Abstract
DNA and RNA are known to have different structural properties. In the present study, molecular dynamics (MD) simulations on a series of RNA and DNA duplexes indicate differential structural flexibility for the two classes of oligonucleotides. In duplex RNA, multiple base pairs experienced local opening events into the major groove on the nanosecond time scale, while such events were not observed in the DNA simulations. Three factors are indicated to be responsible for the base opening events in RNA: solvent–base interactions, 2′OH(n)–O4′(n+1) intra-strand hydrogen bonding, and enhanced rigid body motion of RNA at the nucleoside level. Water molecules in the major groove of RNA contribute to initiation of base pair opening. Stabilization of the base pair open state is due to a ‘conformational switch’ comprised of 2′OH(n)–O4′(n+1) hydrogen bonding and a rigid body motion of the nucleoside moiety in RNA. This rigid body motion is associated with decreased flexibility of the glycosyl linkage and sugar moieties in A-form structures. The observed opening rates in RNA are consistent with the imino proton exchange experiments for AU base pairs, although not for GC base pairs, while structural and flexibility changes associated with the proposed conformational switch are consistent with survey data of RNA and DNA crystal structures. The possible relevance of base pair opening events in RNA to its many biological functions is discussed.
INTRODUCTION
RNA is one of the key biological macromolecules, being involved in a variety of processes that require specific molecular recognition, including transcription and translation (1). Our understanding of the varied roles of RNA has increased rapidly in the past two decades, particularly since the discovery of catalytic RNA (2,3), which has been followed by the elucidation of the extensive structural and catalytic contributions of RNA to the ribosome (4,5). Central to its diverse biological functions, RNA molecules adopt a variety of complex three-dimensional (3D) structures, showing it to be significantly more polymorphic than DNA (6–8). For example, the large ribosomal subunit (9) includes various non-canonical conformations of RNA, including those important for interactions with proteins, while small ribozymes assume four distinct tertiary structural types, with known variations amongst each type (5). Other examples are the structures of transfer RNA (10). In all of these RNA structures, double-helical, duplex regions are combined with various stem–loops, bulges and mismatches. In contrast, DNA assumes primarily double-helical, duplex structures.
The double-helical, duplex forms of RNA and DNA are their most studied 3D structures. Helical, duplex RNA is conformationally more confined than DNA, with duplex RNA primarily being of the A form while DNA duplexes are able to assume the B, A or Z forms, among others (11–15), with the assumed form being influenced by both sequence and the environment (16–20). The more variable nature of duplex DNA is supported by its ability to incorporate non-standard nucleotides without disrupting the overall B-form conformation (21). In addition, DNA can assume the A form in hybrid duplexes with RNA, while RNA usually cannot fit into the B-form context, with such structures being unstable (22–24). Thus, there is an inconsistency in the structural variability of RNA versus DNA. While RNA assumes a wider variety of 3D structures as compared to DNA, at the secondary (i.e. helical) level it is less polymorphic than DNA. Experimental studies have clearly shown this apparent contradiction. However, experimental approaches alone may be limited in understanding this phenomenon.
Understanding of the structure and dynamics of oligonucleotides at the atomic level has been greatly facilitated by theoretical approaches. Multiple examples of the application of both quantum mechanical (QM) (25–30) and molecular dynamic (MD) based studies of nucleic acids are available in the literature (31–48). With respect to RNA, the orientation of the 2′OH group, which is believed to be the dominant factor responsible for the structural and dynamical differences between RNA and DNA, is not readily deduced from experimental data (49,50). Therefore, interpretation of experimental data is equivocal regarding the role of the 2′OH on RNA structure. MD simulations, on the other hand, can provide an atomic detail picture to help elucidate subtle structural effects associated with the 2′OH (51,52). Clearly, MD simulations will make a significant contribution in elucidating oligonucleotide structural and dynamic properties.
In this manuscript, we report results from MD simulations indicating differential structural fluctuations of duplex RNA relative to DNA, leading to increased base pair opening events on the nanosecond time scale in the former. These opening events are shown to be related to altered hydration and a ‘conformational switch’ unique to RNA that is comprised of a rigid body behavior of the nucleoside subunits and 2′OH(n)–O4′(n+1) hydrogen bonds. In addition, published experimental results and data from surveys of the nucleic acid database (NDB) (53) are presented to support the conclusions based on the MD simulations.
MATERIALS AND METHODS
MD simulations were performed using the program CHARMM (54) with the CHARMM27 nucleic acid force field (55,56), the modified TIP3P water model (57) and published sodium and chloride parameters (58). Starting structures were either the canonical A or B form (59) of the respective sequences and were prepared with QUANTA (Accelrys Inc.). For UAAGGAGGUGAU (IX in Table 1) (PDB identifier: 1sdr) (60) the starting structure was the crystal structure. Starting structures were overlaid with a pre-equilibrated solvent box consisting of water and sodium ions or NaCl, as required. The solvent box was extended 8 Å beyond the nucleic acid non-hydrogen atoms. Solvent molecules that were within 1.8 Å of nucleic acid non-hydrogen atoms were deleted. The number of sodium ions in the solvent box was adjusted to yield NaCl concentrations of ∼0.3 and 1 M by deleting sodium ions that were furthest from nucleic acid or adding sodium and chloride ions at random positions. The number of chloride ions added was selected to neutralize the systems for the high salt concentration (1 M). Periodic boundary conditions were simulated using the CRYSTAL module (61) in CHARMM. Each system was minimized for 500 Adopted Basis Newton–Raphson (ABNR) steps with mass-weighted harmonic constraints of 2.0 kcal/mol/Å on the non-hydrogen atoms of the oligonucleotide. The minimized system was then subjected to a 20 ps MD simulation in the constant volume, isothermal (NVT) ensemble in the presence of the harmonic constraints. The integration time step was 2 fs and SHAKE (62) was used to constrain all covalent bonds involving hydrogens. In all calculations, electrostatics were treated via the particle mesh Ewald method (63,64), Lennard–Jones interactions were truncated at 12 Å with a switch smoothing function from 10 to 12 Å and the non-bond atom lists were updated heuristically. Production simulations were performed for 5 ns in the isothermal isobaric (NPT) ensemble at 300K with the Leap-Frog integrator. The NPT ensemble was achieved using Hoover chains (65) for temperature control with a thermal piston mass of 1000 kcal/mol/ps2 at 300K with pressure control to 1 ATM via the Langevin Piston method (66) with a piston mass of 600 a.m.u. and the piston collision frequency set to 0. In selected cases the simulations were extended to 7 ns to ensure that the final 4 ns had converged (see below). The final 4 ns from each trajectory were used for analysis. For the identification of hydrogen bonds, distance cutoffs of 2.5 and 3.5 Å were used for hydrogen–heteroatom and heteroatom–heteroatom pairs, respectively. A base pair is defined as being in the open state when the N1(purine)–N3(pyrimidine) hydrogen bond distance exceeds 3.5 Å.
Table 1. Duplex sequences, starting conformations and salt concentrations for performed simulations.
| IDa | Duplex sequenceb | Starting conformation | NaCl conc. (M) |
|---|---|---|---|
| I | GAGUACUC | A | 0.3, 1 |
| II | GCGAGUACUCGC | A | 0.3, 1 |
| III | d(GAGUACUC) | A | 0.3, 1 |
| IV | d(GCGAGUACUCGC) | A | 0.3, 1 |
| V | d(GAGTACTC) | A | 0.3, 1 |
| VI | d(GAGTACTC) | B | 0.3, 1 |
| VII | CGCGAUCGCG | A | 0.3, 1 |
| VIII | CCUUUCGAAAGG | A | 0.3, 1 |
| IX | UAAGGAGGUGAUc | A | 0.3, 1 |
aThe roman numerals identifiers are used for referring to the corresponding nucleic acid duplexes throughout the manuscript.
bThe central six bases used for analysis are printed in bold.
Experimental survey data of oligonucleotide crystal structures was based on the NDB (53). Structures compiled included duplex structures for RNA, B-form DNA and A-form DNA. In addition to the RNA duplex structures, tRNAs, ribozymes and RNA in the ribosome were analyzed. These structures were selected based on the NDB classifications and only structures with a resolution of 2.0 Å or better were used for the survey of duplex A-DNA, B-DNA and A-RNA. Based on this criteria, 74, 68 and 35 duplex B-DNA, A-DNA and A-RNA duplexes, respectively, were identified. Most of the tRNA, ribosome and ribozyme structures have lower resolutions (>2.0 Å) and therefore all nine tRNA, 65 ribosomal RNAs and nine ribozymes were included in the survey.
