Abstract
The antibiotic chloramphenicol produces modifications in 23S rRNA when bound to ribosomes from the bacterium Escherichia coli and the archaeon Halobacterium halobium and irradiated with 365 nm light. The modifications map to nucleotides m5U747 and C2611/C2612, in domains II and V, respectively, of E.coli 23S rRNA and G2084 (2058 in E.coli numbering) in domain V of H.halobium 23S rRNA. The modification sites overlap with a portion of the macrolide binding site and cluster at the entrance to the peptide exit tunnel. The data correlate with the recently reported chloramphenicol binding site on an archaeal ribosome and suggest that a similar binding site is present on the E.coli ribosome.
INTRODUCTION
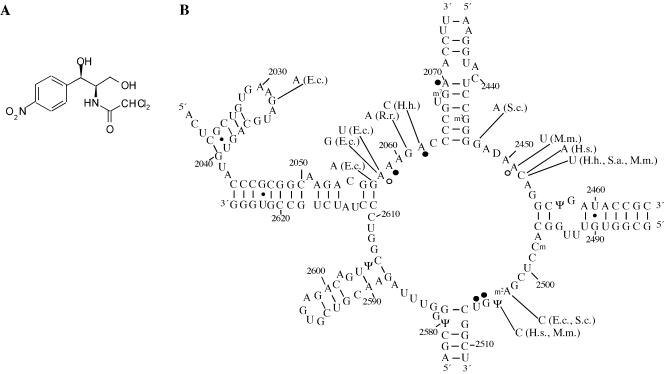
The antibiotic chloramphenicol (Fig. 1A) inhibits protein biosynthesis by targeting the peptidyl transferase center on the large ribosomal subunit. Due to its high specificity for bacteria, chloramphenicol is an important drug that has been widely used clinically, notably in the treatment of eye infections (1). Although archaea are generally less sensitive to ribosomal antibiotics, growth of Halobacterium halobium and Sulfolobus acidocaldarius is inhibited at elevated concentrations of chloramphenicol (2,3). Thirteen point mutations at 11 nucleotides in the peptidyl transferase loop of 23S rRNA that result in decreased sensitivity or resistance to chloramphenicol have been characterized in bacteria, archaea and mitochondria (Fig. 1 and references therein). Chloram phenicol produces a chemical footprint in the peptidyl transferase loop of Escherichia coli ribosomes, comprised of altered reactivities to base-specific modifying chemicals (Fig. 1B) (4,5). In the presence of chloramphenicol, protection effects are observed at nucleotides A2059, A2062, A2070, G2505 and U2506, while enhanced reactivities are observed at nucleotides A2058 and A2451. In contrast, a chemical footprint was not detected in the halophilic archaeon Haloferax mediterranei, even at 0.1 mM chloramphenicol (5).
Figure 1.
Chloramphenicol and the peptidyl transferase loop of 23S rRNA. (A) The chemical structure of chloramphenicol. (B) Secondary structure of the peptidyl transferase loop and adjacent regions of E.coli 23S rRNA. Nucleotides that exhibit altered reactivity in the presence of bound chloramphenicol are indicated with circles (4,23). Filled circles represent protections whereas open circles denote enhancements. Mutations that confer resistance or decreased susceptibility to chloramphenicol are indicated with the relevant organism(s) in parentheses. E.c., E.coli; H.h., H.halobium; H.s., Homo sapiens; M.m., Mus musculus; R.r., Rattus rattus; S.a., S.acidocaldarius; S.c., Saccharomyces cerevisiae. All of the eukaryotic mutations were characterized in mitochondrial rRNAs. Literature references to the indicated mutations are as follows: G2032A (11), G2057A (10), A2058G and A2058U (11), G2061A (36), A2062C (2), G2447A (37), A2451U (38), C2452A (39), C2452U in H.h. (2), M.m. (40) and S.a. (3), A2503C in S.c. (37) and E.c. (41), U2504C in M.m. (39,42) and H.s. (38).
An early investigation provided evidence that chloramphenicol binds to two sites on the 50S subunit from equilibrium dialysis measurements (6). These sites were found to have different binding constants (KD1 = 2 µM and KD2 = 200 µM), where only the stronger binding site was involved in inhibition of the reaction between puromycin and CACCA-Leu-Ac (7). In addition, kinetic analysis of the effect of chloramphenicol on the puromycin reaction reveals two phases of inhibition. Competitive and mixed non-competitive inhibition types with respect to puromycin have been reported at chloramphenicol concentrations up to and greater than 3 µM, respectively (8). Moreover, there is evidence to suggest that chloramphenicol can bind to a site that partially overlaps with that of erythromycin, a macrolide antibiotic that does not interfere directly with peptide bond formation. Erythromycin interferes with chloramphenicol binding to ribosomes (9), and mutations in nucleotides 2057 and 2058 that result in reduced chloramphenicol susceptibility in E.coli confer cross resistance to erythromycin (10,11).
Recently, two chloramphenicol binding sites have been reported in structures of antibiotic-ribosomal subunit complexes solved through X-ray crystallography. In one complex with the Deinococcus radiodurans 50S subunit, chloramphenicol binds to the A site (12). The position of the bound drug suggests that it hinders substrate binding directly by interfering with the positioning of the aminoacyl moiety in the A site. This would correspond to the binding site involved in the direct inhibition of peptide bond formation by chloramphenicol. In the other complex with the Haloarcula marismortui large ribosomal subunit, chloramphenicol binds to a hydrophobic crevice at the entrance to the peptide exit tunnel (13). This binding site suggests that chloramphenicol inhibits protein synthesis by perturbing the egress of nascent polypeptides into the exit tunnel (14).
