Abstract
The first synthesis of 5-amino-3-(2′-deoxy-β-d-ribofuranosyl)imidazo[4,5-b]pyridin-7-one (1-deaza-2′-deoxyguanosine) is described. The compound was converted from the known AICA-deoxyriboside. The tautomeric structure of the base moiety was determined by theoretical calculation to be a hydroxyl form. Although the analog was found to be labile to acidic conditions, 1-deaza-2′-deoxyguanosine was successfully converted into a phosphoramidite derivative, which was incorporated into oligodeoxynucleotides by the standard phosphoramidite method. Thermal stabilities of oligodeoxynucleotides containing 1-deaza-2′-deoxyguanosine were investigated by thermal denaturing experiments. Also, a triphosphate analog of 1-deaza-2′-deoxyguanosine was synthesized for polymerase extension reactions. Single nucleotide insertion reactions using a template containing 1-deaza-2′-deoxyguanosine, as well as 1-deaza-2′-deoxyguanosine triphosphate, were performed using the Klenow fragment (exonuclease minus) polymerase and other polymerases. No hydrogen bonded base pairs, even a 1-deaza-2′-deoxyguanosine:cytidine base pair, were indicated by thermal denaturing studies. However, though less selective and less effective than the natural guanosine counterpart, the polymerase extension reactions suggested the formation of a base pair of 1-deaza-2′-deoxyguanosine with cytidine during the insertion reactions.
INTRODUCTION
Replacement of a single atom of an active molecule with other atoms is a useful method for investigating the structure–function relationships of the molecule. Numerous nucleoside and nucleotide analogs, in which a single atom has been removed or replaced with another atom, have been synthesized (1–4). A series of deazanucleoside analogs are one of the common modified molecules of this kind and have potential applicability in the fields of nucleic acid chemistry and molecular biology (1–3,5). All nitrogen atoms in nucleobase ring systems are involved in a variety of biological interactions, such as interactions for the formation of complement base pairs (4,6) as well as interactions for the formation of DNA/RNA higher dimensional structures (7) and in protein–nucleic acid interactions (8,9). In deazanucleosides, a hydrophilic nitrogen atom had been replaced with a hydrophobic carbon atom but they retain the geometry of the original structure. Therefore, deazanucleoside analogs can provide important information about these biological interactions. A large number of both ribo- and deoxyribo-deazanucleoside analogs have been reported. These analogs have also been incorporated into oligodeoxynucleotides to study the function of a particular nitrogen atom in the DNA strand.
7-Deazapurine nucleosides, especially 7-deaza-2′-deoxyguanosine analogs, are the most well-known compounds of this kind and have been used extensively as sequencing tools (10). Removal of the hydrogen bond acceptor at the 7 position of the guanine base prevents the formation of aggregated polydeoxynucleotides in sequencing reactions. Many analogs of 7-deazapurine nucleosides have been reported and their properties, such as enzymatic behavior (10) and thermal stabilities of duplex (11,12) and/or triplex oligodeoxynucleotides (13), have been extensively investigated. Several 3-deazapurine analogs have also been reported and have been used to investigate the function of the nitrogen atom at the 3 position of purine nucleosides, which has been removed in these analogs (14–17). For example, Spratt and co-workers used 3-deaza-2′-deoxyguanosine to identify the hydrogen bonded interactions between DNA polymerase I (Klenow fragment) and the minor groove of the DNA strand (14,15). Furthermore, 3-deaza-3-halopurine nucleoside analogs are thought to be good model compounds for studying nucleoside–enzyme interactions, since glycosyl torsion angles are fixed in the anti region in these analogs (18).
Despite the usefulness and interesting features of deazanucleosides, little attention has been paid to 1-deazapurine analogs, especially 1-deazaguanine nucleosides. Synthesis of 1-deazaguanosine was first reported by Townsend et al. in 1975 (19,20). However, to the best of our knowledge, no modifications were made to the method for synthesizing this compound until Spratt et al. reported the O-methylated analog of 1-deaza-2′-deoxyguanosine in 1994 (21). They prepared O6-methyl-1-deaza-2′-deoxyguanosine (3) (Fig. 1) by enzymatic trans-glycosylation of the corresponding nucleobase with thymidine and introduced into oligodeoxynucleotides by the phosphoramidite method. The compound was used to investigate interactions between O6-alkylguanine-DNA alkyltransferase and the guanine group (21,22). This analog was also used in DNA replication studies (23). It was only recently that a modified method for synthesizing 1-deazaguanosine was reported (24). However, there has been no report of the synthesis of a deoxynucleoside analog of 1-deazaguanine (1), and the characteristics of this analog are not known.
