Abstract
The Ser/Thr SAD kinases are evolutionarily conserved, critical regulators of neural development. Exciting findings in recent years have significantly advanced our understanding of the mechanism through which SAD kinases regulate neural development. Mammalian SAD-A and SAD-B, activated by a master kinase LKB1, regulate microtubule dynamics and polarize neurons. In C. elegans, the sad-1 gene encodes two isoforms, namely the long and the short, which exhibit overlapping and yet distinct functions in neuronal polarity and synaptic organization. Surprisingly, our most recent findings in C. elegans revealed a SAD-1-independent LKB1 activity in neuronal polarity. We also found that the long SAD-1 isoform directly interacts with a STRADα pseudokinase, STRD-1, to regulate neuronal polarity and synaptic organization. We elaborate here a working model of SAD-1 in which the two isoforms dimer/oligomerize to form a functional complex, and STRD-1 clusters and localizes the SAD-1 complex to synapses. While the mechanistic difference between the vertebrate and invertebrate SAD kinases may be puzzling, a recent discovery of the functionally distinct SAD-B isoforms predicts that the difference likely arises from our incomplete understanding of the SAD kinase mechanism and may eventually be reconciled as the revelation continues.
Key words: SAD kinases, SAD-1, C. elegans, neural development, neuronal polarity, synapse, isoform
The transformation of a nascent amorphous cell to a mature polarized neuron requires a concerted interplay of various factors. These include extracellular morphogens and growth factors providing cues; intracellular messengers relaying the information; and cytoskeletal networks effecting neurite extension and directional transport of axon- or dendrite-specific organelles.1–5
SAD Kinases Regulate Neural Development through Distinct Mechanisms
C. elegans SAD-1 kinase (Synapses of the Amphid Defective) was the first regulator of neuronal polarity identified in vivo.6 It restricts the localization of synaptic vesicle proteins to axons and also organizes them at synapses.6,7 Its mammalian counterparts, SAD-A and SAD-B, together also play essential roles in neuronal polarization. 8,9 SAD-A and SAD-B are activated by LKB1,9 a tumour suppressor10,11 and master kinase12 implicated in various processes including cell cycle regulation13 and cell polarity.14,15 LKB1 functions in a complex with a STRAD pseudokinase that stabilizes,16 activates12 and properly localizes the kinase.16,17 Activated SAD kinases phosphorylate a microtubule-associated protein, Tau, and modulate microtubule dynamics during neuronal polarization.9 SAD-B also regulates neurotransmitter release.18 SAD kinases therefore are conserved regulators of neural development and functions.
Notwithstanding their functional conservation, our recent study revealed an unexpected difference in the mechanism between the vertebrate and invertebrate SAD kinases.19 Unlike the linear mammalian pathway of the LKB1-STRAD complex functioning through SAD kinases, in C. elegans, the sole LKB1 ortholog, PAR-4,14,20 displayed SAD-1-independent activities in neuronal polarity. Instead, PAR-4 regulates neuronal polarity by activating another kinase, PAR-1.19,21 SAD-1, on the other hand, directly associates with and functions exclusively through a C. elegans ortholog of STRADα, STRD-1. These findings also challenge the common notion that STRADα functions exclusively through LKB1. Do these findings simply denote evolutionary divergence in the function of LKB1 and SAD kinases? Or do they also reflect the complexity in the regulation of, and interplay between, these signaling components in vivo? These questions warrant further commentary presented here and investigations to follow.