RESULTS AND DISCUSSION
A variety of duplexes have been investigated in the proposed study, with the majority of sequences studied in two salt concentrations. Sequence I was initially selected as it represents the control sequence in an experimental study on GU mismatches in RNA (67); our long-term goal is to elucidate the physical origins of the unique stability properties of these mismatches. During these efforts we came upon the interesting results presented in this manuscript. Results of the GU mismatch study will be published elsewhere (Pan and MacKerell, in preparation). Sequence II was chosen to study the impact of end effects on the observed structural and dynamic properties. ‘DNA’ sequences III and IV have the same base pair sequence and composition as I and II and were selected to test how the RNA 2′OH group impacts the structural and dynamic properties while maintaining the uracil bases. DNA sequence V was selected as it includes both the omission of the 2′OH groups and the U→T base switch. Sequence VI is the same as V except the simulation was initiated from the B conformation, allowing for convergence based on the starting structure to be tested. Sequences VII and VIII were included as they have been the subject of imino proton exchange experiments (68). Finally, RNA duplex IX was included as its sequence differs significantly from the other oligonucleotides and a crystal structure is available (60). This collection of oligonucleotides allows for the influence of sequence and chemical differences between RNA and DNA on the observed differences in the fluctuations between RNA and DNA to be investigated, as well as allowing for direct comparisons with experimental data.
Structural and dynamical properties
Average root mean square differences (RMSD) for the simulations are presented in Table 2. In all cases both the canonical A and B conformations are used as reference states, despite the fact that only two of the simulations were started from the B form (Table 1). The DNAs starting from the A form, including those containing U bases, all experienced an A→B transition, although the longer sequences took more time to converge (see Supplementary Material, Fig. S1). These simulations (sequence IV) were extended to 7 ns to ensure that the last 4 ns of the trajectories were properly converged for analysis. It can be seen that in both low (0.3 M) and high (1 M) salt concentrations, the RMSDs of RNA simulations are distinctly smaller when the reference state was the A form versus the B form. In addition, for sequence IX the average RMS differences with respect to the crystal structure were 1.5 and 1.3 Å at low and high salt concentrations, respectively, further indicating that the presented calculations yield the experimentally appropriate conformations.
Table 2. Average RMS differences (Å).
| Duplex | 0.3 M NaCl | 1 M NaCl | ||||||
|---|---|---|---|---|---|---|---|---|
| Heavy | Base | Sugar | Phos | Heavy | Base | Sugar | Phos | |
| Versus A form | ||||||||
| I | 2.7 ± 0.5 | 2.6 ± 0.5 | 3.2 ± 0.7 | 2.6 ± 0.5 | 2.5 ± 0.7 | 2.4 ± 0.7 | 3.0 ± 0.9 | 2.5 ± 0.7 |
| II | 2.0 ± 0.6 | 1.8 ± 0.5 | 2.6 ± 0.7 | 2.0 ± 0.6 | 1.6 ± 0.6 | 1.5 ± 0.6 | 1.9 ± 0.7 | 1.6 ± 0.5 |
| III | 2.4 ± 0.3 | 1.8 ± 0.2 | 3.8 ± 0.5 | 2.4 ± 0.4 | 2.5 ± 0.3 | 1.9 ± 0.3 | 3.8 ± 0.5 | 2.4 ± 0.4 |
| IV | 2.4 ± 0.4 | 1.7 ± 0.3 | 3.6 ± 0.5 | 2.5 ± 0.4 | 2.6 ± 0.3 | 1.9 ± 0.3 | 3.8 ± 0.5 | 2.7 ± 0.4 |
| V | 2.4 ± 0.3 | 1.7 ± 0.3 | 3.9 ± 0.5 | 2.4 ± 0.4 | 2.3 ± 0.3 | 1.6 ± 0.2 | 3.7 ± 0.5 | 2.3 ± 0.4 |
| VI | 2.6 ± 0.3 | 1.7 ± 0.3 | 4.3 ± 0.4 | 2.5 ± 0.3 | 2.4 ± 0.3 | 1.6 ± 0.2 | 4.1 ± 0.3 | 2.4 ± 0.3 |
| VII | 2.3 ± 0.6 | 2.2 ± 0.4 | 3.2 ± 1.1 | 2.0 ± 0.6 | 2.4 ± 0.4 | 2.8 ± 0.6 | 2.2 ± 0.3 | 2.0 ± 0.3 |
| VIII | 1.4 ± 0.4 | 1.2 ± 0.4 | 2.1 ± 0.6 | 1.3 ± 0.4 | 1.7 ± 0.5 | 1.5 ± 0.4 | 2.4 ± 0.8 | 1.6 ± 0.5 |
| IX | 1.5 ± 0.4 | 1.7 ± 0.5 | 1.7 ± 0.4 | 1.3 ± 0.3 | 1.3 ± 0.4 | 1.1 ± 0.4 | 1.8 ± 0.6 | 1.2 ± 0.4 |
| Versus B form | ||||||||
| I | 4.2 ± 0.5 | 3.4 ± 0.5 | 5.3 ± 0.7 | 4.6 ± 0.5 | 4.2 ± 0.7 | 3.3 ± 0.7 | 5.3 ± 0.9 | 4.6 ± 0.6 |
| II | 3.6 ± 0.4 | 2.5 ± 0.4 | 4.8 ± 0.6 | 4.0 ± 0.4 | 3.4 ± 0.3 | 2.3 ± 0.3 | 4.6 ± 0.3 | 3.8 ± 0.3 |
| III | 1.6 ± 0.3 | 1.1 ± 0.3 | 1.9 ± 0.4 | 1.9 ± 0.4 | 1.7 ± 0.4 | 1.2 ± 0.3 | 1.9 ± 0.5 | 2.0 ± 0.5 |
| IV | 1.7 ± 0.3 | 1.1 ± 0.3 | 2.8 ± 0.4 | 1.7 ± 0.4 | 1.6 ± 0.3 | 1.0 ± 0.2 | 2.6 ± 0.3 | 1.6 ± 0.4 |
| V | 1.8 ± 0.3 | 1.6 ± 0.2 | 2.8 ± 0.3 | 1.7 ± 0.4 | 1.9 ± 0.3 | 1.6 ± 0.2 | 2.8 ± 0.4 | 1.7 ± 0.4 |
| VI | 1.4 ± 0.3 | 1.1 ± 0.2 | 1.8 ± 0.4 | 1.7 ± 0.3 | 1.5 ± 0.3 | 1.1 ± 0.2 | 1.9 ± 0.4 | 1.8 ± 0.3 |
| VII | 3.7 ± 0.4 | 3.1 ± 0.3 | 4.7 ± 0.7 | 3.9 ± 0.5 | 3.7 ± 0.3 | 3.6 ± 0.5 | 4.4 ± 0.4 | 3.6 ± 0.3 |
| VIII | 6.0 ± 0.6 | 5.1 ± 0.7 | 7.4 ± 0.7 | 6.3 ± 0.5 | 5.7 ± 0.4 | 4.9 ± 0.4 | 7.0 ± 0.6 | 6.1 ± 0.5 |
| IX | 3.3 ± 0.3 | 2.4 ± 0.2 | 4.2 ± 0.4 | 3.7 ± 0.3 | 3.3 ± 0.3 | 2.3 ± 0.3 | 4.3 ± 0.5 | 3.8 ± 0.3 |
The canonical A- and B-form structures were used as the reference states. Averages over the final 4 ns and only non-hydrogen atoms were included. Errors represent the RMS fluctuations.
Average phosphodiester backbone dihedrals (Table 3) and sugar pseudorotation angle distributions (data not shown) confirm the conformational difference between RNA and DNA. The calculated dihedral angles are in good agreement with experimental survey data for DNA and RNA (69–71), respectively, and the sugar pucker distributions are also typical for their respective canonical conformations (25,72). In addition, analysis of helicoid parameters via the program CURVES (73) yielded results (not shown) consistent with the conclusions based on the RMS difference and dihedral angle analysis. These results indicate that the present simulations yield the expected A-like ensembles for the RNAs and B-like ensembles for the DNAs (31,74). Moreover, these results confirm the dominant role of the 2′OH group on the conformational preference for RNA (11), since all the DNA sequences lacking the 2′OH but still containing U bases underwent the transition from the A to B forms.
