment with SCPVU-TAT in ATM positive MRC5 (Fig. 5C) as well as in the ATM negative AT-5 (Fig. 5D) cell line. The same was true for HU treatment that resulted in comparable, although consistently higher induction of p53-Ser15 phosphorylation. This p53 in vivo phosphorylation was sensitive towards the ATM/ATR inhibitor caffeine, AT-5 cells incubated with 2 mM caffeine during SCPVU-TAT and also HU treatment did not exhibit significant p53-Ser15 phosphorylation (Fig. 5E).
ATM was previously shown to induce histoneH2AX (H2AX)-Ser139 phosphorylation as an early hallmark event in response to irradiation (35). U2OS cells were transfected with vector controls or with a plasmid for the expression of a dominant loss of function ATM construct (kdATM) (21) together with an EGFP-histone2B transfection marker. Twenty hours after transfections, the cells were treated with SCPVU-TAT for 60 min, fixed and H2AX-phosphorylation was assessed. For this, objects were stained with H2AX-phospho-serine-139 specific antiserum and analyzed with a microscope on a single cell basis. An example and summary of the results are shown in Figure 5F and G. SCPVU-TAT treatment induces H2AX phosphorylation in an ATM-specific manner, because the ATM inhibitor (but not ATR) Wortmanin (50 µM) is antagonistic. Consistently, cells transfected with a dominant negative kdATM did not show significant H2AX phosphorylation in response to SCPVU-TAT treatment. Similar results were also obtained with MCF7 epithelial cells (data not shown).
DISCUSSION
In this communication, we have analyzed the contribution of a minimal DSB on DNA repair and checkpoint controls. We argued that in case a repair and/or checkpoint component is already required for the repair of a low complexity DSB lesion, then these factors should have a role in the repair of all types of DSB in general. For the production of a low complexity DSB, enzymatic hydrolysis seemed suitable. In a vast body of repair and recombination studies in mammalian cells, chromosomal DSBs were introduced by the expression of the I-SceI endonuclease (reviewed in 11). A drawback to this frequently used prototype experiment for highly defined break-populations is the fact that the I-SceI nuclease strongly binds to the cleaved DNA ends and does not dissociate from the hydrolyzed ends which may cause persistence of the cleaved products (reviewed in 18). This feature is believed to be the basis for a significant increase in homologous recombination by gene targeting to I-SceI cleavage sites (17). Similarly, during V(D)J signal joint formation, the recombining nucleases RAG1/2 show low turnover rates and consequently cap the cleaved ends in a non-covalent fashion (16). It seems reasonable that the I-SceI approach in many respects resembles the joining step in signal joint formation where entry of KU has to compete with end-bound nucleases. Consistently, a frequent repair outcome of I-SceI DSBs is precise religation (36). Thus I-SceI DSBs are clearly more complex and do not represent a minimal DSB. We extended these experiments and have developed a novel procedure to induce specific DSBs in whole cell populations. These experiments are supposed to resemble more generally the minimal substrate for DSB repair. The introduction of a high turnover endonuclease which produces free accessible ends, the choice of blunt end cleavage and the fast and efficient protein delivery, advantageously helped us to define the impact of a pure minimal DSB on NHEJ and checkpoint controls.
This was possible by combining the specificity of the scPvuII endonuclease (19) with the TAT-mediated protein transduction technology (28). The introduction of in vivo ‘PvuII breaks’ by means of protein transduction works in the nanomolar range, is rapid, technically simple to perform and due to its high efficiency, does not need positive selection for a read out. The additional use of a nuclease impaired but cognate DNA-binding mutant of PvuII as a control, clearly demonstrates that the nuclease activity of SCPVU-TAT per se causes the checkpoint and repair responses observed in the experiments presented. We conclude that these dsDNA breaks are not biased by side effects and SCPVU-TAT activity in vivo is equivalent to DNA damage induced by 3′-OH and 5′-phosphate containing blunt end DSBs.