In the present investigation, a crosslinking approach has been used to examine binding of chloramphenicol to both archaeal and bacterial ribosomes. We find that the drug affects nucleotides that are also part of the macrolide binding site. The results correlate with the chloramphenicol binding site reported recently at the entrance to the peptide exit tunnel on an archaeal ribosome and suggest that a similar binding site is present on the E.coli ribosome.
MATERIALS AND METHODS
Crosslinking of chloramphenicol to E.coli and H.halobium ribosomes
Ribosomes from E.coli MRE600 and H.halobium R1 cells were prepared as described previously (15,16). Chloram phenicol was incubated with 70S ribosomes (0.15–0.5 µM) in 10–200 µl of 20 mM Tris–HCl (pH 7.5), 50 mM NH4Cl and 10 mM MgCl2 (E.coli) or 70 mM HEPES–KOH (pH 7.8), 60 mM magnesium acetate, 3 M KCl and 1 mM DTT (H.halobium) for 20 min at 37°C. The concentration of chloramphenicol was 1.2 mM in all experiments except those examining chloramphenicol concentration dependence, where a 10 µM to 10 mM concentration range was investigated. In some experiments, deacylated tRNA or N-Ac-Phe-tRNA (1.3 mol/mol ribosomes) was added in the presence of poly(U) (1 µg/pmol ribosomes) (17). In the antibiotic interference experiments, other drugs (the final concentration of the drugs was 200 µM except for puromycin, which was used at 1 mM) were added separately either to chloramphenicol complexed with 70S ribosomes or directly to 70S ribosomes before addition of chloramphenicol. After formation of ribosomal complexes, samples were placed in a microtiter tray, on top of an ice-water bath, and irradiated at 254, 312 or 365 nm in a Stratalinker 1800 (with five 8 W bulbs; Stratagene) for 2–60 min. Samples were extracted with phenol, (1:1) phenol:chloroform and chloroform, followed by ethanol precipitation to remove protein and non-crosslinked antibiotics. Samples from H.halobium were washed extensively (four to five times) to remove excess salt. The isolated rRNA was subjected to primer extension analysis with AMV reverse transcriptase (Finnzymes) and different 5′ 32P-labeled deoxyoligonucleotides complementary to 23S rRNA (Table 1) (18). The extension products were separated in denaturing polyacrylamide sequencing gels and autoradiographed.
Table 1. DNA oligonucleotides used for primer extension and RNase H analyses.
| Name | Sequence (5′ to 3′) | Complementary 23S rRNA nucleotides |
|---|---|---|
| Ec617 | GGT TTC CCT TCG GCT CCC C | 617–635 |
| Ec770 | GGC CTT TCA CCC CCA GCC | 770–787 |
| Ec821 | GGC GCT ACC TAA ATA GCT | 821–838 |
| Ec2563 | TCG CGT ACC ACT TTA | 2563–2577 |
| Ec2654 | TCC GGT CCT CTC GTA CT | 2654–2670 |
| Hh1928 | GTC ATA GTT ACT CCC GCC G | 1928–1947 |
| Hh2133 | TCC TAC CTA CTC TGC ACA TC | 2133–2152 |
| Hh2255 | CCG CCC CAG TCA AAC TCC CC | 2255–2274 |
Isolation of 23S rRNA fragments
Chloramphenicol (1.2 mM) was complexed to ribosomes (0.15–0.5 µM) in 100–200 µl of the appropriate buffer as described above for 20 min at 37°C. Samples were irradiated at 365 nm for 30 min, extracted with phenol, and precipitated with ethanol as described above. The rRNA (25 pmol) was resuspended in 10 µl of 20 mM Tris–HCl (pH 7.8) and 63.5 mM NH4Cl, followed by the addition of 37.5 pmol of each complementary oligonucleotide (Table 1 and Figs 2 and 4). The samples were incubated for 5 min at 55°C, followed by the addition of 1.5 µl of 10 mM magnesium acetate and 0.25 U of RNase H (Amersham-Pharmacia). After a second incubation at 37°C for 5 min, rRNA was precipitated with ethanol. The released fragments were purified on 5% polyacrylamide–7 M urea denaturing gels and localized by staining with toluidine blue (0.1% in 7.5% acetic acid). Excised gel slices were transferred to 300 µl of 0.3 M sodium acetate (pH 6.0) and 2 mM EDTA, and were extracted overnight with an equal volume of phenol at room temperature. The rRNA fragments were recovered through ethanol precipitation, and resuspended in double distilled water and subjected to primer extension analysis as described above. The RNase H fragment sizes given in the text are approximate and based on the fact that RNase H is known to cleave the RNA strand near the 3′ end of an RNA–DNA duplex, followed by exonucleolytic trimming that stops ∼4 nt from the 5′ end of the hybrid duplex (19). An extensive effort was made to obtain evidence for direct chloramphenicol–rRNA crosslinks by isolating a radioactive RNase H fragment using 14C-labeled chloramphenicol. The experiments did not yield conclusive results due to both the low crosslinking yield and specific activity of the drug.
Figure 2.