Table 3. Average backbone dihedral angles and their fluctuations for the central six residues (degrees)a.
| Duplex | α | β | γ | δ | ε | ζ | χ |
|---|---|---|---|---|---|---|---|
| 0.3 M NaCl | |||||||
| I | 282 ± 20 | 173 ± 16 | 63 ± 11 | 77 ± 5 | 205 ± 19 | 290 ± 24 | 200 ± 9 |
| II | 285 ± 21 | 172 ± 18 | 66 ± 12 | 78 ± 5 | 210 ± 18 | 285 ± 29 | 200 ± 8 |
| III | 298 ± 24 | 165 ± 20 | 52 ± 14 | 129 ± 18 | 194 ± 20 | 250 ± 31 | 249 ± 18 |
| IV | 299 ± 18 | 170 ± 18 | 51 ± 14 | 130 ± 19 | 192 ± 19 | 253 ± 28 | 248 ± 19 |
| V | 299 ± 18 | 163 ± 23 | 52 ± 14 | 129 ± 19 | 197 ± 24 | 250 ± 45 | 250 ± 18 |
| VI | 297 ± 27 | 166 ± 24 | 53 ± 16 | 126 ± 22 | 198 ± 27 | 245 ± 42 | 250 ± 21 |
| VII | 283 ± 22 | 173 ± 17 | 63 ± 11 | 78 ± 6 | 204 ± 23 | 285 ± 22 | 200 ± 10 |
| VIII | 283 ± 20 | 173 ± 17 | 65 ± 12 | 77 ± 5 | 207 ± 17 | 287 ± 20 | 200 ± 9 |
| IX | 283 ± 17 | 174 ± 18 | 64 ± 11 | 77 ± 5 | 208 ± 19 | 290 ± 19 | 199 ± 8 |
| 1 M NaCl | |||||||
| I | 285 ± 21 | 173 ± 18 | 64 ± 13 | 77 ± 5 | 208 ± 18 | 288 ± 19 | 200 ± 8 |
| II | 285 ± 19 | 170 ± 18 | 64 ± 10 | 77 ± 5 | 210 ± 17 | 285 ± 22 | 200 ± 8 |
| III | 296 ± 25 | 165 ± 20 | 52 ± 14 | 126 ± 21 | 197 ± 22 | 249 ± 35 | 246 ± 22 |
| IV | 300 ± 20 | 169 ± 20 | 52 ± 14 | 133 ± 15 | 193 ± 21 | 249 ± 31 | 251 ± 16 |
| V | 298 ± 22 | 166 ± 22 | 53 ± 14 | 127 ± 21 | 193 ± 23 | 243 ± 48 | 247 ± 19 |
| VI | 299 ± 21 | 168 ± 21 | 52 ± 15 | 128 ± 20 | 191 ± 17 | 256 ± 24 | 247 ± 19 |
| VII | 280 ± 25 | 173 ± 19 | 62 ± 13 | 77 ± 6 | 206 ± 22 | 285 ± 22 | 202 ± 13 |
| VIII | 283 ± 19 | 172 ± 17 | 64 ± 12 | 77 ± 5 | 208 ± 18 | 286 ± 27 | 200 ± 9 |
| IX | 284 ± 20 | 173 ± 19 | 63 ± 12 | 77 ± 6 | 210 ± 19 | 291 ± 25 | 199 ± 9 |
| Expt (A form)b | 291 | 175 | 57 | 80 | 205 | 287 | 199 |
| Expt (B form)b | 298 | 168 | 51 | 134 | 187 | 262 | 252 |
aEach dihedral was averaged over the last 4 ns for all the central six nucleosides. Errors represent RMS fluctuations.
bSee Foloppe and MacKerell (69).
While the expected conformations were obtained from the simulations, inspection of the data in Table 2 reveals some interesting differences between RNA and DNA. The RMSDs of the bases and phosphates from the RNA simulations (I, II, VII, VIII and IX) with respect to the A form were similar, with the differences between those moieties being <0.3 Å, while with the DNA simulations (III, IV, V and VI), the differences between base and phosphate RMSDs with respect to the B form were at least 0.4 Å, with the base variations less than the phosphates. This behavior is opposite to that observed in the RNAs. This observation indicates that the structural properties of the backbone, relative to the bases, differ in RNA versus DNA.
Consistent with the structural differences in RNA and DNA are the RMS fluctuations of the bases and the backbone moieties, as shown in Table 4. In RNA the RMS fluctuations for base, sugar and phosphate moieties are similar. In contrast, the phosphates have the largest fluctuations and the bases fluctuate the least in the DNA simulations. Thus, the present results indicate that there are inherent differences in the structural variations and flexibility in RNA versus DNA, with the motions of the individual moieties in DNA being relatively independent, while in RNA those motions are more coordinated, with the nucleosides and nucleotides moving more as a rigid body.
Table 4. RMS structural fluctuations (Å) for the central six residues from the last 4 ns of the trajectories.
| Duplex | All | Base | Sugar | Phosphate |
|---|---|---|---|---|
| 0.3 M NaCl | ||||
| I | 1.5 | 1.6 | 1.3 | 1.6 |
| II | 1.3 | 1.4 | 1.2 | 1.4 |
| III | 0.9 | 0.7 | 0.9 | 1.2 |
| IV | 0.9 | 0.7 | 0.9 | 1.2 |
| V | 0.9 | 0.7 | 0.9 | 1.2 |
| VI | 0.9 | 0.7 | 0.9 | 1.2 |
| VII | 1.2 | 1.2 | 1.4 | 1.1 |
| VIII | 1.1 | 1.0 | 1.2 | 1.0 |
| IX | 1.0 | 1.0 | 1.1 | 0.9 |
| 1 M NaCl | ||||
| I | 1.8 | 1.8 | 1.6 | 1.8 |
| II | 1.0 | 1.0 | 0.9 | 1.1 |
| III | 0.9 | 0.8 | 1.0 | 1.3 |
| IV | 0.9 | 0.7 | 0.9 | 1.1 |
| V | 0.9 | 0.7 | 0.9 | 1.1 |
| VI | 0.8 | 0.7 | 0.9 | 1.1 |
| VII | 0.9 | 0.9 | 0.9 | 0.8 |
| VIII | 1.4 | 1.4 | 1.6 | 1.3 |
| IX | 1.0 | 0.9 | 1.2 | 1.0 |
To validate the differences in the relative mobilities of the bases and backbone moieties in RNA and DNA a survey was undertaken of duplex DNA and RNA in the NDB. Reported in Table 5 are the average B-factors for the different moieties in B-form DNA, A-form DNA and RNA duplexes along with the average differences of the B-factors for the individual crystal structures. In B-form DNA, the difference in the average B-factors upon going from the base to the phosphate is close to 11, while in A-form DNA it decreases to ∼8 and is slightly larger than 6 in RNA. Comparison of the sugar and base B-factors shows a similar pattern, where the differences are 6.2 in B DNA, 4.6 in A DNA and 3.6 in RNA. These differences are beyond the standard errors as well as being above 99% confidence levels in the two-tail t-test, with the exception of the average sugar–phosphate difference between A DNA and RNA (not shown). Thus, the survey data are consistent with the simulation results, indicating the moieties in B DNA to have the most independent flexibilities, with the differences decreased in A-form DNA and the lowest in RNA, supporting the present observation that RNA at the nucleoside and nucleotide levels moves more like a rigid body than DNA.
Table 5. Average B factors and B-factor differences of DNA and RNA duplexes from NDB database.
| Average B factors | |||
|---|---|---|---|
| Base | Sugar | Phosphate | |
| B-DNA | 15.7 ± 0.9 | 20.4 ± 1.1 | 24.9 ± 1.3 |
| A-DNA | 16.1 ± 0.8 | 19.8 ± 1.0 | 23.5 ± 1.2 |
| RNA |
19.9 ± 1.6 |
23.1 ± 1.5 |
25.5 ± 1.6 |
| Average B-factor differences | |||
| |
Phosphate–Base |
Sugar–Base |
Phosphate–Sugar |
| B-DNA | 10.8 ± 0.7 | 6.2 ± 0.4 | 4.6 ± 0.3 |
| A-DNA | 8.4 ± 0.5 | 4.6 ± 0.3 | 3.8 ± 0.2 |
| RNA | 6.3 ± 0.6 | 3.6 ± 0.4 | 2.6 ± 0.3 |
Averages are over the non-hydrogen atom B factors for the respective moieties in each structure included in the survey. Average B-factor differences represent the average over all crystal structures for the respective duplex types of the differences over the respective moieties in each crystal. Errors represent the standard errors (88). Units in Å2.