SAD Isoforms Perform Distinct Functions through Different Partners
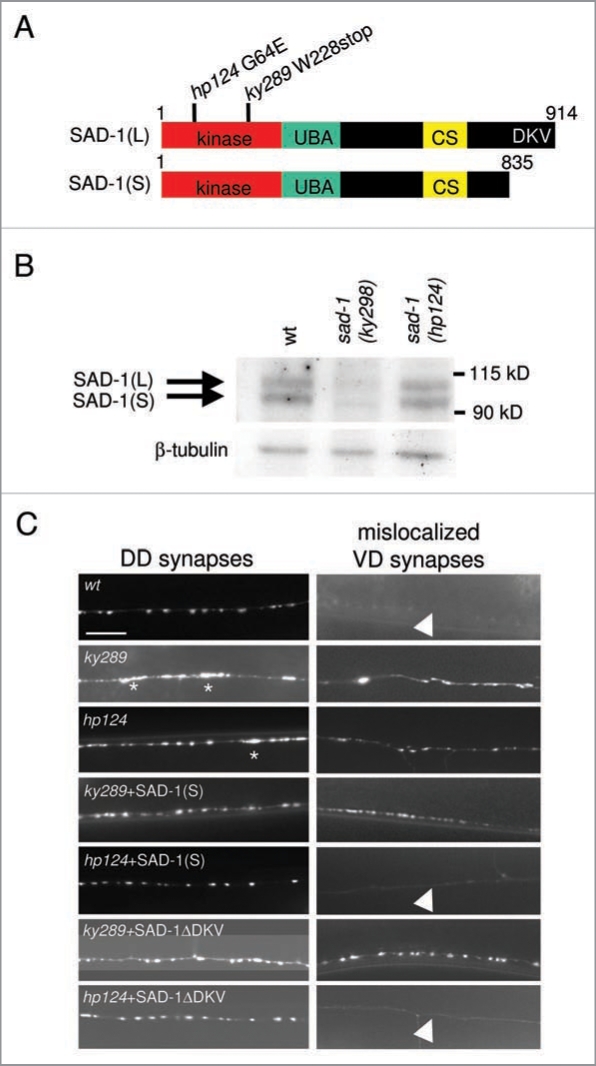
Not all SAD kinases are created equal. Previously, we reported the identification of two SAD-1 isoforms7 (Fig. 1A). The long isoform (SAD-1(L)) rescued both neuronal polarity and synaptic organization defects7 of sad-1(ky289) proteinnull mutants (Fig. 1B). In contrast, the short isoform (SAD-1(S)), truncated at the C-terminus, restored synaptic organization but not neuronal polarity.7 We showed that SAD-1(S) failed to interact with NAB-1, the sole C. elegans ortholog of an F-actin binding scaffold protein, Neurabin, which regulates neuronal polarity through its interaction with the C-terminus of SAD-1(L)7 (Fig. 1A). The SAD-1 isoforms therefore serve overlapping and yet distinct functions through different effectors.
Figure 1.
SAD-1 dimer/oligomerizes. (A) Schematic representations of SAD-1(L) and SAD-1(S) protein structures. Each isoform comprises a kinase domain, ubiquitin-associated (UBA) domain, and a unique C-terminal sequence (CS) conserved amongst SAD kinases. In addition, SAD-1(L) has a PDZ domain-binding consensus sequence at the C-terminus (DKV) to which NA B-1 binds. The molecular lesions of two sad-1 mutants, ky289 and hp124, are shown. The ky289 mutation causes an early stop codon whereas hp124 changes a conserved glycine residue in the kinase domain to glutamic acid. (B) Biochemical characterization of sad-1 mutants. SAD-1 protein levels were examined in wild-type, ky289, and hp124 animals by immunoblotting using anti-SAD-1 (top) and anti-β-tubulin for loading control (bottom). While no SAD-1 was detected in ky289 null mutants, both isoforms were detected in hp124 mutants. (C) Differential neuronal phenotype rescues by SAD-1(S) or SAD-1ΔDKV in ky289 and hp124 mutants. Neuronal phenotypes of the GABAergic neurons along the dorsal nerve cord (DN C) were examined using a pre-synaptic vesicle marker, SNB-1::GFP. For synaptic organization, SNB-1::GFP signals in the axons of the DD class GABAergic neurons were examined (left). In wild-type animals, SNB-1::GFP exhibited uniform shape, size, and spacing. Both alleles of sad-1 displayed uneven and diffuse SNB-1::GFP morphology (asterisks) which was fully rescued by either SAD-1(S) or SAD-1ΔDKV. For neuronal polarity, SNB-1::GFP signals mis-localizing to the dendrites of the VD class GABAergic neurons were examined (right). In wild-type animals, no pre-synaptic SNB-1::GFP was mis-localized (arrowhead). In both alleles of sad-1, ectopic SNB-1::GFP signals were observed. This defect was rescued only when SAD-1(S) or SAD-1ΔDKV was expressed in SAD-1(KD)-producing hp124 mutants and not in the protein-null ky289 mutants. Scale bar, 5 µm.