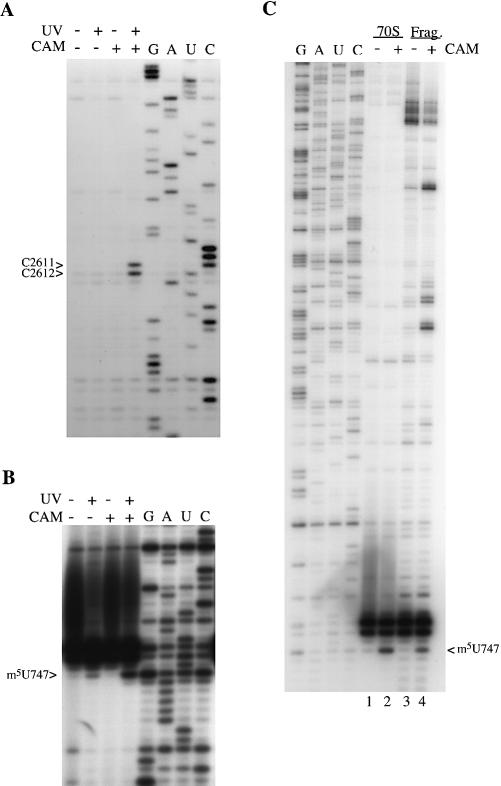
Chloramphenicol-dependent, UV-induced modifications in E.coli 23S rRNA. (A and B) Ribosomes (0.15 µM) were incubated with (+) and without (–) chloramphenicol (1.2 mM) for 20 min at 37°C, and with (+) and without (–) UV irradiation at 365 nm for 30 min. Ribosomal RNA was isolated and analyzed by primer extension with primers Ec2654 (A) and Ec770 (B). (C) RNase H analysis of the modification at 747. Ribosomes (0.5 µM) were incubated with (+) and without (–) chloramphenicol (1.2 mM) for 20 min at 37°C, irradiated at 365 nm for 30 min, followed by isolation of rRNA. A small portion of each sample (70S) was analyzed directly by extension from primer Ec770. The remainder of each sample was treated with RNase H in the presence of oligonucleotides Ec617 and Ec820. The released fragment (Frag.) was gel-purified and subjected to primer extension using primer Ec770. In (B) and (C), 23S rRNA from the E.coli IB10 strain lacking the m1G745 methyltransferase and the strong stop at G745 was used for the sequencing tracks (43). The rRNA modification-induced stop is displaced by one base below the corresponding stop in the sequencing tracks (G, A, U and C).
Figure 4.
A chloramphenicol-dependent, UV-induced modification in H.halobium 23S rRNA. (A) Complexes were formed between ribosomes (0.15 µM) and chloramphenicol (1.2 mM) by incubating at 37°C for 20 min. The samples were irradiated for 30 min at 365 nm, followed by isolation of rRNA and primer extension with primer Hh2133. (B) Ribosomes (0.5 µM) were incubated with (+) and without (–) chloramphenicol (1.2 mM) for 20 min at 37°C, irradiated at 365 nm for 30 min, followed by isolation of rRNA. A small portion of each sample (70S) was analyzed directly by extension from primer Hh2133. The remainder of each sample was treated with RNase H in the presence of oligonucleotides Hh1928 and Hh2255. The resulting fragment (Frag.) was gel-purified and subjected to primer extension using primer Hh2133. The rRNA modification-induced stops are displaced by one base below the corresponding stop in the sequencing tracks (G, A, U and C).
RESULTS
Identification of chloramphenicol-dependent modification sites on E.coli 23S rRNA
Chloramphenicol was added to E.coli 70S ribosomes and the resulting complexes were irradiated with 365 nm light. Total rRNA was isolated and examined for chloramphenicol-induced stops using primer extension analysis with reverse transcriptase. The entire domain V region was scanned, as well as a region of domain II centered around helix 35, which has been connected to the 2058 region of the peptidyl transferase loop in the chemical footprints of various macrolide and ketolide antibiotics (20). Chloramphenicol causes reverse transcriptase stops at nucleotide m5U747 in domain II and at nucleotides C2611/C2612 in domain V of E.coli 23S rRNA. The modifications at C2611/C2612 are not observed when the experiments are performed in the presence of either chloramphenicol or UV light (Fig. 2A, compare lanes 2 and 3 with lane 4). A weak reverse transcriptase stop at m5U747 is observed in the presence of UV irradiation (Fig. 2B, lane 2), but increases strongly in the presence of chloramphenicol and UV light (Fig. 2B, lane 4). Thus, the stops are dependent on both UV irradiation and chloramphenicol.
The observed reverse transcriptase stops can arise as a result of several events, namely formation of a direct drug–rRNA crosslink, a drug-induced rRNA–rRNA or rRNA–protein crosslink, or a drug-induced cleavage of the rRNA backbone. To distinguish between some of these possibilities, the modifications were examined further using oligonucleotide-directed RNase H cleavage. Smaller fragments of 23S rRNA containing each modification were excised using RNase H and a pair of DNA oligonucleotides complementary to rRNA flanking the fragments. Fragments were purified on acrylamide gels and examined by primer extension analysis. A modification isolated on a small RNA fragment is due either to a direct drug–rRNA crosslink or a rRNA–rRNA crosslink internal to the fragment. On the other hand, a modification that is not observed on a smaller RNA fragment is due either to a crosslink to protein or rRNA external to the isolated fragment, or alternatively, to drug-induced cleavage of the rRNA backbone.
The modification at m5U747 was isolated on a 200 nt fragment with oligonucleotides Ec617 and Ec820. Primer extension analysis showed that the modification is present on the isolated fragment (Fig. 2C, lanes 2 and 4). As no additional primer extension stops from a potential rRNA crosslinking partner were detected on the fragment, the data suggests that the modification is a direct drug–rRNA crosslink.