Further support for the more rigid body like motion in RNA versus DNA at the nucleoside and nucleotide levels comes from comparison of the RMS fluctuations of the phosphodiester backbone dihedrals (Table 3) versus the cartesian coordinate RMS fluctuations (Table 4). In RNA, the RMS fluctuations of the dihedrals are generally smaller than in DNA, with the difference being most significant with δ and χ. In contrast, the RMS fluctuations of the cartesian coordinates of DNA are typically lower than those of the RNA. Such a difference is consistent with a model where RNA at the nucleoside and nucleotide level is moving more as a rigid body than in DNA, leading to lower dihedral fluctuations, while larger fluctuations of the nucleoside and nucleotide moieties are occurring in RNA leading to the larger cartesian RMS fluctuations.
Base pair opening
Further investigations of the differential mobility in RNA versus DNA involved monitoring the Watson–Crick (WC) base pairing. Analysis of the N1–N3 hydrogen bond distances revealed that the WC hydrogen bonds in RNA were frequently broken. Supplementary Material Table S1 summarizes the number and duration of the opening events for the central six base pairs based on the N1–N3 distance. The corresponding time series details are also shown in the Supplementary Material (Table S2 and Fig. S2). For the central six base pairs in the RNA octamers, the N1–N3 H-bonds were broken at least once, some of which were reformed over the 4 ns production trajectory. For duplex II, of which the central eight residues are the same as duplex I, four of the six central base pairs show the same property. The duration of these events covered a wide range, from a few picoseconds to a couple of nanoseconds. Less base opening events were observed at the high salt versus low salt concentration, consistent with salt stabilization of RNA (75). However, opening events did occur in the high salt conditions. Interestingly, all the base pair opening events occurred into the major groove. In contrast, DNA did not have a significant tendency toward base pair opening (Supplementary Material, Fig. S2). In all of the DNA simulations there were only four short opening events, the duration of which were all <10 ps (Table S1).
To see whether the base pair opening may be a common phenomenon in RNA, additional simulations of RNA with different sequences were undertaken (sequences VII, VIII and IX, Table 1). As may be seen in Table S1 a variety of opening events occurred in these systems, suggesting that the observed opening events in RNA are not limited to selected sequences. Of the additional sequences studied, VII and VIII have been subjected to NMR-based imino proton exchange experiments (68). In the NMR study it was observed that the lifetime of several of the AU base pairs are quite short, with most being ≤0.1 ms. In several cases the lifetimes were too short to be measured at 15°C. This contrasts lifetimes for d(AT) base pairs in DNA, which are typically 10 ms or longer (76,77). In addition, it was shown that the dissociation constants of AU pairs were similar to that of d(AT) pairs, indicating that the lifetimes of both the open and closed states are reduced in RNA versus DNA, resulting in open lifetimes of ∼1 ns being reported. This timescale is consistent with opening observed in the present calculations. Thus, the present results are in agreement with experimental data indicating enhanced opening of AU base pairs in RNA versus AT base pairs in DNA, with the timescales of those motions being on the nanosecond level.
However, several additional points require discussion. Snoussi and Leroy (68) did not see a significant increase in the opening of GC base pairs in RNA, while the present simulation data does indicate increased opening of GCs. Such a discrepancy indicates a possible bias by the force field used in the present study towards favoring opening, although other confounding factors may contribute. These include temperature differences between the experimental and MD studies, differences in ionic strengths and ions used, limited sampling in the MD simulations as well as inherent limitations in the force field, including lack of electronic polarization. Moreover, it has been shown experimentally that base pair lifetimes in the millisecond time range occur in polyA–polyU (78) and in AU pairs stacked between GC pairs (68), indicating significant sequence-specific effects. Consistent with the latter are computational studies on base flipping in RNA, where the conversion from DNA to RNA for flipping of a U stacked between two Gs (versus a T in the DNA) did not significantly change the flipping energy surface (79). Despite these limitations, the significant increase in base pair opening in AU base pairs is consistent with the experimental data, indicating the calculated observations to be representative of the experimental regimen.
2′OH(n)–O4′(n+1) hydrogen bonding
Simply switching the thymines to uracils in the context of DNA does not lead to enhanced opening, as evidenced by the results for III and IV. This is consistent with experimental studies (68) and, again, points to the dominant role of the 2′OH in the altered behavior of DNA versus RNA. To examine the role of the 2′OH group in RNA base pair opening, hydrogen bonding between adjacent sugars via the 2′OH(n) and the O4′(n+1) was analyzed for each base opening event (Supplementary Material, Table S2). Notably, at least one of the two possible 2′OH–O4′ hydrogen bonds from the two strands was formed in 133 out of the 172 opening events. This was based on the average 2′OH(n)–O4′(n+1) distance being less than 2.5 Å while the average N1–N3 distance was >3.5 Å. In 65 cases, the 2′OH(n)–O4′(n+1) distances for both hydrogen bonds were <2.5 Å. Compared with the average distance (3.3–3.5 Å) between 2′OH(n) and O4′(n+1) for the all the RNA trajectories (see Supplementary Material), it is evident that 2′OH(n) and O4′(n+1) groups had significant interactions during the base pair open states. These results indicate that 2′OH(n)–O4′(n+1) hydrogen bonding is playing an important role in the base pair opened state.
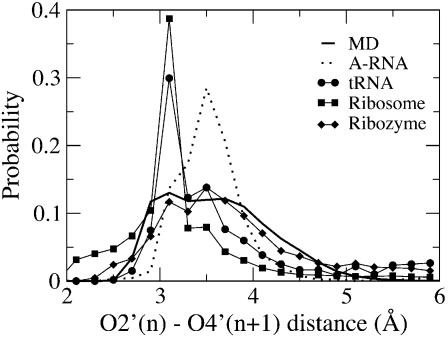
To verify the relevance of the 2′OH(n)–O4′(n+1) hydrogen bonds to the experimental regimen, probability distributions of the O2′(n)–O4′(n+1) distances were obtained from the RNA MD simulations and from the NDB survey data. Presented in Figure 1 are O2′(n)–O4′(n+1) distance probability distributions for all the RNA simulations along with those from the NDB survey for the different structural types of RNA. In the distributions two peaks are evident. The shorter peak at ∼3 Å corresponds to a O2′(n)–O4′(n+1) hydrogen bond while the longer peak represents a non-hydrogen bond state. In the MD simulations the presence of the hydrogen bond peak is evident. This peak is also present in the survey data for tRNA, ribozymes and ribosomal RNA, while in the duplex RNA structures only a shoulder is observed in this region. Such a discrepancy is interesting. First, the observed opening events are consistent with imino proton exchange data for AU base pairs, however, in RNA A-form duplex structures the base pairs are well maintained and only a minor amount of O2′(n)–O4′(n+1) hydrogen bonding is evident from the survey data. This suggests that either the crystal environment is stabilizing the AU base pairs, thereby inhibiting the opening events, or that the opening events are short-lived such that they are not observed in the crystal structures. The latter is consistent with the fast AU opening and closing rates in the experimental study, suggesting that base pair opening events may be occurring in the crystals, however, they are not observed due to the timescale of the experiments. Consistent with our simulation results are the sugar B factors for RNA versus DNA (Table 5), where larger B-factors occur in RNA versus DNA, while the B-factor difference between structural moieties is more dramatic in DNA than in RNA. However, in the non-canonical RNA structures, where a significant number of bases are not involved in WC interactions, a high percentage of the O2′(n)–O4′(n+1) pairs are hydrogen bonded. This suggests a scenario where opening events can occur in all forms of RNA, but are typically not stable enough to be observed via crystallography in A-form duplex RNA. Only when the WC interactions are not present are O2′(n)–O4′(n+1) intrastrand pairs in hydrogen bonding orientations experimentally observed. While not conclusive, NMR data did not exclude the presence of such hydrogen bonds (49), supporting the feasibility of the present hypothesis.
Figure 1.
O2′(n)–O4′(n+1) probability distribution from all the RNA simulations (solid line) and from a survey of the NDB. Survey results for A-RNAs (dotted line), tRNAs (circles), ribosomes (squares) and the ribozyme (diamonds) are shown.