The presence of multiple isoforms of SAD kinases is not unique to C. elegans. Recently, isoforms of mouse SAD-B have also been reported, and one isoform was implicated in centrosome duplication during cell cycle progression.22 This newlyidentified role of the SAD-B isoform has an interesting connection to the neural functions of SAD kinases, revealed in a previous study which implicated a role of centrosome localization in determining the axonal fate.23 Consistently, in both neuronal polarization and cell cycle regulation, SAD kinases regulate microtubule dynamics through Tau or tubulin, respectively. It is then not inconceivable that different isoforms of SAD kinases regulate neural development in parallel, via distinct mechanisms and effectors, in different cellular contexts. In view of this, the STRD-1-dependent activity of SAD-1 in C. elegans19 and the LKB1-dependent activation of SAD-A and SAD-B in mammals9 may simply represent our limited understanding of all aspects of the mechanism governing SAD activities.
SAD-1 Dimer/Oligomerizes
In our recent work, we reported an additional difference between the two SAD-1 isoforms in their organization along the axon.19 When fluorescently-tagged and expressed separately, SAD-1(L) organized into tight clusters along the axon whereas SAD-1(S) appeared more diffuse.19 Co-expressed SAD-1(L) and SAD-1(S) resembled the tight clustering pattern of SAD-1(L),19 implying that the two isoforms might interact.
Indeed, our new in vivo and in vitro data strongly support this possibility. SAD-1(S) or SAD-1(L) lacking the C-terminus (SAD-1ΔDKV) cannot interact with NAB-1 and fails to rescue the neuronal polarity defect of sad-1(ky289) protein-null mutants.7 However, when expressed in sad-1(hp124) loss-of-function mutants, which produce a kinase-dead but otherwise intact SAD-1 (SAD-1(KD)) protein (Fig. 1B), both SAD-1(S) and SAD-1ΔDKV fully restored both neuronal polarity and synaptic organization (Fig. 1C). As neither SAD-1(KD) nor SAD-1(S)/SAD-1ΔDKV alone can rescue the neuronal polarity defect,7 a plausible explanation for this observation is that a fully functional complex comprised of the SAD-1(KD) long isoform and SAD-1(S) or SAD-1ΔDKV was formed.
We further confirmed that SAD-1 proteins interact with each other using the yeast-two-hybrid system in which SAD-1(S) and SAD-1(L) exhibited robust interactions through their ubiquitin-associated (UBA) domain (Fig. 1A; data not shown). The UBA domain has been shown to interact with kinase domains,24 suggesting that the protein-protein interaction within a SAD-1 complex may be mediated by the UBA domain of one SAD-1 molecule and the kinase domain of another. Together, these findings are consistent with our co-expression data in which SAD-1(S) assumed the tight clustering pattern of SAD-1(L).19
SAD-1 Clusters at and Localizes to Synapses through SAD-1(L) and STRD-1 Interaction
The two SAD-1 isoforms also differed in their interaction with STRD-1.19 The strd-1 gene shares the same genetic pathway with sad-1 to regulate neuronal polarity and synaptic organization, and STRD-1 physically interacts with SAD-1(L).19 In strd-1 loss-of-function mutants, co-expressed SAD-1(L) and SAD-1(S) failed to cluster along the axon, suggesting that STRD-1 regulates the sub-cellular organization and localization of the SAD-1(L)/SAD-1(S) complex.19 On the other hand, when expressed separately, only SAD-1(L) displayed abnormal clustering and localization in strd-1 mutants whereas SAD-1(S) remained unaffected.19 Taken together, these data suggest that the clustering and localization of the SAD-1(L)/SAD-1(S) complex at synapses are mediated by STRD-1 through its direct interaction with SAD-1(L).