Figure 1.
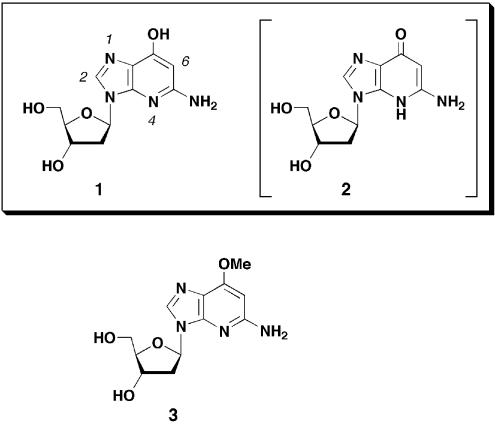
Structures of 1-deaza-2′-deoxyguanosine (1), with the tautomeric structure 2 in parentheses, and O6-methyl-1-deaza-2′-deoxyguanosine (3). The base numberings are indicated in italic.
Here, we report the first synthesis of 1-deaza-2′-deoxyguanosine (1) (1-deaza-dG) (Fig. 1). The synthesized compound was converted into a phosphoramidite unit (14), which was used in oligodeoxynucleotide (ODN) synthesis. Using this ODN, a thermal denaturing experiment was performed to evaluate the base pairing abilities of 1-deaza-dG (1). Furthermore, 1-deaza-dG was transformed into a triphosphate analog (16). Single nucleotide insertion reactions by DNA polymerases using a template containing 1-deaza-dG (1) and a triphosphate analog (16) are also reported in this manuscript.
MATERIALS AND METHODS
General information
Physical data were measured as follows: 1H and 13C NMR spectra were recorded on a JEOL 270 MHz FT-NMR spectrometer in DMSO-d6 as the solvent with tetramethylsilane as an internal standard. 85% D3PO4 was used as an internal standard for 31P spectrum. Chemical shifts are reported in parts per million (δ) and signals are expressed as s (singlet), d (doublet), t (triplet), m (multiplet) or br (broad). All exchangeable protons were detected by disappearance on the addition of D2O. Mass spectra were recorded on a JEOL JMS-FABmate (FAB-MS) or JEOL JMS-HX110 (EI-MS) spectrometer at the Center for Instrumental Analysis, Hokkaido University. TLC was done on Merck silica gel 60F254 precoated plates (Merck, Germany). Silica gels used for column chromatography were Wakogel C-200 (particle size 75–150 µm) (Wako Pure Chemical Industries, Japan) or ICN Silica 60Å (particle size 63–200 µm) (ICN Biomedicals, Belgium). RP-HPLC analysis and purifications were performed on a Gilson HPLC system. Klenow fragment (exo–) and Therminator™ DNA polymerase were purchased from New England BioLabs (Beverly, MA). AMV reverse transcriptase, M-MLV reverse transcriptase and dNTPs were purchased from Takara Biomedicals (Japan).