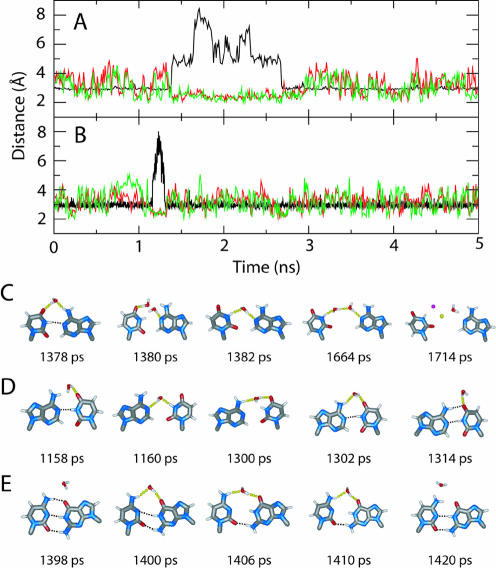
To more closely investigate the relationship of 2′OH(n)–O4′(n+1) hydrogen bonding to base pair opening, we selected two representative events, one from the sequence II simulation at 1 M NaCl and the second from the sequence VIII at 0.3 M NaCl, for further analysis. The event from duplex II lasted ∼1.4 ns (from 1.38 to 2.78 ns) and the second from duplex VIII lasted 0.14 ns (from 1.16 to 1.30 ns). The N1–N3 and corresponding 2′OH–O4′ distance time series for these events are shown in Figure 2. The 2′OH–O4′ distances fluctuate within the range of 2.5–4.5 Å when the base pair WC hydrogen bonds are present for both events. However, during base pair opening, not only do the 2′OH–O4′(n+1) distances decrease significantly, but the fluctuations of those distances become smaller, indicating how 2′OH(n)–O4′(n+1) hydrogen bond formation and the breakage of the N1–N3 hydrogen bond are correlated.
Figure 2.
Selected N1–N3 and 2′OH(n)–O4′(n+1) distances (A and B) and snapshots of base pairs with local waters or ions in the major groove (C, D and E) for the (A and C) U6-A7 base pair from the duplex II simulation at 1 M NaCl and (B and D) A9-U4 base pair from the duplex VIII simulation at 0.3 M NaCl. Also included (E) is the DNA C6-G3 base pair with local waters from the duplex V simulation at 0.3 M NaCl. In (A) and (B), black is the N1–N3 distance, and red and green are the 2′OH–O4′ distances for strands one and two, respectively. For (C), (D) and (E), hydrogen bonds are shown with dotted lines and the two spheres at 1714 ps in (C) are Na+ (yellow) and Cl– (magenta) ions.
To understand if the 2′OH(n)–O4′(n+1) hydrogen bond was initiating base pair opening, the N1-N3 and 2′OH(n)–O4′(n+1) distances from strand one were followed chronologically for the two events (Supplementary Material, Table S3). During initial base pair opening, breakage of the N1–N3 hydrogen bond (distance changed from <3.5 to >3.5 Å) and formation of the 2′OH(n)–O4′(n +1) hydrogen bond (distance changed from >2.5 to <2.5 Å) occurred simultaneously at 1380 ps for one event and at 1160 ps for the other (Table S3). In the reverse process (i.e. base pair closing), the N1–N3 distance decreased from 4.2 to 3.4 Å and then to 3.1 Å from 2670 to 2674 ps for event one. However, the hydrogen bond between 2′OH and O4′(n+1) was maintained during that period, at 1.9, 1.8 and 2.0 Å, respectively. Similar results were observed for the second event. Since 2′OH–O4′(n+1) hydrogen bond formation prior to base pair opening and loss of the 2′OH–O4′(n+1) hydrogen bond prior to the base pair re-closing were not observed, it is concluded that the formation and breakage of the 2′OH(n)–O4′(n+1) hydrogen bonds are not pre-requisites for base pair opening and closing, respectively. Thus, the role of the 2′OH(n)–O4′(n+1) hydrogen bond appears to be stabilization of the open state of the base pairs rather than being responsible for initiation of such events.
The role of solvent molecules in base pair opening
Solvent–solute interactions are an indispensable part of the structure and function of macromolecules. Accordingly, it may be anticipated that solvent may contribute to the differential base pair opening behavior in RNA versus DNA. To investigate this possibility, the number of water molecules within 3.5 Å of the N1 and N3 atoms of each base pair was calculated for the closed states of the simulations (Supplementary Material, Table S4). For the central six base pairs, more water molecules were adjacent to the N1 and N3 atoms in RNA as compared to DNA, supporting the possibility that major groove hydration in RNA may facilitate the opening events.
Details of solvent–base interactions prior to selected base pair opening events were examined to better determine if the increased presence of water molecules near RNA base pairs facilitate base pair opening. The same two events in duplexes II and VIII discussed in the previous section were analyzed. Since the base pair opening pathway was through the major groove for all events, we only focus on the major groove side of the base pairs. For the first opening event (Fig. 2C), at 1378 ps, a water hydrogen bonds with the O4 atom (1.75 Å) of U6 from strand one and with H62 (1.83 Å) of A7 of strand two. At this point, the N1–N3 hydrogen bond is still formed. At 1380 ps, as the N1–N3 hydrogen bond breaks, the water molecule forms a hydrogen bond (1.94 Å) with N1 of A7, followed by an additional hydrogen bond with N3 of U6 at 1382 ps. After this, the base pair remained open until 2780 ps, during which at least one water or sodium chloride ions were between the two bases, forming a hydrogen bond network (see below). For the second event (Fig. 2D), a water molecule competes with H62 of A9 for hydrogen bonding with O4 of U4, disrupting the H62–O4 hydrogen bonding right before the N1–N3 hydrogen bond breakage (1158 ps). This water molecule further hydrogen bonds with both the N1 of A9 and the H3N3 of U4, marking the beginning of the base pair opening event at 1160 ps. There was at least one water molecule hydrogen bonding with the bases during the base opened state (data not shown). The WC hydrogen bond was restored for this base pair from 1300 to 1314 ps when the water molecule was released from between the two bases.
Similar interactions between water and base pairs were found for the DNA simulations. For example, in the d(GAGTACTC)2 A-form simulation at 0.3 M NaCl (V), which involves a 10 ps base pair opening event (Table S1), the longest among all of the DNA simulations, a water molecule formed a hydrogen bond with O6 of G3 in strand one at 1398 ps (Fig. 2C). This water molecule then formed an additional hydrogen bond with the N4 of C6 in strand 2 at 1400 ps, when the base pair opened (N1–N3 distance of 3.6 Å). The base pair remained open until 1410 ps, during which the water–base hydrogen bonds were maintained. The water–base interactions were lost by 1420 ps, at which time the base pair reverted to a fully closed state. Thus, water molecules appear to be able to compete with the WC interactions via the major groove in both DNA and RNA. However, the increased presence of water molecules in the vicinity of the N1–N3 atoms of RNA may facilitate water-mediated base opening initiation events as compared to DNA. Once a base pair opening initiation does occur, additional factors in RNA lead to the actual base pair opening events. These factors include the increased rigid body motion of the individual nucleosides and the 2′OH–O4′(n+1) hydrogen bonds.
Further analysis of the interactions between water molecules and the opened base pairs reveal the dynamic nature of hydrogen bonding between water molecules and the bases during the base pair open state. For example, in the event shown in Figure 2C, a total of 12 water molecules were within 3.5 Å of N1 or N3 during the 1380–2670 ps period (Supplementary Material, Fig. S3), with different waters interacting with the open bases during that time frame. This dynamic behavior further suggests that the rigid body behavior and the 2′OH(n)–O4′(n+1) interactions in RNA stabilize the base pair open state allowing for exchange of water molecules or ions without base pair closing.
Of note is the relationship between the number of water molecules interacting with the bases and the extent of base pair opening. When one water molecule bridges the N1 and N3 atoms, the N1 and N3 separation is ∼5 Å (Fig. S2), which is typical in many of the opening events. Further separation of N1 and N3 to ∼6.5 Å or more was associated with two water molecules or ions participating in the hydrogen bond network (1664 ps, Fig. 2C). Moreover, for the event depicted in Figure 2A, no water–base hydrogen bonds were found during the 1676–1858 ps period. Instead, sodium and chloride acted as the bridging elements (1714 ps, Fig. 2C). From these results it is clear that solvent molecules play important roles, not only in triggering base pair opening, but also in maintaining the open state and influencing the extent of base pair opening.