The chloramphenicol modifications at C2611/C2612 were first observed in a previous investigation of the sparsomycin binding site on the ribosome and determined to be independent of sparsomycin (21). In the present study, the modifications were isolated on an 80 nt fragment using oligonucleotides Ec2563 and Ec2654. The modifications were not observed upon primer extension analysis of the isolated fragment. This result was confirmed through isolation of the 3′ end of 23S rRNA by cleavage with Ec2563, followed by primer extension analysis with a different primer located further from the modification site. The chloramphenicol-dependent modifications at C2611/C2612 are, therefore, not due to direct chloramphenicol–rRNA crosslinks. In addition, the fact that the modification at m5U747 could be isolated on a smaller fragment of 23S rRNA argues against the possibility of a chloramphenicol-induced rRNA–rRNA crosslink between positions 747 and 2611/2612.
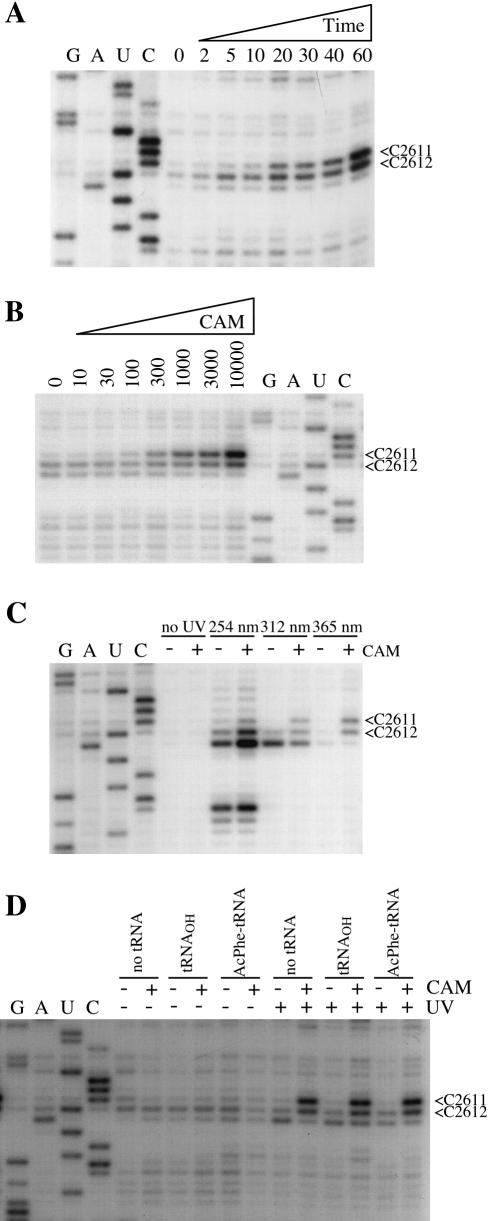
The effects of irradiation time and chloramphenicol concentration on the modification yields at C2611/C2612 were examined. The yields of the modifications increased linearly over at least 60 min of irradiation at 365 nm, suggesting that the UV irradiation employed neither reduced the capacity of the ribosomes to bind the drug nor damaged the drug itself (Fig. 3A). Monitoring the modification yields as a function of chloramphenicol concentration (Fig. 3B) enabled the dissociation constant to be estimated at ∼300 µM. This correlates with chloramphenicol binding to the weaker of the two binding sites (KD2 = 200 µM) characterized in E.coli (6). A similar dependence of modification yield on irradiation time and drug concentration was observed for the modification at m5U747 (data not shown).
Figure 3.
Chemical characteristics of the chloramphenicol-dependent modifications at C2611/C2612 of E.coli 23S rRNA. Complexes between 70S ribosomes (0.15 µM) and chloramphenicol (1.2 mM, unless otherwise indicated) were irradiated with 365 nm light (30 min, unless otherwise indicated). Ribosomal RNA was isolated and analyzed by primer extension from EC2654. (A) Time dependence. Complexes were irradiated for the time (in minutes) indicated above the gels. (B) Chloramphenicol concentration dependence. Complexes were irradiated with increasing concentrations of chloramphenicol (in µM) indicated above the gels. A dissociation constant was estimated by quantifying the modification yields at C2611/C2612 relative to the natural stop at C2613 using a phosphoimager. (C) Wavelength dependence. Complexes were incubated in the absence (–) or presence (+) of chloramphenicol and either untreated or irradiated with 254, 312 or 365 nm light. (D) The effect of P-site-bound tRNA. Deacylated tRNAPhe or N-Ac-Phe-tRNA was included in marked samples. The lanes marked G, A, U and C represent sequencing tracks.
The effect of irradiation wavelength on the modifications at C2611/C2612 was tested by performing the experiment with 254, 312 and 365 nm light (Fig. 3C). Irradiation at 365 nm produces modifications that are entirely chloramphenicol-dependent at positions 2611 and 2612, whereas chloramphenicol-independent stops are also produced at positions 2612 and 2613 upon irradiation at shorter wavelengths (Fig. 3C). As an earlier study showed that chloramphenicol binding to ribosomes is enhanced in the presence of peptidyl–tRNA bound in the P site (22), the effect of a P-site tRNA substrate on the modifications was investigated. The presence of either deacylated tRNA or N-Ac-Phe-tRNA substrates bound in the P site did not significantly alter the modification yields at C2611/C2612 (Fig. 3D).