The core repair proteins involved in the NHEJ pathway have been extensively investigated. Essential proteins for proper NHEJ function include the XRCC4/DNA-LigaseIV and KU70/KU80 complexes. In vertebrates, in addition, DNA-PKcs has an essential function in certain aspects of end-joining. NHEJ proteins have essential functions in the joining steps of immunoglobulin V(D)J recombination, which has been thoroughly investigated (37–39). The KU70/80 and DNA-PKcs components are also involved in telomere capping, which is supposed to prevent ‘end-to-end fusions’ of the chromosomes (40,41). Initially, we argued that XRCC4/DNA-Ligase IV but probably also other DNA ligases could be sufficient to seal blunt end breaks in the chromatin scaffold context and part of the NHEJ proteins might be redundant for these ‘low complexity’ DNA lesions. This appeared not to be the case. Cell lines containing mutations in XRCC4 or in one of the DNA-PK components were sensitive to SCPVU-TAT treatment. Involvement of the KU70/80 complex in blunt end sealing was confirmed. DNA-PKcs however is only involved in coding join formation during hairpin opening (i.e. formed by transposases including RAG1, 2) (42,43) and overhang processing together with the Artemis proteins (6) and DNA-PKcs orthologues are not found in lower eukaryotes, which taken together suggests a specialized function in vertebrates (38). The fact that cell lines that carry mutations in DNA-PKcs are hypersensitive to SCPVU-TAT treatment in colony formation assays, but show normal checkpoint responses suggested a more general direct involvement in the repair of blunt end DSBs. We analyzed this in more detail and found a strong increase of beyond 4n DNA content in DNA-PKcs impaired cells. Moreover, it was shown previously that the DNA-PKcs deficient SCID mice and also DNA-PKcs (–/–) MEF cells exhibit an increased frequency of spontaneous telomeric fusions and anaphase bridges, which suggests that beside NHEJ functions the DNA-PKcs has its role in protecting telomeres from fusions during normal growth (44,45). Strikingly, due to the high specificity of SCPVU-TAT, we were able to extend these findings and demonstrated that introduction of blunt end breaks into DNA-PKcs impaired human as well as rodent cell lines in vivo and in general results in a significantly higher frequency of anaphase bridges than scored for wild-type or not treated DNA-PKcs mutated cells. These findings suggest a genome-wide function for DNA-PK in preventing heterologous fusions of ligateable DSBs in chromosomes. Finally, chromosomal aberrations like the occurrence of anaphase bridges during mitosis are believed to be important for tumor progression (39,46). The DNA-PKcs proteins from the mutant cell lines used exhibit low kinase activity (47), thus suggesting a rate limiting function for the DNA-PKcs kinase activity not only in telomere function but more generally for the maintenance of chromosomal stability exposed to DSBs. Preliminary experiments with SCPVU-TAT treated XRCC4 mutated cells did not show any significant increase in anaphase bridges which might suggest a separate function for DNA-PK. Future experiments in isogenic backgrounds are needed to finally prove functional differences for inter-chromosomal ligation by the various essential NHEJ components.
Blunt end DSBs are sufficient to induce H2AX phosphorylation and this mainly depends on ATM function. SCPVU-TAT treatment in contrast induces p53 Ser15 phosphorylation also in the absence of ATM, suggesting an overlapping function for ATR in response to blunt end breaks. Consistently, ATM/ATR specific inhibitor concentrations of caffeine abolish this p53 Ser15 phosphorylation completely. So far the exact substrate requirements for activation of ATM or ATR by DSBs in vivo are elusive (5). Results obtained with Saccharomyces cerevisiae suggest that single-strand overhangs are necessary for recognition in vivo (48) and are essential for ATR recruitment to sites of DNA damage in human cells (49). In a cell-free system from Xenopus laevis eggs, an ATM repair response is sufficiently induced by linear, blunt end containing plasmids (50). The SCPVU-TAT experiments demonstrate an activation of ATM/ATR also by blunt end DSBs in human cells in vivo. However, so far we cannot rule out the need for the generation of single-strand overhangs by exonucleolytic activity before checkpoint induction and repair for this low complex lesion. The unique specificity of SCPVU-TAT will allow us to focus on this issue. The simplicity of this technique further provides for high throughput screening for virtually any system accessible for protein transduction, including animal models (28) and favors its use in the analysis of DNA repair pathology profiles.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank K. Raj, P. Beard, L. Zentilin, G. del Sal, B. Vogelstein, P. Jeggo, M. Kastan, S. Lees-Miller and A. Ciccio for cell lines and plasmids, and G. Taccioli for comments on the manuscript. M.-L.T. and A.S. are on leave from Stockholm University, The Arrhenius Laboratories for Natural Sciences, Stockholm, Sweden. C.K. wishes to thank F. Baralle for invaluable help and support.