Mechanism for RNA conformational switch
Based on the data presented above, it is proposed that structural fluctuations of both DNA and RNA combined with interactions with the surrounding solvent allow water molecules to compete for WC hydrogen bonding, initiating base pair opening. In the case of RNA, this can lead to base pair opening on the nanosecond time scale; such opening does not occur on that time scale in DNA. This additional opening in RNA is suggested to be facilitated by the increased rigid body nature of RNA at the nucleoside level as compared to DNA and hydrogen bonding between 2′OH(n) and O4′(n+1) atoms in RNA. This combination of rigid-body behavior and the 2′OH(n)–O4′(n+1) hydrogen bond therefore acts as a conformational switch allowing for increased structural variability in RNA as compared to DNA on a nanosecond timescale.
How the rigid body behavior and the 2′OH(n)–O4′(n+1) hydrogen bond combine to create a conformational switch is worth additional discussion. First, for the interactions associated with the 2′OH moiety to be transmitted to the base there are only two flexible regions that must be considered; the sugar pucker and the χ dihedral (i.e. glycosyl bond). Crystal surveys of the distributions of these terms show them to be narrow for A-form conformations (25,69). With the pseudorotation angle, in A-form DNA structures the distribution is centered in the vicinity of 18° and is quite narrow, while in B-form DNA structures it is centered in the vicinity of 160° and is quite broad. Importantly, the narrow distribution of pseudorotation angles also occurs in RNA crystal structures (55). Similarly, with χ, the distributions are wider for B-DNA crystal structures as compared to A-form structures, including RNA. Thus, the decreased flexibility of the sugar pucker and χ in A-form RNA structures will lead to a more rigid-body-like behavior as compared to DNA, which is dominated by the B form. This rigid body behavior is suggested to contribute to the correlation between base opening and the 2′OH–O4′(n+1) hydrogen bonds.
Importantly, the more restrained nature of χ and sugar pucker in RNA is supported by QM calculations on nucleic acid related models compounds. QM studies on energies associated with rotation about χ show the global minimum in the energy surface to coincide with the region sampled in A-form RNA, while B-form DNA is sampling a higher energy region (69,80). Since in the A form, χ is near its global minimum, decreased fluctuations would be expected as compared to the B form, where χ is sampling a higher energy region. With the pseudorotation angle, QM energy surfaces have been performed on a series of model compounds (25,55,69). These results also suggest that conformational energies associated with the sugar pucker lead to the narrow distribution seen in the north conformation in A-form crystal structures as compared to the south conformation typical to the B form. These QM results further support a model where intrinsic conformational energies of the glycosyl linkage and the sugar pucker in the A form lead to more rigid-body motions of the nucleoside in RNA as compared to DNA. The consistent picture from the present simulations, imino protein exchange experiments, crystal database surveys and the QM data support the validity of the proposed conformational switch in RNA. An image of the closed and open states from the duplex II simulation is shown in Figure 3.
Figure 3.
Snapshots of base pair closed (1.2 ns) and open (2.0 ns) states from duplex II simulation at 1 M NaCl, showing the conformational switch. Only three base pairs (G5U6A7 from strand one and U6A7C8 from strand two) are shown with the central pair involving the conformational switch colored in red. O2′H2′–O4′(n+1) hydrogen bonds present in the open state are shown as green lines. The major groove faces toward the reader in the upper panel and towards the top in the lower panel.
Conclusions
In the present study it is shown that DNA and RNA duplexes display different flexibility properties associated with the 2′OH group. The presence of the 2′OH group in RNA, allowing for the formation of 2′OH(n)–O4′(n+1) hydrogen bonds, combined with decreased intrinsic flexibility of the sugar and the χ dihedral in A-form structures leads to a proposed conformational switch in RNA that is not present in DNA. The presence of this conformational switch allows for local base pair opening events in RNA on the nanosecond timescale. Importantly, the increased base opening events in RNA versus DNA are consistent with imino proton exchange experiments for AU base pairs (68); however, increases in GC opening seen in the present calculations are not consistent with experimental data, indicating a possible bias towards the open state in the applied force field. In addition, 2′O(n)–O4′(n+1) hydrogen bonds occur in tRNA, ribozymes and ribosomal RNA crystal structures, while decreased flexibility of the sugar pucker and χ is consistent with NDB survey data for duplex DNA and RNA as well as conformational energies of model compounds based on QM calculations. The combination of simulation and experimental data strongly indicate the presence of a conformational switch in RNA that is responsible for increased base pair opening in RNA on the nanosecond time scale.
The proposed conformational switch in RNA may be important for its structural diversity and biological function. For instance, RNA folding is known to often involve kinetic traps associated with misfolded states (81,82). These misfolded states include mispairings which have been suggested to undergo pairing rearrangements prior to attaining the correctly folded state. Such conformational malleability is evidenced by the P5abc region from the Tetrahymena thermophilia self-splicing intron, which assumes different tertiary structures in different environments (81,83). Conformational heterogeneity also appears to be important for catalytic RNA. For example, the hairpin ribozyme undergoes alterations in hydrogen bonds during catalysis (84) and the hammerhead ribozyme is thought to undergo significant conformational changes in its catalytic cycle (5,85,86). Interestingly, in the hammerhead ribozyme, a 2′OH(n)–O4′(n+1) hydrogen bond has been indicated to stabilize an intermediate, possibly important for the catalytic mechanism (87). While speculative, it may be suggested that the conformational switch involving rigid-body motions at the nucleoside level and the 2′OH(n)–O4′(n+1) hydrogen bond may act to lower the barrier to these conformational transitions. For example, if the opening events observed in the present calculations occur in the vicinity of additional hydrogen bonding moieties (e.g. other bases or phosphate moieties) adjacent to the major groove, possible interactions between the opened base and those moieties may lead to further opening and larger conformational changes. Alternatively, although opening beyond 8 Å is only observed once in the present nanosecond MD simulations, such events may lead to more open states on the millisecond timescale associated with RNA folding; states that may be important for exiting misfolded conformations leading to the correctly folded structure. Further experimental studies are required to better understand the proposed conformational switch in RNA.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank DOD ASC Major Shared Resource Computing and PSC Pittsburgh Supercomputing Center for their generous CPU allocations. This project was financially supported by the NIH (GM-51501).