Identification of a chloramphenicol-dependent modification site on H.halobium 23S rRNA
Chloramphenicol was added to H.halobium ribosomes and the resulting complexes were irradiated with 365 nm light. After isolation of rRNA, the peptidyl transferase loop region of domain V and the region of domain II including helix 35 of 23S rRNA was examined for chloramphenicol-induced stops using primer extension analysis. A reverse transcriptase stop was observed in domain V of 23S rRNA at G2084 (2058 in E.coli numbering) that was dependent on both UV irradiation and chloramphenicol (Fig. 4A). No chloramphenicol- dependent reverse transcriptase stops were observed in the investigated region of domain II of 23S rRNA.
The modification at G2084 (2058) was isolated on a 320 nt fragment by RNase H digestion and oligonucleotides Hh1928 and Hh2255. The fragment was purified on a polyacrylamide gel and subjected to primer extension analysis (Fig. 4B). The primer extension stop at G2084 (2058) is also observed on the isolated rRNA fragment (Fig. 4B, lanes 2 and 4). No evidence for additional primer extension stops from a potential crosslinking partner was detected within the isolated fragment (data not shown). The data suggest that chloramphenicol is crosslinked directly to G2084 (2058) of H.halobium 23S rRNA.
Interference of the E.coli and H.halobium modifications by antibiotics
Altered nucleotide reactivities to base-specific modifying chemicals have been observed at nucleotides 2058 and adjacent regions of the peptidyl transferase loop with other antibiotics (23). Thus, the ability of a group of antibiotics to interfere with the chloramphenicol-dependent rRNA modifications at nucleotides C2611/C2612 of E.coli and G2084 (2058) of H.halobium 23S rRNAs was tested. Antibiotics were added individually to pre-incubated chloramphenicol– ribosome complexes, followed by a second incubation period and irradiation with 365 nm light. Ribosomal RNA was isolated and the peptidyl transferase loop region was analyzed by primer extension. Identical results were obtained irrespective of the order of addition of chloramphenicol (data not shown).
For the modifications at C2611/C2612 of E.coli 23S rRNA, strong reductions in modification yields are observed with the macrolide antibiotics carbomycin and erythromycin (Fig. 5A). Pristinamycin IIA could compete with the modifications to a lesser degree (Fig. 5A). The other drugs tested, blasticidin S, gougerotin, celesticetin and lincomycin, did not interfere significantly with the modifications (Fig. 5A). The antibiotics puromycin and pristinamycin IIA interfered with the modification at G2084 (2058) of H.halobium 23S rRNA, whereas blasticidin S, gougerotin, carbomycin, erythromycin, celesticetin and anisomycin produced no significant changes in modification yields (Fig. 5B).
Figure 5.

Interference of antibiotics with the chloramphenicol-dependent modifications. Complexes were formed between ribosomes (0.15 µM) and chloramphenicol (1.2 mM) by incubating at 37°C for 10 min. A second antibiotic was added and incubation was continued at 37°C for another 20 min. The samples were irradiated for 30 min at 365 nm, followed by isolation of rRNA. Primer extension analysis was carried out using primers (A) Ec2654 for the modifications at C2611/C2612 in E.coli and (B) Hh2133 for the modification at G2084 (2058) in H.halobium. The drugs used were anisomycin (Ani), blasticidin S (Bla), carbomycin (Car), celesticetin (Cel), erthyromycin (Ery), gougerotin (Gou), lincomycin (Lin), pristinamycin IIA (PIIA) and puromycin (Pur). All antibiotics were used at a concentration of 200 µM with the exception of puromycin, which was used at a concentration of 1 mM. The lanes labeled G, A, U and C represent sequencing tracks.
The streptogramin A antibiotic pristinamycin IIA is able to interfere with the modifications at both C2611/C2612 and 2058. This is consistent with the fact that a A2059G mutation in H.halobium confers resistance to virginiamycin M1, and also that a range of nucleotide protection and enhancements have been documented at positions 2058 and 2059 in the chemical footprints of streptogramin A antibiotics in E.coli, Bacillus megaterium, H.halobium and Hf.mediterranei (5,24). In a streptogramin A antibiotic-large ribosomal subunit complex, the drug binds to the A and P sites, inducing a conformational change of 2062 (13). This nucleotide is positioned between two hydrophobic crevices at the active site and the exit tunnel and can interact with antibiotics bound to either crevice.
Puromycin interferes with the modification at G2058. Puromycin yields a protection effect at position G2058 in Haloferax gibbonsii and Hf.mediterranei ribosomes (25). Unlike other nucleotides affected in the chemical footprint of puromycin (A2439, A2503, G2505, G2553), G2058 is not immediately adjacent to the puromycin binding site in the complex of the CCdAp–puromycin inhibitor bound to the large ribosomal subunit (26). As antibiotics produce diverse organism-dependent effects in this region (24), the data indicate that drug binding to the peptidyl transferase center leads to changes in the conformation of 2058.
DISCUSSION
Two distinct chloramphenicol binding sites have been identified on drug-large ribosomal subunit complexes solved by X-ray crystallography, one in the bacterium D.radiodurans (12) and the other in the archaeon H.marismortui (13). The binding site on the bacterial ribosome was observed in the presence of 0.1 mM chloramphenicol, in both soaking and co-crystallization experiments (12), and correlates with the higher affinity site (KD1 = 2 µM) reported in the literature (6). The antibiotic is bound in the A site and positioned <4.4 Å away from nucleotides G2061, A2451, C2452, U2500, U2504, G2505 and U2506 (12). The position of the bound drug is consistent with the results of chemical footprinting experiments on E.coli ribosomes (4,5). The altogether different binding site on the archaeal ribosome was observed at higher chloramphenicol concentrations (up to 20 mM) and corresponds to the lower affinity site (KD2 = 200 µM) described previously (6). Here chloramphenicol is bound in a hydrophobic pocket at the entrance to the peptide exit tunnel (13). The apparent lack of a high affinity binding site on archaeal ribosomes is in agreement with both the lack of a chemical footprint on Hf.mediterannei ribosomes with 0.1 mM chloramphenicol (5) and the decreased susceptibility of archaeal versus bacterial ribosomes to the drug.