A Working Model for SAD-1
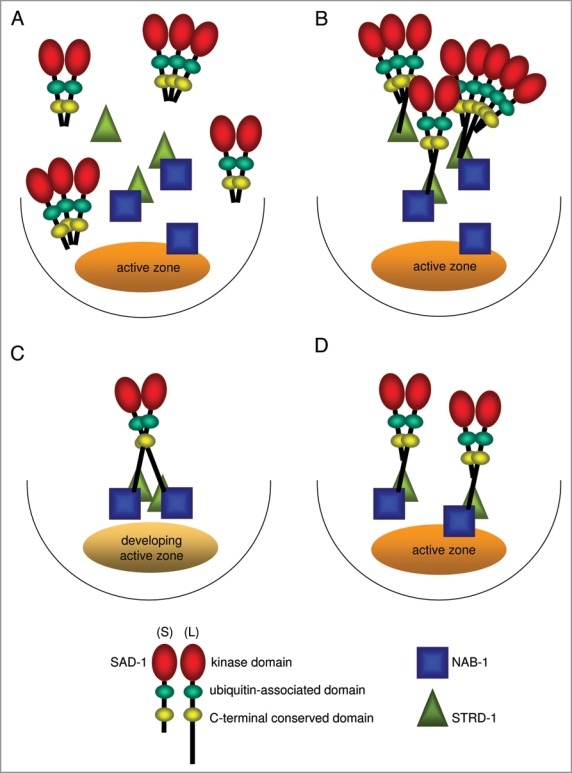
In view of the presented data, we propose the following model (Fig. 2). Along the axon, SAD-1(L) interacts with other SAD-1 through its UBA domain. The transformation of the diffuse SAD-1(S) localization pattern to tight clusters resembling that of SAD-1(L) when the two isoforms are co-expressed suggests that SAD-1(S) preferentially interacts with SAD-1(L) over another SAD-1(S) (Fig. 2A and B). The SAD-1(L)/SAD-1(S) complex interacts with STRD-1 via SAD-1(L), and this association promotes the clustering and localization of the SAD-1 complex at synapses. The complex subsequently interacts with NAB-1 to establish neuronal polarity.
Figure 2.
SAD-1 working model. (A and B) Formation of the SAD-1 complex. Expressed alone, SAD-1(S) fails to interact and cluster with STRD-1 or NAB-1 (A). When co-expressed, SAD-1(L) recruits SAD-1(S), and the SAD-1 complex binds STRD-1 and NAB-1 through the C-terminus of the long isoform (B). The interaction with STRD-1 is crucial for clustering and localizing the SAD-1 complex along the axon. The localization of STRD-1 and NAB-1 is unaffected by SAD-1 as evidenced in our previous studies. (C and D) Developmental regulation of the isoform expression. During neuronal polarization, SAD-1(L) is predominantly expressed (C). Once polarity is established, SAD-1(S) is expressed and dimer/oligomerizes with SAD-1(L) to perform a ‘surveillance’ role in synaptic organization (D).
We have confirmed that both isoforms are indeed co-expressed in the same neurons (data not shown). What, then, is the physiological role of SAD-1(S) when SAD-1(L) alone can fully rescue the neural defects of sad-1 complete loss-of-function mutants? The simplest explanation is that there may be additional, distinct functions for SAD-1(S) yet to be discovered. Here, we propose an alternative scenario in which the expression of the SAD-1 isoforms is regulated by a developmental switch during neural development.
Previously, we demonstrated that the establishment of neuronal polarity and synaptic organization has distinct temporal requirements for SAD-1 kinase activity.25 While SAD-1 activity is strictly required during a narrow window of time to establish neuronal polarity, synaptic organization could be established or even corrected at flexible developmental time points.25 Furthermore, whereas establishing neuronal polarity depends strictly on SAD-1(L), either isoform suffices for synaptic organization, suggesting that neuronal polarization is a much more tightly-controlled process. We thus speculate that a temporal switch for SAD-1(S) expression is activated once neuronal polarity is established and when SAD-1(L) is no longer necessary (Fig. 2C and D). In polarized neurons, newly-synthesized SAD-1(S) associates with the existing SAD-1(L) and STRD-1 to perform ‘surveillance’ functions, correcting any abnormal synaptic organization.