Synthesis of 1-deaza-2′-deoxyguanosine
1-(2′-Deoxy-3′,5′-di-O-triisopropylsilyl-β-d-ribofuranosyl)- 5-[N,N-di-(t-butoxycarbonyl)]aminoimidazole-4-[N,N-di-(t- butoxycarbonyl)]carboxamide (5). To a solution of 4 (2.47 g, 4.45 mmol), trietylamine (2.50 ml, 17.8 mmol) and DMAP (110 mg, 0.90 mmol) in 1,2-dichloroethane (60 ml) was added di-t-butyl dicarbonate (6.13 ml, 26.7 mmol) at room temperature. The reaction mixture was heated at 75°C for 1 h. The solvent was removed in vacuo and the residue was purified by silica gel column chromatography (4.0 × 14 cm, 15% AcOEt in hexane) to give 5 (3.30 g, 78%) as a white foam. FAB-LRMS m/z 955 (MH+); FAB-HRMS calculated for C47H87N4O12Si2 (MH+) 955.5859, found 955.5859. 1H NMR (270 MHz, DMSO-d6) δ: 7.99 (s, 1 H, 2-H), 5.78 (t, 1 H, 1′-H, J = 6.3 Hz), 4.64 (dt, 1 H, 3′-H, J = 3.6, 4.3 Hz), 3.94 (ddd, 1 H, 4′-H, J = 3.3, 3.6, 4.0 Hz), 3.85 (dd, 1 H, 5′-Ha, J = 4.0, 12.9 Hz), 3.83 (dd, 1 H, 5′-Hb, J = 3.3, 12.9 Hz), 2.35 (dd, 2 H, 2′-H2, J = 4.3, 6.3 Hz), 1.39 (s, 18 H, t-Bu2), 1.34 (s, 18 H, t-Bu2), 1.05∼1.00 (m, 42 H, i-Pr6). 13C NMR (67.8 MHz, DMSO-d6) δ: 162.05 (C), 149.46 (C), 148.45 (C), 148.19 (C), 132.55 (CH), 131.69 (C), 127.58 (C), 87.21 (CH), 83.48 (CH), 83.37 (C), 83.30 (C), 83.21 (C), 71.03 (CH), 62.45 (CH2), 41.63 (CH2), 27.19 (CH3), 27.16 (CH3), 17.77 (CH3), 17.72 (CH3), 11.55 (CH), 11.31 (CH).
5-Amino-4-cyanoacetyl-1-(2′-deoxy-3′,5′-di-O-triisopropylsi lyl-β-d-ribofuranosyl)imidazole (7). To a solution of n-BuLi (1.63 M in hexane, 2.52 ml, 4.10 mmol) in THF (15 ml), mixed at –78°C, was added acetonitrile (0.24 ml, 4.50 mmol) over 15 min at –78°C. After the mixture was stirred at the same temperature for 30 min, a THF (7.0 ml) solution of 5 (780 mg, 0.82 mmol) was added dropwise and the whole was stirred at –78°C for another 1.5 h. The reaction was quenched by addition of saturated aqueous NH4Cl (4 ml), which was then diluted with AcOEt (80 ml). The organic layer was washed with H2O (30 ml × 2) and brine (30 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography (3.0 × 13 cm, 20% AcOEt in hexane) to give crude 6 as a colorless oil [FAB-LRMS m/z 679 (MH+)]. Silica gel (12 g) was added to a solution of crude 6 in CHCl3 (5 ml), which was dried under vacuum using a rotary evaporator. The dried silica gel mixture was heated at 80°C under reduced pressure for 2.5 h. The mixture was purified by silica gel column chromatography (3.0 × 12 cm, 40% AcOEt in hexane) to give 7 (194 mg, 41%, two steps) as a white solid. FAB-LRMS m/z 579 (MH+); FAB-HRMS calculated for C29H55N4O4Si2 (MH+) 579.3762, found 579.3743. 1H NMR (270 MHz, DMSO-d6) δ: 7.38 (s, 1 H, 2-H), 6.99 (br s, 2 H, NH2), 6.01 (dd, 1 H, 1′-H, J = 5.9, 7.6 Hz), 4.60 (m, 1 H, 3′-H), 4.13 (s, 2 H, CH2), 3.91 (m, 1 H, 4′-H), 3.80 (dd, 1 H, 5′-Ha, J = 4.6, 11.2 Hz), 3.74 (dd, 1 H, 5′-Hb, J = 4.3, 11.2 Hz), 2.51 (ddd, 1 H, 2′-Ha, J = 6.3, 7.6, 13.3 Hz), 2.31 (ddd, 1 H, 2′-Hb, J = 2.6, 5.9, 13.3 Hz), 1.08∼0.99 (m, 42 H, i-Pr6). 13C NMR (67.8 MHz, DMSO-d6) δ: 179.96 (C), 146.48 (C), 128.75 (CH), 117.88 (C), 116.32 (C), 87.35 (CH), 82.29 (CH), 71.91 (CH), 62.92 (CH2), 39.74 (CH2), 27.21 (CH2), 17.69 (CH3), 17.63 (CH3), 11.42 (CH), 11.19 (CH).