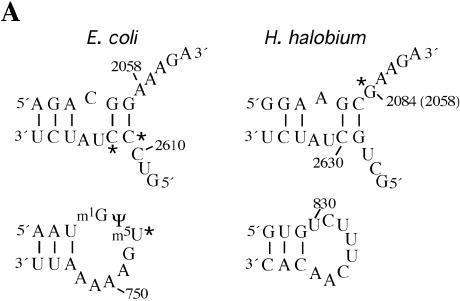
A UV crosslinking approach has been used to identify chloramphenicol-dependent modifications on the 23S rRNAs of organisms from two domains of life, the bacterium E.coli and the archaeon H.halobium. The modification sites occur at different nucleotide positions in each organism (Fig. 6A). As crosslinking reactions are dependent on the position and identity of specific functional groups, this result is not unexpected since the sequence at positions 2058 and 2611 of 23S rRNA (using E.coli numbering) is not conserved between E.coli and H.halobium (Fig. 6A). The modification sites identified in this work are clustered at the entrance to the peptide channel on the large ribosomal subunit (Fig. 6B).
Figure 6.
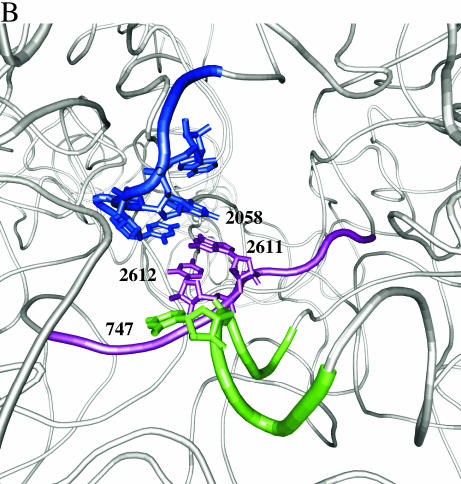
Chloramphenicol-dependent modification sites on E.coli and H.halobium 23S rRNAs. (A) The modification sites are indicated with asterisks on the secondary structures of relevant portions of 23S rRNA. Segments of domains V (top) and II (bottom) of E.coli (left) and H.halobium (right) 23S rRNAs are illustrated. On the domain V segment of H.halobium 23S rRNA, E.coli numbering is given in parenthesis. (B) The location of chloramphenicol-dependent modification sites on the H.marismortui 50S subunit. The view shown is from the back of the subunit looking through the peptide exit tunnel towards the subunit interface. The coordinates of the H.marismortui 50S subunit are used (PDB accession no. 1JJ2). The positions of the modifications are labeled (using E.coli numbering) and shown in blue (2058), magenta (2611, 2612) and green (747). Other nucleotides in the hydrophobic crevice (13), 2057 and 2059, are depicted in blue. The respective segments of 23S rRNA flanking these nucleotides are depicted as tubes of the same color. Other regions of 23S rRNA are shown as light gray ribbons. This image was made with VMD. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign (http://www.ks.uiuc.edu/).
The modification sites identified in the present work correlate strongly with chloramphenicol binding to the site observed on the H.marismortui ribosome. Importantly, the data suggest that this chloramphenicol binding site also exists on the E.coli ribosome. In the H.marismortui 50S subunit–chloramphenicol complex, the drug is bound in a hydrophobic crevice composed of nucleotides 2058 and 2059 and the top of the G2057–C2611 base pair (11). The modifications at 2058 in H.halobium and 2611/2612 in E.coli involve nucleotides that form the hydrophobic crevice (Fig. 6). The modification at 747 can also be explained by chloramphenicol binding in the hydrophobic crevice. The closest distance between nucleotides 747 and 2611 is 4.56 Å, measured between the O2P atom of 747 and the 2′-hydroxyl group of C2611 (Fig. 6B). The proximity of 747 to the chloramphenicol binding site is supported by a crosslink between domains II and V of 23S rRNA, induced under conditions of mild UV irradiation and without antibiotics, between positions G748 and U2613/A2614 (27–29). Moreover, the conformation of another nucleotide in helix 35 of domain II of 23SrRNA, A752, is affected by binding of the macrolide antibiotic erythromycin (20,30).
The modification sites overlap with a portion of the macrolide binding site. The macrolide antibiotics interfere with the modifications at C2611/C2612. This is consistent with the structures of macrolide-large ribosomal subunit complexes where C2611 forms part of a hydrophobic crevice into which one edge of the lactone ring of macrolide antibiotics inserts (14). The macrolide antibiotics did not interfere with the modification at 2058 in H.halobium under the investigated conditions. This is likely due to the reduced affinity of macrolide antibiotics to archaeal ribosomes, which has been rationalized by the presence of a G instead of an A at position 2058. The two hydrogen-bond donors in the exocyclic N2 amino moiety of G2058 interrupt the hydrophobic binding surface for the lactone ring, upon which there are no hydrogen-bond acceptors (14).