REFERENCES
- 1.Watson J.D., Hopkins,N.H., Roberts,J.W., Steitz,J.A. and Seiner,A.M. (1987) Molecular Biology of the Gene. Benjamin Cummings, Inc., Menlo Park, CA. [Google Scholar]
- 2.Cech T.R. and Bass,B.L. (1986) Biological catalysis by RNA. Annu. Rev. Biochem., 55, 599–629. [DOI] [PubMed] [Google Scholar]
- 3.Bustamante C. and Tinoco,I.,Jr (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 4.Walberer B.J., Cheng,A.C. and Frankel,A.D. (2003) Structural diversity and isomorphism of hydrogen-bonded base interactions in nucleic acids. J. Mol. Biol., 327, 767–780. [DOI] [PubMed] [Google Scholar]
- 5.Butcher S.E. (2001) Structure and function of the small ribozymes. Curr. Opin. Struct. Biol., 11, 315–320. [DOI] [PubMed] [Google Scholar]
- 6.Doherty E.A. and Doudna,J.A. (2001) Ribozyme structures and mechanisms. Annu. Rev. Biophys. BioMol. Struct., 30, 457–475. [DOI] [PubMed] [Google Scholar]
- 7.Serra M.J., Axenson,T.J. and Turner,D.H. (1994) A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry, 33, 14289–14296. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder S., Kim,J. and Turner,D.H. (1996) G.A and U.U mismatches can stabilize RNA internal loops of three nucleotides. Biochemistry, 35, 16105–16109. [DOI] [PubMed] [Google Scholar]
- 9.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 10.Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys-tRNA Cys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure Fold Des., 7, 143–156. [DOI] [PubMed] [Google Scholar]
- 11.Saenger W. (1984) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY. [Google Scholar]
- 12.Pohl F.M. and Jovin,T.M. (1972) Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly(dG-dC). J. Mol. Biol., 67, 375–396. [DOI] [PubMed] [Google Scholar]
- 13.Leslie A.G.W., Arnott,S., Chandrasekaran,R. and Ratliff,R.L. (1980) Polymorphism of DNA double helices. J. Mol. Biol., 143, 49–72. [DOI] [PubMed] [Google Scholar]
- 14.Dickerson R.E., Drew,H.R., Conner,B.N., Wing,R.M., Fratini,A.V. and Kopka,M.L. (1982) The anatomy of A-, B- and Z-DNA. Science, 216, 475–485. [DOI] [PubMed] [Google Scholar]
- 15.Guzikevich-Guerstein G. and Shakked,Z. (1996) A novel form of the DNA double helix imposed on the TATA-box by the TATA-binding protein. Nat. Struct. Biol., 3, 32–37. [DOI] [PubMed] [Google Scholar]
- 16.Gryaznov S.M., Lloyd,D.H., Chen,J.K., Schultz,R.G., DeDionisio,L.A., Ratmeyer,L. and Wilson,W.D. (1995) Oligonucleotide N3′→P5′ phosphoramidates. Proc. Natl Acad. Sci. USA, 92, 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz R.G. and Gryaznov,S.M. (1996) Oligo-2′-fluoro-2′-deoxynucleotide N3′→P5′ phosphoramidates: synthesis and properties. Nucleic Acids Res., 24, 2966–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Thomas,G.A. and Peticolas,W.L. (1987) Sequence dependence of the B to Z transition in crystals and aqueous NaCl solutions for deoxyoligonucleotides containing all four canonical DNA bases. Biochemistry, 26, 5178–5186. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Thomas,G.A. and Peticolas,W.L. (1987) Sequence dependent conformations of oligomeric DNAs in aqueous solutions and in crystals. J. Biomol. Struct. Dyn., 5, 249–274. [DOI] [PubMed] [Google Scholar]
- 20.Mazur J., Sarai,A. and Jernigan,R.L. (1989) Sequence dependence of the B-A conformational transition of DNA. Biopolymers, 28, 1223–1233. [DOI] [PubMed] [Google Scholar]
- 21.Wilds C.J., Wawrzak,Z., Krishnamurthy,R., Eschenmoser,A. and Egli,M. (2002) Crystal structure of a B-form DNA duplex containing (l)-α-threofuranosyl (3′-2′) nucleosides: A four-carbon sugar is easily accommodated into the backbone of DNA. J. Am. Chem. Soc., 124, 13716–13721. [DOI] [PubMed] [Google Scholar]
- 22.Roberts R.W. and Crothers,D.M. (1992) Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science, 258, 1463–1466. [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto N., Nakano,M. and Nakano,S. (2000) Thermodynamics–structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry, 39, 11270–11281. [DOI] [PubMed] [Google Scholar]
- 24.Gupta G., Sarma,M.H. and Sarma,R.H. (1985) Secondary structure of the hybrid poly(rA).poly(dT) in solution. Studies involving NOE at 500 MHz and stereochemical modelling within the constraints of NOE data. J. Mol. Biol., 186, 463–469. [DOI] [PubMed] [Google Scholar]
- 25.Foloppe N. and MacKerell,A.D.,Jr (1998) Conformational properties of the deoxyribose and ribose moieties of nucleic acids: A quantum mechanical study. J. Phys. Chem. B, 102, 6669–6678. [Google Scholar]
- 26.Mishra S.K. and Mishra,P.C. (2002) An ab initio theoretical study of electronic structure and properties of 2′-deoxyguanosine in gas phase and aqueous media. J. Comput. Chem., 23, 530–540. [DOI] [PubMed] [Google Scholar]
- 27.Hanus M., Ryjacek,F., Kabelac,M., Kubar,T., Bogdan,T.V., Trygubenko,S.A. and Hobza,P. (2003) Correlated ab initio study of nucleic acid bases and their tautomers in the solution. J. Am. Chem. Soc., 125, 7678–7688. [DOI] [PubMed] [Google Scholar]
- 28.Strajbl M. and Florian,J. (1996) Ab initio investigation of the molecular structure of methyl methoxymethyl phosphonate, a promising nuclease-resistant alternative of the phosphodiester linkage. J. Biomol. Struct. Dyn., 13, 687–694. [DOI] [PubMed] [Google Scholar]
- 29.Reha D., Kabelac,M., Ryjacek,F., Sponer,J., Sponer,J.E., Elstner,M., Suhai,S. and Hobza,P. (2002) Intercalators. 1. Nature of stacking interactions between intercalators (ethidium, daunomycn, ellipticine and 4′,6-diaminide-2-phenylindole) and DNA base pairs. Ab initio quantum chemical, density functional theory and empirical potential study. J. Am. Chem. Soc., 124, 3366–3376. [DOI] [PubMed] [Google Scholar]
- 30.Hocquet A., Leulliot,N. and Ghomi,M. (2000) Ground-state properties of nucleic acid constituents studied by density functional calculatins. 3. Role of sugar puckering and base orientation on the energetics and geometry of 2′-deoxyribonucleosides and ribonucleosides. J. Phys. Chem. B, 104, 4560–4568. [Google Scholar]
- 31.Giudice E. and Lavery,R. (2002) Simulations of nucleic acids and their complexes. Acc. Chem. Res., 35, 350–357. [DOI] [PubMed] [Google Scholar]
- 32.Giudice E., Varnai,P. and Lavery,R. (2001) Energetic and conformational aspects of A:T base-pair opening within the DNA double helix. Chem. Phys. Chem., 2, 673–677. [DOI] [PubMed] [Google Scholar]
- 33.Banavali N.K. and MacKerell,A.D.,Jr (2002) Free energy and structural pathways of base flipping in a DNA GCGC containing sequence. J. Mol. Biol., 319, 141–160. [DOI] [PubMed] [Google Scholar]
- 34.MacKerell A.D. Jr (1997) Influence of magnesium ions on duplex DNA structural, dynamic and solvation properties. J. Phys. Chem. B, 101, 646–650. [Google Scholar]
- 35.Cheatham T.E.I. and Kollman,P.A. (1997) Insight into the stabilization of A-DNA by specific ion association: spotaneous B-DNA to A-DNA transitions observed in molecular dynamics simulations of d[ACCCGCGGGT]2 in the presence of hexaamminecobalt(III). Structure, 5, 1297–1311. [DOI] [PubMed] [Google Scholar]
- 36.Jayaram B., Sprous,D., Young,M.A. and Beveridge,D.L. (1998) Free energy analysis of the conformational preferences of A and B forms of DNA in solution. J. Am. Chem. Soc., 120, 10629–10633. [Google Scholar]
- 37.Auffinger P. and Westhof,E. (2000) Water and ion binding around RNA and DNA (C,G) oligomers. J. Mol. Biol., 300, 1113–1131. [DOI] [PubMed] [Google Scholar]
- 38.Bandyopadhyay D. and Bhattacharyya,D. (2000) Effect of neighboring bases on base-pair stacking orientation: a molecular dynamics study. J. Biomol. Struct. Dyn., 18, 29–43. [DOI] [PubMed] [Google Scholar]
- 39.Derreumaux S. and Fermandjian,S. (2000) Bending and adaptability to proteins of the cAMP DNA-responsive element: molecular dynamics contrasted with NMR. Biophys. J., 79, 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C., Brandl,M. and Suhnel,J. (2001) Molecular dynamics simulation reveals conformational switching of water-mediated uracil-cytosine base-pairs in an RNA duplex. J. Mol. Biol., 305, 659–667. [DOI] [PubMed] [Google Scholar]
- 41.Hamelberg D., Williams,L.D. and Wilson,W.D. (2002) Effect of a neutralized phosphate backbone on the minor groove of B-DNA: molecular dynamics simulation studies. Nucleic Acids Res., 30, 3615–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheatham T.E.I. and Kollman,P.A. (1996) Observation of the A-DNA to B-DNA transition during unrestrained molecular dynamics in aqueous solution. J. Mol. Biol., 259, 434–444. [DOI] [PubMed] [Google Scholar]