5-Amino-3-(2′-deoxy-3′,5′-di-O-triisopropylsilyl-β-d-ribofur anosyl)imidazo[4,5-b]pyridin-7-one (8). A solution of 7 (520 mg, 0.90 mmol) in a mixture of EtOH and 5% aqueous Na2CO3 (3:1, 40 ml) was heated at 90°C. After the mixture was stirred at the same temperature for 3.5 h, the solvent was removed in vacuo. The residue was dissolved in AcOEt (80 ml), which was washed with H2O (30 ml × 2) and brine (30 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography (3.0 × 13 cm, 40% AcOEt in hexane) to give 8 (447 mg, 86%) as a pale yellow solid. FAB-LRMS m/z 579 (MH+); FAB-HRMS calculated for C29H55N4O4Si2 (MH+) 579.3762, found 579.3790. 1H NMR (270 MHz, DMSO-d6) δ: 10.53 (br s, 1 H, 7-OH), 7.94 (s, 1 H, 2-H), 6.27 (dd, 1 H, 1′-H, J = 5.9, 6.6 Hz), 5.79 (s, 1 H, 6-H), 5.66 (br s, 2 H, 5-NH2), 4.66 (ddd, 1 H, 3′-H, J = 1.9, 2.3, 5.3 Hz), 3.91 (ddd, 1 H, 4′-H, J = 1.9, 4.3, 5.6 Hz), 3.86 (dd, 1 H, 5′-Ha, J = 5.6, 10.6 Hz), 3.72 (dd, 1 H, 5′-Hb, J = 4.3, 10.6 Hz), 2.81 (ddd, 1 H, 2′-Ha, J = 5.3, 8.6, 13.2 Hz), 2.26 (ddd, 1 H, 2′-Hb, J = 2.3, 5.9, 13.2 Hz), 1.10∼0.99 (m, 42 H, i-Pr6). 13C NMR (67.8 MHz, DMSO-d6) δ: 157.91 (C), 156.50 (C), 146.99 (C), 135.20 (CH), 118.40 (C), 89.40 (CH), 86.97 (CH), 81.97 (CH), 72.51 (CH), 63.25 (CH2), 38.95 (CH2), 17.72 (CH3), 17.65 (CH3), 11.44 (CH), 11.22 (CH).
5-Amino-4-cyanoacetyl-1-(2′-deoxy-β-D-ribofuranosyl)imi dazole (9). To a solution of 7 (503 mg, 0.87 mmol) in THF (10 ml) was added TBAF (1.0 M in THF, 2.17 ml, 2.17 mmol) at 0°C. After the mixture was stirred at room temperature for 1 h, acetic acid (124 µl, 2.17 mmol) was added and the solvent was removed in vacuo. The residue was purified by silica gel column chromatography (3.5 × 15 cm, 25% EtOH in CHCl3) to give 9 (174 mg, 75%) as a pale yellow foam. FAB-LRMS m/z 267 (MH+); FAB-HRMS calculated for C11H15N4O4 (MH+) 267.1093, found 267.1104; 1H NMR (270 MHz, DMSO-d6) δ: 7.43 (s, 1 H, 2-H), 7.04 (br s, 2 H, NH2), 6.00 (dd, 1 H, 1′-H, J = 6.1, 7.6 Hz), 5.29 (br m, 2 H, 5′-OH and 3′-OH), 4.32 (ddd, 1 H, 3′-H, J = 2.6, 3.0, 5.9 Hz), 4.13 (s, 2 H, CH2), 3.84 (m, 1 H, 4′-H), 3.55 (m, 2 H, 5′-H2), 2.38 (ddd, 1 H, 2′-Ha, J = 5.9, 7.6, 13.2 Hz), 2.15 (ddd, 1 H, 2′-Hb, J = 2.6, 6.1, 13.2 Hz). 13C NMR (67.8 MHz, DMSO-d6) δ: 179.85 (C), 146.30 (C), 129.93 (CH), 117.95 (C), 116.39 (C), 87.37 (CH), 83.42 (CH), 70.40 (CH), 61.10 (CH2), 39.26 (CH2), 27.16 (CH2).