In addition to biochemical and genetic data discussed earlier linking the binding sites of chloramphenicol and erythromycin, other evidence indicates that the two antibiotics inhibit protein synthesis through a similar mechanism. An accumulation of short peptides has been observed with both chloramphenicol and the macrolide antibiotic erythromycin in in vitro poly(A)-directed polylysine translation assays (31,32). Both drugs have also been reported to enhance the release of oligopeptidyl–tRNAs by interfering with positioning of the nascent peptide (33,34). A possible biological role for a chloramphenicol binding site at the entrance to the peptide exit tunnel could be in translation attenuation regulation of chloramphenicol resistance in bacteria. Current models suggest that leader peptides of a defined sequence and length and the inducer chloramphenicol function together in ribosome stalling, leading to conformational changes in the 5′ leader regions of cat or cmlA mRNA transcripts and subsequent translation initiation of the resistance determinants (35).
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. V. Kirillov for E.coli ribosomes and tRNAs, R. Garrett for discussions in the early stages of the work, S. Douthwaite for E.coli strain IB10, J. Bøsling for assistance with preparation of Figure 6, and B. Vester for helpful advice and comments on the manuscript. K.S.L. was supported by a fellowship from Alfred Benzons Fund and a grant from the European Commission’s 5th Framework Program (grant QLK2-CT-2002-00892).
REFERENCES
- 1.Lam R.F., Lai,J.S.M., Ng,J.S.K., Rao,S.K., Law,R.W.K. and Lam,D.S.C. (2002) Topical chloramphenicol for eye infections. Hong Kong Med. J., 8, 44–47. [PubMed] [Google Scholar]
- 2.Mankin A.S. and Garrett,R.A. (1991) Chloramphenicol resistance mutations in the single 23S rRNA gene of the archaeon Halobacterium halobium. J. Bacteriol., 173, 3356–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aagaard C., Phan,H., Trevisanato,S. and Garrett,R.A. (1994) A spontaneous point mutation in the single 23S rRNA gene of the thermophilic archaeon Sulfolobus acidocaldarius confers multiple drug resistance. J. Bacteriol., 176, 7744–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moazed D. and Noller,H.F. (1987) Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S rRNA. Biochimie, 69, 879–884. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Fonseca C., Amils,R. and Garrett,R.A. (1995) Fine structure of the peptidyl transferase centre on 23S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol., 247, 224–235. [DOI] [PubMed] [Google Scholar]
- 6.Lessard J.L. and Pestka,S. (1972) Studies on the formation of transfer ribonucleic acid-ribosome complexes. XXIII. Chloramphenicol, aminoacyl-oligonucleotides and Escherichia coli ribosomes. J. Biol. Chem., 247, 6909–6912. [PubMed] [Google Scholar]
- 7.Fernandez-Muñoz R. and Vazquez,D. (1973) Kinetic studies of peptide bond formation. Effect of chloramphenicol. Mol. Biol. Rep., 1, 75–79. [DOI] [PubMed] [Google Scholar]
- 8.Drainas D., Kalpaxis,D.L. and Coutsogeorgopoulos,C. (1987) Inhibition of ribosomal peptidyltransferase by chloramphenicol. Kinetic studies. Eur. J. Biochem., 164, 53–58. [DOI] [PubMed] [Google Scholar]
- 9.Pestka S. (1977) Inhibitors of protein synthesis. In Weissbach,H. and Pestka,S. (eds), Molecular Mechanisms of Protein Biosynthesis. Academic Press, New York, pp. 467–553. [Google Scholar]
- 10.Ettayebi M., Prasad,S.M. and Morgan,E.A. (1985) Chloramphenicol–erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J. Bacteriol., 162, 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douthwaite S. (1992) Functional interactions within 23S rRNA involving the peptidyltransferase center. J. Bacteriol., 174, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlünzen F., Zarivach,R., Harms,J., Bashan,A., Tocilj,A., Albrecht,R., Yonath,A. and Franceschi,F. (2001) Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature, 413, 814–821. [DOI] [PubMed] [Google Scholar]
- 13.Hansen J.L., Moore,P.B. and Steitz,T.A. (2003) Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol., 330, 1061–1075. [DOI] [PubMed] [Google Scholar]
- 14.Hansen J.L., Ippolito,J.A., Ban,N., Nissen,P., Moore,P.B. and Steitz,T.A. (2002) The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell, 10, 117–128. [DOI] [PubMed] [Google Scholar]
- 15.Makhno V.I., Peshin,N.N., Semenkov,Y.P. and Kirillov,S,V. (1988) Modified procedure of producing ‘tight’ 70S ribosomes from E. coli, highly active in individual stages of the elongation cycle. Mol. Biol., 22, 528–537. [PubMed] [Google Scholar]
- 16.Leviev I.G., Rodriguez-Fonseca,C., Phan,H., Garrett,R.A., Heilek,G., Noller,H.F. and Mankin,A.S. (1994) A conserved secondary structural motif in 23S rRNA defines the site of interaction of amicetin, a universal inhibitor of peptide bond formation. EMBO J., 13, 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirillov S.V., Makhno,V.I. and Semenkov,Y.P. (1978) The mechanism of codon–anticodon interaction in ribosomes: quantitative study of codon-dependent binding of tRNA to the 30S ribosomal subunits of E. coli. Eur. J. Biochem., 89, 297–304. [DOI] [PubMed] [Google Scholar]
- 18.Christiansen J., Egebjerg,J., Larsen,N. and Garrett,R.A. (1990) Analysis of RNA structure: Experimental and theoretical considerations. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: A Practical Approach. Oxford University Press, Oxford, pp. 229–252. [Google Scholar]