- 43.Soliva R., Luque,F.J., Alhambra,C. and Orozco,M. (1999) Role of sugar re-puckering in the transition of A and B forms of DNA in solution. A molecular dynamics study. J. Biomol. Struct. Dyn., 17, 89–99. [DOI] [PubMed] [Google Scholar]
- 44.Sprous D., Young,M.A. and Beveridge,D.L. (1998) Molecular dynamics studies of the conformational preferences of a DNA double helix in water and ethanol/water mixture: Theoretical considerations of the A-B transition. J. Phys. Chem. B, 102, 4658–4667. [Google Scholar]
- 45.Cieplak P., Cheatham,T.E.I. and Kollman,P.A. (1997) Molecular dynamics simulations find that 3′ phosphoramidate modified DNA duplexes undergo a B to A transition and normal DNA duplexes an A to B transition. J. Am. Chem. Soc., 119, 6722–6730. [Google Scholar]
- 46.Bertrand H., Ha-Duong,T., Fermandjian,S. and Hartmann,B. (1998) Flexibility of the B-DNA backbone: effects of local and neighbouring sequences on pyrimidine–purine steps. Nucleic Acids Res., 26, 1261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacKerell A.D. Jr and Nilsson,L. (2001) In Becker,O.M., MacKerell,A.D.,Jr, Roux,B. and Watanabe,M. (eds), Computational Biochemistry and Biophysics. Marcel Dekker, Inc., New York, NY, pp. 441–463. [Google Scholar]
- 48.Varnai P., Djuranovic,D., Lavery,R. and Hartmann,B. (2002) α/γ Transitions in the B-DNA backbone. Nucleic Acids Res., 30, 5398–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auffinger P., Louise-May,S. and Westhof,E. (1995) Multiple molecular dynamics simulations of the anticodon loop of tRNAasp in aqueous solution with counterions. J. Am. Chem. Soc., 117, 6720–6726. [Google Scholar]
- 50.Egli M., Portmann,S. and Usman,N. (1996) RNA hydration: a detailed look. Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 51.Auffinger P. and Westhof,E. (1998) Simulations of the molecular dynamics of nucleic acids. Curr. Opin. Struct. Biol., 8, 227–236. [DOI] [PubMed] [Google Scholar]
- 52.Auffinger P. and Westhof,E. (1997) Rules governing the orientation of the 2′-hydroxyl group in RNA. J. Mol. Biol., 274, 54–63. [DOI] [PubMed] [Google Scholar]
- 53.Berman H.M., Olson,W.K., Beveridge,D.L., Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.-H., Srinivasan,A.R. and Schneider,B. (1992) The Nucleic Acid Database: A Comprehensive Relational Database of Three-Dimensional Structures of Nucleic Acids. Biophys. J., 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks B.R., Bruccoleri,R.E., Olafson,B.D., States,D.J., Swaminathan,S. and Karplus,M. (1983) CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem., 4, 187–217. [Google Scholar]
- 55.Foloppe N. and MacKerell,A.D.,Jr (2000) All-atom empirical force field for nucleic acids. 1. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem., 21, 86–104. [Google Scholar]
- 56.MacKerell A.D. Jr and Banavali,N.K. (2001) All-atom empirical force field for nucleic acids. 2. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem., 21, 105–120. [Google Scholar]
- 57.Jorgensen W.L., Chandrasekhar,J., Madura,J.D., Impey,R.W. and Klein,M.L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys., 79, 926–935. [Google Scholar]
- 58.Beglov D. and Roux,B. (1997) Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys., 100, 9050–9063. [Google Scholar]
- 59.Arnott S. and Hukins,D.W. (1973) Refinement of the structure of B-DNA and implications for the analysis of X-ray diffraction data from fibers of biopolymers. J. Mol. Biol., 81, 93–105. [DOI] [PubMed] [Google Scholar]
- 60.Schindelin H., Zhang,M., Bald,R., Furste,J.P., Erdmann,V.A. and Heinemann,U. (1995) Crystal structure of an RNA dodecamer containing the Escherichia coli Shine–Dalgarno sequence. J. Mol. Biol., 249, 595–603. [DOI] [PubMed] [Google Scholar]
- 61.Field M.J. and Karplus,M. (1992) Crystal simulation module in CHARMM. Harvard University, Cambridge, MA. [Google Scholar]
- 62.Ryckaert J., Ciccotti,G. and Berendsen,H.J.C. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J. Comput. Phys., 23, 327–341. [Google Scholar]
- 63.Essmann U., Perera,L., Berkowitz,M.L., Darden,T.A., Lee,H. and Pedersen,L.G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys., 103, 8577–8593. [Google Scholar]
- 64.Darden T., Perera,L., Li,L. and Pedersen,L. (1999) New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure Fold Des., 7, R55–R60. [DOI] [PubMed] [Google Scholar]
- 65.Hoover W.G. (1985) Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A, 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- 66.Feller S.E., Zhang,Y., Pastor,R.W. and Brooks,R.W. (1995) Constant pressure molecular dynamics simulations: the Langevin piston method. J. Chem. Phys., 103, 4613–4621. [Google Scholar]
- 67.Chen X., McDowell,J.A., Kierzek,R., Krugh,T.R. and Turner,D.H. (2000) Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2). Biochemistry, 39, 8970–8982. [PubMed] [Google Scholar]
- 68.Snoussi K. and Leroy,L.-L. (2001) Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry, 40, 8898–8904. [DOI] [PubMed] [Google Scholar]
- 69.Foloppe N. and MacKerell,A.D.,Jr (1999) Contribution of the phosphodiester backbone and glycosyl linkage intrinsic tosional energetics to DNA structure and dynamics. J. Phys. Chem. B, 103, 10955–10964. [Google Scholar]
- 70.Hartmann B. and Lavery,R. (1996) DNA structural forms. Q. Rev. Biophys., 29, 309–368. [DOI] [PubMed] [Google Scholar]
- 71.Dickerson R.E. (1992) DNA structure from A to Z. Methods Enzymol., 211, 67–111. [DOI] [PubMed] [Google Scholar]
- 72.Djuranovic D. and Hartmann,B. (2003) Conformational characteristics and correlations in crystal structures of nucleic acid oligonuleotides: evidence for sub-states. J. Biomol. Struct. Dyn., 20, 771–788. [DOI] [PubMed] [Google Scholar]
- 73.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 74.Cui G. and Simmerling,C. (2002) Conformational heterogeneity observed in simulations of a pyrene-substituted DNA. J. Am. Chem. Soc., 124, 12154–12164. [DOI] [PubMed] [Google Scholar]
- 75.Pan J., Thirumalai,D. and Woodson,S.A. (1999) Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics and kinetics. Proc. Natl Acad. Sci. USA, 96, 6149–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gueron M., Kochoyan,M. and Leroy,J.-L. (1987) A single mode of DNA base-pair opening drivers imino proteon exchange. Nature, 328, 89–92. [DOI] [PubMed] [Google Scholar]
- 77.Moe J.G., Folta-Stogniew,E. and Russu,I.M. (1995) Enertetics of base pair opening in a DNA dodecamer containing an A3T3 tract. Nucleic Acids Res., 23, 1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leroy J.-L., Broseta,D. and Gueron,M. (1985) Proton exchange and base-pair kinetics of poly(rA).poly(rU) and poly(rI).poly(rC). J. Mol. Biol., 184, 165–178. [DOI] [PubMed] [Google Scholar]
- 79.Giudice E. and Lavery,R. (2003) Nucleic acid base pair dynamics: the impact of sequence and structure using free-energy calculations. J. Am. Chem. Soc., 125, 4998–4999. [DOI] [PubMed] [Google Scholar]
- 80.Foloppe N., Nilsson,L. and MacKerell,A.D.,Jr (2001) Ab initio conformational analysis of nucleic acid components: intrinsic energetic contributions to nucleic acid structure and dynamics. Biopolymers, 61, 61–76. [DOI] [PubMed] [Google Scholar]
- 81.Tinoco I. Jr and Bustamante,C. (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 82.Treiber D.K. and Williamson,J.R. (2001) Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol., 11, 309–314. [DOI] [PubMed] [Google Scholar]
- 83.Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 84.Rupert P.B., Massey,A.P., Sigurdsson,S.T. and Ferre-D’Amare,A.R. (2002) Transition state stabilization by a catalytic RNA. Science, 298, 1421–1424. [DOI] [PubMed] [Google Scholar]
- 85.Wang S., Karbstein,K., Peracchi,A., Beigelman,L. and Herschlag,D. (1999) Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry, 38, 14363–14378. [DOI] [PubMed] [Google Scholar]
- 86.Murray J.B. and Scott,W.G. (2000) Does a single metal ion bridge the A-9 and scissile phosphates in the catalytically active hammerhead ribozyme structure? J. Mol. Biol., 296, 33–41. [DOI] [PubMed] [Google Scholar]
- 87.Scott W.G., Murray,J.B., Arnold,J.R., Stoddard,B.L. and Klug,A. (1996) Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science, 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- 88.Loncharich R.J., Brooks,B.R. and Pastor,R.W. (1992) Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers, 32, 523–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.