5-Amino-3-(2′-deoxy-β-D-ribofuranosyl)imidazo[4,5-b]pyri din-7-one (1-deaza-2′-deoxyguanosine) (1). A solution of 9 (170 mg, 0.64 mmol) in a mixture of EtOH and 5% aqueous Na2CO3 (2:1, 15 ml) was heated at 90°C. After the mixture was stirred at the same temperature for 3 h, the solvent was removed in vacuo. The residue was dissolved in H2O (50 ml), which was neutralized with saturated aqueous NH4Cl to pH 8.0. To the solution was added activated charcoal (3.0 g) and the mixture was stirred at room temperature for 15 min, and then poured into a glass chromatography tube. The activated charcoal was washed with H2O (200 ml) and then the target compound was eluted with a mixed solvent of EtOH and aqueous 28% NH4OH (7:3, 100 ml). The solvent was removed to give 1 (98 mg, 58%) as a pale yellow foam. FAB-LRMS m/z 267 (MH+); FAB-HRMS calculated for C11H15N4O4 (MH+) 267.1093, found 267.1196; UV λmax (pH 7.0) 279.4 nm (ε = 8600), 256.3 nm (ε = 9000); λmax (pH 4.8) 284.8 nm (ε = 9000), 255.0 nm (ε = 7600); λmax (pH 8.9) 263.8 nm (ε = 11 300); 1H NMR (270 MHz, DMSO-d6) δ: 10.57 (br s, 1 H, 7-OH), 7.98 (s, 1 H, 2-H), 6.26 (dd, 1 H, 1′-H, J = 5.9, 8.3 Hz), 5.81 (s, 1 H, 6-H), 5.64 (br s, 2 H, 5-NH2), 5.31∼5.25 (br m, 2 H, 5′-OH and 3′-OH), 4.36 (ddd, 1 H, 3′-H, J = 2.3, 3.2, 5.6 Hz), 3.84 (ddd, 1 H, 4′-H, J = 2.7, 3.2, 4.3 Hz), 3.59 (dd, 1 H, 5′-Ha, J = 4.3, 11.9 Hz), 3.51 (dd, 1 H, 5′-Hb, J = 2.7, 11.9 Hz), 2.61 (ddd, 1 H, 2′-Ha, J = 5.6, 8.3, 13.1 Hz), 2.16 (ddd, 1 H, 2′-Hb, J = 2.3, 5.9, 13.1 Hz). 13C NMR (67.8 MHz, DMSO-d6) δ: 157.87 (C), 156.83 (C), 146.80 (C), 136.02 (CH), 118.72 (C), 89.43 (CH), 87.50 (CH), 83.06 (CH), 71.07 (CH), 62.04 (CH2), 39.34 (CH2).
Theoretical calculations
Ab initio calculations were carried out by the program GAUSSIAN 94 (Gaussian Inc., Pittsburgh, PA). The tautomeric conformations were calculated at the [MP2/6-31G(d,p)//MP2/6-31G+(2d′,p′)] level. The hydrogen bond energies were calculated at the [MP2/6-31+G(2d′,p′)//HF/6-31G(d,p)] level. In the latter case, Cs symmetry was assumed: all atoms, except for hydrogen atoms in the methyl groups, were placed on the plane of symmetry.
Stabilities of 1-deaza-2′-deoxyguanosine in acidic conditions
A solution containing 1-deaza-dG (1) (0.1 mM) and thymine (0.04 mM), as an internal standard, in 0.1 M sodium phosphate (pH 2.2–7.2) were incubated at 37 or 60°C. At appropriate time points, aliquots of the reaction mixture were sampled and analyzed by RP-HPLC (Waters µ-Bondasphere, 3.9 × 150 mm, 0.1 M TEAA buffer and CH3CN solvent system, pH 7.0). Disappearance of 1 was accompanied by the appearance of a new single peak, which was identified as 1-deazaguanine by co-injection with authentic sample. Authentic sample was prepared as follows. A solution of 8 (58 mg, 0.1 mmol) in 80% aqueous acetic acid (2.5 ml) was heated at 60°C for 2.5 h. The solvent was removed in vacuo and the residue was co-evaporated with EtOH. The resulting white precipitate was suspended in EtOH and collected to give 1-deazaguanine (12 mg, 80%) as a white powder. EI-LRMS m/z 150 (M+); EI-HRMS calculated for C6H6N4O (M+) 150.0542, found 150.0549. 1H NMR (270 MHz, DMSO-d6) δ: 11.75 and 11.24 (each br s, each 1 H, 3-NH and 7-OH), 7.70 (s, 1 H, 2-H), 5.57 (s, 1 H, 6-H), 5.34 (br s, 2 H, 5-NH2).