- 19.Crooke S.T., Lemonidis,K.M., Neilson,L., Griffey,R., Lesnik,E.A. and Monia,B.P. (1995) Kinetic characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide–RNA duplexes. Biochem. J., 312, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen L.H., Mauvais,P. and Douthwaite,S. (1999) The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol., 31, 623–631. [DOI] [PubMed] [Google Scholar]
- 21.Porse B.T., Kirillov,S.V., Awayez,M.J., Ottenheijm,H.C.J. and Garrett,R.A. (1999) Direct crosslinking of the antitumour antibiotic sparsomycin and its derivatives, to A2602 in the peptidyl transferase center of 23S-like rRNA within ribosome–tRNA complexes. Proc. Natl Acad. Sci. USA, 96, 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contreras A. and Vazquez,D. (1977) Cooperative and antagonistic interactions of peptidyl–tRNA and antibiotics with bacterial ribosomes. Eur. J. Biochem., 74, 539–547. [DOI] [PubMed] [Google Scholar]
- 23.Garrett R.A. and Rodríguez-Fonseca,C. (1995) The peptidyl transferase center. In Zimmermann,R.A. and Dahlberg,A. (eds), Ribosomal RNA: Structure, Evolution, Processing and Function. CRC Press, Boca Raton, FL, pp. 327–355. [Google Scholar]
- 24.Porse B.T. and Garrett,R.A. (1999) Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23S rRNA and the synergism of their inhibitory mechanisms. J. Mol. Biol., 286, 375–387. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Fonseca C., Phan,H., Long,K.S., Porse,B.T., Kirillov,S.V., Amils,R. and Garrett,R.A. (2000) Puromycin–rRNA interactions at the peptidyl transferase center. RNA, 6, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 27.Stiege W., Glotz,C. and Brimacombe,R. (1983) Localisation of a series of intra-RNA cross-links in the secondary and tertiary structure of 23S RNA, induced by ultraviolet irradition of Escherichia coli 50S ribosomal subunits. Nucleic Acids Res., 11, 1687–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiege W., Atmadja,J., Zobawa,M. and Brimacombe,R. (1986) Investigation of the tertiary folding of Escherichia coli ribosomal RNA by intra-RNA cross-linking in vivo. J. Mol. Biol., 191, 135–138. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell P., Osswald,M., Schueler,D. and Brimacombe,R. (1990) Selective isolation and detailed analysis of intra-RNA cross-links induced in the large ribosomal subunit of E. coli: a model for the tertiary structure of the tRNA binding domain in 23S RNA. Nucleic Acids Res., 18, 4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulsen S.M., Kofoed,C. and Vester,B. (2000) Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol., 304, 471–481. [DOI] [PubMed] [Google Scholar]
- 31.Julian G.R. (1965) [14C]Lysine peptides synthesized in an in vitro Escherichia coli system in the presence of chloramphenicol. J. Mol. Biol., 12, 9–16. [DOI] [PubMed] [Google Scholar]
- 32.Mao J.C.H. and Robishaw,E.E. (1971) Effects of macrolides on peptide-bond formation and translocation. Biochemistry, 10, 2054–2061. [DOI] [PubMed] [Google Scholar]
- 33.Menninger J.R. and Otto,D.P. (1982) Erythromycin, carbomycin and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl–tRNA from ribosomes. Antimicrob. Agents Chemother., 21, 810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rheinberger H.J. and Nierhaus,K,H. (1990) Partial release of AcPhe-Phe-tRNA from ribosomes during poly(U)-dependent poly(Phe) synthesis and the effects of chloramphenicol. Eur. J. Biochem., 193, 643–650. [DOI] [PubMed] [Google Scholar]
- 35.Lovett P.S. (1996) Translation attenuation regulation of chloramphenicol resistance in bacteria—a review. Gene, 179, 157–162. [DOI] [PubMed] [Google Scholar]
- 36.Koike K., Taira,M., Kuchino,Y., Yaginuma,K., Sekiguchi,T. and Kobayashi,M. (1983) In Schweyen,R.J., Wolf,K. and Kudewitz,F. (eds), Mitochondria. Walter de Gruyter, Berlin, pp. 372–387. [Google Scholar]
- 37.Dujon B. (1980) Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell, 20, 185–197. [DOI] [PubMed] [Google Scholar]
- 38.Kearsey S.E. and Craig,I.W. (1981) Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature, 290, 607–608. [DOI] [PubMed] [Google Scholar]
- 39.Blanc H., Adams,C.W. and Wallace,D.C. (1981) Different nucleotide changes in the large rRNA genes of the mitochondrial DNA confer chloramphenicol resistance on two human cell lines. Nucleic Acids Res., 9, 5785–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slott E.F. Jr, Shade,R.O. and Lansman,R.A. (1983) Sequence analysis of mitochondrial DNA in a mouse cell line resistant to chloramphenicol and oligomycin. Mol. Cell. Biol., 3, 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vester B. and Garrett,R.A. (1988) The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S ribosomal RNA. EMBO J., 7, 3577–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanc H., Wright,C.T., Bibb,M.J., Wallace,D.C. and Clayton,D.A. (1981) Mitochondrial DNA of chloramphenciol-resistant mouse cells contains a single nucleotide change in the region encoding the 3′ end of the large ribosomal RNA. Proc. Natl Acad. Sci. USA, 78, 3789–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustafsson C. and Persson,B.C. (1998) Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol., 180, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]