REFERENCES
- 1.Jeggo P., Singleton,B., Beamish,H. and Priestley,A. (1999) Double strand break rejoining by the Ku-dependent mechanism of non-homologous end-joining. C. R. Acad. Sci. III, 322, 109–112. [DOI] [PubMed] [Google Scholar]
- 2.Featherstone C. and Jackson,S.P. (1999) DNA double-strand break repair. Curr. Biol., 9, R759–761. [DOI] [PubMed] [Google Scholar]
- 3.Walker J.R., Corpina,R.A. and Goldberg,J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 4.Jeggo P.A. (1997) DNA-PK: at the cross-roads of biochemistry and genetics. Mutat. Res., 384, 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Durocher D. and Jackson,S.P. (2001) DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol., 13, 225–231. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y., Pannicke,U., Schwarz,K. and Lieber,M.R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell, 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 7.Gennery A.R., Cant,A.J. and Jeggo,P.A. (2000) Immunodeficiency associated with DNA repair defects. Clin. Exp. Immunol., 121, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins E.J., Nair,A., Cowley,D.O., Van Dyke,T., Chang,Y. and Ramsden,D.A. (2002) Sensing of intermediates in V(D)J recombination by ATM. Genes Dev., 16, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeggo P.A. (1998) Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res., 150, S80–91. [PubMed] [Google Scholar]
- 10.Povirk L.F. (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res., 355, 71–89. [DOI] [PubMed] [Google Scholar]
- 11.Richardson C., Elliott,B. and Jasin,M. (1999) Chromosomal double-strand breaks introduced in mammalian cells by expression of I-Sce I endonuclease. Methods Mol. Biol., 113, 453–463. [DOI] [PubMed] [Google Scholar]
- 12.Obe G., Johannes,C. and Schulte-Frohlinde,D. (1992) DNA double-strand breaks induced by sparsely ionizing radiation and endonucleases as critical lesions for cell death, chromosomal aberrations, mutations and oncogenic transformation. Mutagenesis, 7, 3–12. [DOI] [PubMed] [Google Scholar]
- 13.Manivasakam P., Aubrecht,J., Sidhom,S. and Schiestl,R.H. (2001) Restriction enzymes increase efficiencies of illegitimate DNA integration but decrease homologous integration in mammalian cells. Nucleic Acids Res., 29, 4826–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel J., Fritsche,M., Mai,S., Brandner,G. and Hess,R.D. (1995) Enhanced p53 activity and accumulation in response to DNA damage upon DNA transfection. Oncogene, 11, 1363–1370. [PubMed] [Google Scholar]
- 15.Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- 16.Jones J.M. and Gellert,M. (2001) Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA, 98, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epinat J.C., Arnould,S., Chames,P., Rochaix,P., Desfontaines,D., Puzin,C., Patin,A., Zanghellini,A., Paques,F. and Lacroix,E. (2003) A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res., 31, 2952–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier B.S. and Stoddard,B.L. (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res., 29, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simoncsits A., Tjornhammar,M.L., Rasko,T., Kiss,A. and Pongor,S. (2001) Covalent joining of the subunits of a homodimeric type II restriction endonuclease: single-chain PvuII endonuclease. J. Mol. Biol., 309, 89–97. [DOI] [PubMed] [Google Scholar]
- 20.Simoncsits A., Chen,J., Percipalle,P., Wang,S., Toro,I. and Pongor,S. (1997) Single-chain repressors containing engineered DNA-binding domains of the phage 434 repressor recognize symmetric or asymmetric DNA operators. J. Mol. Biol., 267, 118–131. [DOI] [PubMed] [Google Scholar]
- 21.Lim D.S., Kim,S.T., Xu,B., Maser,R.S., Lin,J., Petrini,J.H. and Kastan,M.B. (2000) ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature, 404, 613–617. [DOI] [PubMed] [Google Scholar]
- 22.Thompson L.H. and Jeggo,P.A. (1995) Nomenclature of human genes involved in ionizing radiation sensitivity. Mutat. Res., 337, 131–134. [DOI] [PubMed] [Google Scholar]
- 23.Lees-Miller S.P., Godbout,R., Chan,D.W., Weinfeld,M., Day,R.S.R., Barron,G.M. and Allalunis-Turner,J. (1995) Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science, 267, 1183–1185. [DOI] [PubMed] [Google Scholar]
- 24.Chan T.A., Hwang,P.M., Hermeking,H., Kinzler,K.W. and Vogelstein,B. (2000) Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev., 14, 1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 25.Kühne C. and Banks,L. (1998) E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem., 273, 34302–34309. [DOI] [PubMed] [Google Scholar]