Synthesis of phosphoramidite unit
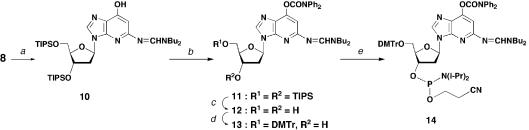
3-(2′-Deoxy-3′-5′-di-O-triisopropylsilyl-β-d-ribofuranosyl)- 5-(N,N-di-t-butylformamidino)amino-7-(N,N-diphenylcarbamoyl)oxyimidazo[4,5-b]pyridine (11). A mixture of 8 (1.00 g, 1.73 mmol) and N,N-di-t-butylformamide dimethyl acetal (0.83 ml, 3.46 mmol) in DMF (30 ml) was stirred at room temperature for 3 h. The mixture was diluted with AcOEt (300 ml), which was washed with H2O (100 ml × 4) and brine (100 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was dried under vacuum to give 10 as a crude product [FAB-LRMS m/z 718 (MH+)]. To a solution of crude 10 in pyridine (30 ml) was added diisopropylethylamine (0.45 ml, 2.60 mmol) and diphenylcarbamoyl chloride (1.20 g, 5.20 mmol) at room temperature. The mixture was stirred at the same temperature for 2 h. EtOH (2.0 ml) was added to the mixture and the solvent was removed in vacuo. The residue was dissolved in AcOEt (150 ml), which was washed with H2O (50 ml × 2), brine (50 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography (neutral silica gel, 3.5 × 16 cm, 20% AcOEt in hexane) to give 11 (1.67 g, 97%, 2 steps) as a pale yellow oil. FAB-LRMS m/z 913 (MH+); FAB-HRMS calculated for C51H81N6O5Si2 (MH+) 913.5807, found 913.5817. 1H NMR (270 MHz, DMSO-d6) δ: 8.39, 8.33 and 6.64 (each s, each 1 H, 2-H, 6-H and N=CH-N), 7.50∼7.25 (m, 10 H, Ph2), 6.45 (dd, 1 H, 1′-H, J = 5.9, 8.2 Hz), 4.70 (ddd, 1 H, 3′-H, J = 1.8, 2.0, 5.4 Hz), 3.96 (ddd, 1 H, 4′-H, J = 1.8, 4.9, 6.0 Hz), 3.83 (dd, 1 H, 5′-Ha, J = 6.0, 10.9 Hz), 3.75 (dd, 1 H, 5′-Hb, J = 4.9, 10.9 Hz), 3.44 (t, 2 H, CH2α, J = 7.3 Hz), 3.30 (t, 2 H, CH2α, J = 7.3 Hz), 3.00 (ddd, 1 H, 2′-Ha, J = 5.4, 8.2, 13.5 Hz), 2.35 (ddd, 1 H, 2′-Hb, J = 2.0, 5.9, 13.5 Hz), 1.59∼1.51 (m, 4 H, (CH2)2), 1.35∼1.22 (m, 4 H, (CH2)2), 1.07∼0.98 (m, 48 H, Me2 and i-Pr6); 13C NMR (67.8 MHz, DMSO-d6) δ: 159.53 (C), 154.32 (CH), 151.03 (C), 149.47 (C), 146.83 (C), 141.80 (C), 140.63 (CH), 128.88 (CH), 126.75 (CH), 123.73 (C), 105.83 (CH), 87.36 (CH), 82.96 (CH), 72.83 (CH), 63.39 (CH2), 50.44 (CH2), 44.27 (CH2), 38.67 (CH2), 30.58 (CH2), 28.80 (CH2), 19.45 (CH2), 19.07 (CH2), 17.70 (CH3), 17.68 (CH3), 17.62 (CH3), 17.59 (CH3), 13.68 (C), 13.39 (C), 11.47 (CH), 11.20 (CH).