Implications of the SAD-1 Working Model on Other SAD Kinases
Clear species differences exist between the vertebrate and invertebrate SAD kinases. For instance, while the two SAD kinases function redundantly in mammals,8 only a single kinase suffices in C. elegans.6 Also, the SAD-1—NAB-1 interaction may not be conserved given the poor sequence similarities in the C-terminus amongst SAD kinases.6
However, SAD kinases may in fact share much in common as demonstrated above. As SAD-1—SAD-1 interaction is mediated through the conserved UBA domain, SAD kinases may also dimer/oligomerize in mammals. In future studies, it will be important to test and refine our model across species to better understand the complex mechanism of SAD kinases in the regulation of neural development.
Acknowledgements
This work was supported by an NSERC fellowship to J.S.M.K. and a CIHR and an NSERC grant to M.Z.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/11455
References
- 1.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arimura N, Kaibuchi K. Key regulators in neuronal polarity. Neuron. 2005;48:881–884. doi: 10.1016/j.neuron.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Wiggin GR, Fawcett JP, Pawson T. Polarity proteins in axon specification and synaptogenesis. Dev Cell. 2005;8:803–816. doi: 10.1016/j.devcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 6.Crump JG, Zhen M, Jin Y, Bargmann CI. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 2001;29:115–129. doi: 10.1016/s0896-6273(01)00184-2. [DOI] [PubMed] [Google Scholar]
- 7.Hung W, Hwang C, Po MD, Zhen M. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development. 2007;134:237–249. doi: 10.1242/dev.02725. [DOI] [PubMed] [Google Scholar]
- 8.Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science. 2005;307:929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- 9.Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, et al. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 11.Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 12.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiainen M, Ylikorkala A, Makela TP. Growth suppression by Lkb1 is mediated by a G(1) cell cycle arrest. Proc Natl Acad Sci USA. 1999;96:9248–9251. doi: 10.1073/pnas.96.16.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts JL, Morton DG, Bestman J, Kemphues KJ. The C. elegans par-4 gene encodes a putative serinethreonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- 15.Martin SG, St. Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421:379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 16.Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, et al. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorfman J, Macara IG. STRADalpha regulates LKB1 localization by blocking access to importinalpha, and by association with Crm1 and exportin-7. Mol Biol Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue E, Mochida S, Takagi H, Higa S, Deguchi-Tawarada M, Takao-Rikitsu E, et al. SAD: a presynaptic kinase associated with synaptic vesicles and the active zone cytomatrix that regulates neurotransmitter release. Neuron. 2006;50:261–275. doi: 10.1016/j.neuron.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Kim JS, Hung W, Narbonne P, Roy R, Zhen M. C. elegans STRADalpha and SAD cooperatively regulate neuronal polarity and synaptic organization. Development. 2010;137:93–102. doi: 10.1242/dev.041459. [DOI] [PubMed] [Google Scholar]
- 20.Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 21.Narbonne P, Hyenne V, Li S, Labbe JC, Roy R. Differential requirements for STRAD in LKB1-dependent functions in C. elegans. Development. 2010;137:661–670. doi: 10.1242/dev.042044. [DOI] [PubMed] [Google Scholar]
- 22.Alvarado-Kristensson M, Rodriguez MJ, Silio V, Valpuesta JM, Carrera AC. SADB phosphorylation of gamma-tubulin regulates centrosome duplication. Nat Cell Biol. 2009;11:1081–1092. doi: 10.1038/ncb1921. [DOI] [PubMed] [Google Scholar]
- 23.de Anda FC, Pollarolo G, Da Silva JS, Camoletto PG, Feiguin F, Dotti CG. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- 24.Murphy JM, Korzhnev DM, Ceccarelli DF, Briant DJ, Zarrine-Afsar A, Sicheri F, et al. Conformational instability of the MARK3 UBA domain compromises ubiquitin recognition and promotes interaction with the adjacent kinase domain. Proc Natl Acad Sci USA. 2007;104:14336–14341. doi: 10.1073/pnas.0703012104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JS, Lilley BN, Zhang C, Shokat KM, Sanes JR, Zhen M. A chemical-genetic strategy reveals distinct temporal requirements for SAD-1 kinase in neuronal polarization and synapse formation. Neural Dev. 2008;3:23. doi: 10.1186/1749-8104-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]