- 26.Kühne C., Gardiol,D., Guarnaccia,C., Amenitsch,H. and Banks,L. (2000) Differential regulation of human papillomavirus E6 by protein kinase A: conditional degradation of human discs large protein by oncogenic E6. Oncogene, 19, 5884–5891. [DOI] [PubMed] [Google Scholar]
- 27.Vives E., Brodin,P. and Lebleu,B. (1997) A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem., 272, 16010–16017. [DOI] [PubMed] [Google Scholar]
- 28.Wadia J.S. and Dowdy,S.F. (2002) Protein transduction technology. Curr. Opin. Biotechnol., 13, 52–56. [DOI] [PubMed] [Google Scholar]
- 29.Nagahara H., Vocero-Akbani,A.M., Snyder,E.L., Ho,A., Latham,D.G., Lissy,N.A., Becker-Hapak,M., Ezhevsky,S.A. and Dowdy,S.F. (1998) Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nature Med., 4, 1449–1452. [DOI] [PubMed] [Google Scholar]
- 30.Jeggo P.A. (1985) Genetic analysis of X-ray-sensitive mutants of the CHO cell line. Mutat. Res., 146, 265–270. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez G.S., Bryntesson,F., Torres-Arzayus,M.I., Priestley,A., Beeche,M., Saito,S., Sakaguchi,K., Appella,E., Jeggo,P.A., Taccioli,G.E. et al. (1999) DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature, 400, 81–83. [DOI] [PubMed] [Google Scholar]
- 32.Prives C. (1998) Signaling to p53: breaking the MDM2-p53 circuit. Cell, 95, 5–8. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B.B. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 34.Sarkaria J.N., Busby,E.C., Tibbetts,R.S., Roos,P., Taya,Y., Karnitz,L.M. and Abraham,R.T. (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res., 59, 4375–4382. [PubMed] [Google Scholar]
- 35.Burma S., Chen,B.P., Murphy,M., Kurimasa,A. and Chen,D.J. (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem., 276, 42462–42467. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y., Lukacsovich,T. and Waldman,A.S. (1999) Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol. Cell. Biol., 19, 8353–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai Y., Kysela,B., Hanakahi,L.A., Manolis,K., Riballo,E., Stumm,M., Harville,T.O., West,S.C., Oettinger,M.A. and Jeggo,P.A. (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc. Natl Acad. Sci. USA, 100, 2462–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson S.P. and Jeggo,P.A. (1995) DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem. Sci., 20, 412–415. [DOI] [PubMed] [Google Scholar]
- 39.Khanna K.K. and Jackson,S.P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet., 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 40.Allen C., Kurimasa,A., Brenneman,M.A., Chen,D.J. and Nickoloff,J.A. (2002) DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl Acad. Sci. USA, 99, 3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAinsh A.D., Scott-Drew,S., Murray,J.A. and Jackson,S.P. (1999) DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol., 9, 963–966. [DOI] [PubMed] [Google Scholar]
- 42.Taccioli G.E., Amatucci,A.G., Beamish,H.J., Gell,D., Xiang,X.H., Torres Arzayus,M.I., Priestley,A., Jackson,S.P., Marshak Rothstein,A., Jeggo,P.A. et al. (1998) Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity, 9, 355–366. [DOI] [PubMed] [Google Scholar]
- 43.McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- 44.Bailey S.M., Meyne,X., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goytisolo F.A., Samper,E., Edmonson,S., Taccioli,G.E. and Blasco,M.A. (2001) The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol., 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferguson D.O. and Alt,F.W. (2001) DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene, 20, 5572–5579. [DOI] [PubMed] [Google Scholar]
- 47.Blunt T., Finnie,N.J., Taccioli,G.E., Smith,G.C., Demengeot,J., Gottlieb,T.M., Mizuta,R., Varghese,A.J., Alt,F.W., Jeggo,P.A. et al. (1995) Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell, 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 48.Lee S.E., Moore,J.K., Holmes,A., Umezu,K., Kolodner,R.D. and Haber,J.E. (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell, 94, 399–409. [DOI] [PubMed] [Google Scholar]
- 49.Zou L. and Elledge,S.J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science, 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
- 50.Costanzo V., Robertson,K.D., Ying,C.Y., Kim,E., Avvedimento,E., Gottesman,M., Grieco,D. and Gautier,J. (2000) Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell, 6, 649–659. [DOI] [PubMed] [Google Scholar]