3-(2′-Deoxy-β-d-ribofuranosyl)-5-(N,N-di-t-butylformamidi no)amino-7-(N,N-diphenylcarbamoyl)oxyimidazo[4,5-b]pyri dine (12). To a solution of 11 (1.45 g, 1.59 mmol) in THF (50 ml) was added TBAF (1.0 M in THF, 4.77 ml, 4.77 mmol) at 0°C. After the mixture was stirred at room temperature for 1 h, acetic acid (0.27 ml, 4.77 mmol) was added and the solvent was removed in vacuo. The residue was purified by silica gel column chromatography (neutral silica gel, 3.5 × 10 cm, 6% EtOH in CHCl3) to give 12 (830 mg, 87%) as a pale yellow foam: FAB-LRMS m/z 601 (MH+); FAB-HRMS calculated for C33H41N6O5 (MH+) 601.3138, found 601.3112. 1H NMR (270 MHz, DMSO-d6) δ: 8.42, 8.39 and 6.63 (each s, each 1 H, 2-H, 6-H and N=CH-N), 7.50∼7.27 (m, 10 H, Ph2), 6.42 (dd, 1 H, 1′-H, J = 5.9, 6.6 Hz), 5.32 (br s, 1 H, 3′-OH), 4.97 (br s, 1 H, 5′-OH), 4.43 (br ddd, 1 H, 3′-H, J = 1.9, 3.3, 5.9 Hz), 3.86 (ddd, 1 H, 4′-H, J = 1.9, 4.6, 5.0 Hz), 3.60 (br dd, 1 H, 5′-Ha, J = 4.6, 11.5 Hz), 3.52 (br dd, 1 H, 5′-Hb, J = 5.0, 11.5 Hz), 3.44 (t, 2 H, CH2α, J = 7.3 Hz), 3.35 (t, 2 H, CH2α, J = 7.2 Hz), 2.78 (ddd, 1 H, 2′-Ha, J = 5.9, 7.2, 13.2 Hz), 2.28 (ddd, 1 H, 2′-Hb, J = 3.3, 6.3, 13.2 Hz), 1.63∼1.50 (m, 4 H, (CH2)2), 1.38∼1.22 (m, 4 H, (CH2)2), 0.92 (t, 3 H, Me, J = 7.3 Hz), 0.91 (t, 3 H, Me, J = 7.3 Hz). 13C NMR (67.8 MHz, DMSO-d6) δ: 159.58 (C), 154.61 (CH), 151.06 (C), 149.44 (C), 146.80 (C), 141.78 (C), 140.84 (CH), 128.89 (CH), 126.72 (CH), 126.49 (CH), 123.66 (C), 105.72 (CH), 87.48 (CH), 83.04 (CH), 70.74 (CH), 61.72 (CH2), 50.42 (CH2), 44.11 (CH2), 39.26 (CH2), 30.57 (CH2), 28.57 (CH2), 19.56 (CH2), 19.07 (CH2), 13.67 (CH3), 13.50 (CH3).
3-[2′-Deoxy-5′-O-(4,4′-dimethoxytrityl)-β-d-ribofuranosyl]- 5-(N,N-di-t-butylformamidino)amino-7-(N,N-diphenylcarb amoyl)oxyimidazo[4,5-b]pyridine (13). A mixture of 12 (490 mg, 0.82 mmol) and dimethoxytrityl chloride (560 mg, 1.64 mmol) in pyridine (15 ml) was stirred at room temperature for 1 h. After EtOH (3.0 ml) was added to the mixture, the solvent was removed in vacuo. The residue was dissolved in AcOEt (120 ml), which was washed with aqueous NaHCO3 (saturated, 40 ml), H2O (40 ml × 2), brine (30 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography (neutral silica gel, 3.0 × 12 cm, 70% AcOEt in hexane) to give 13 (606 mg, 82%) as a pale yellow foam. FAB-LRMS m/z 903 (MH+); FAB-HRMS calculated for C54H59N6O7 (MH+) 903.4445, found 903.4445. 1H NMR (270 MHz, DMSO-d6) δ: 8.36, 8.29 and 6.64 (each s, each 1 H, 2-H, 6-H and N=CH-N), 7.50∼7.40 (m, 8 H, Ph2), 7.33∼7.26 (m, 4 H, DMTr and Ph2), 7.22∼7.14 (m, 7 H, DMTr), 6.80∼6.72 (m, 4 H, DMTr), 6.45 (t, 1 H, 1′-H, J = 6.6 Hz), 5.35 (d, 1 H, 3′-OH, J = 4.6 Hz), 4.43 (ddt, 1 H, 3′-H, J = 4.3 4.6, 6.6 Hz), 3.95 (ddd, 1 H, 4′-H, J = 3.3, 4.3, 6.6 Hz), 3.67 (s, 3 H, OMe), 3.65 (s, 3 H, OMe), 3.43 (t, 2 H, CH2α, J = 7.3 Hz), 3.25 (t, 2 H, CH2α, J = 7.2 Hz), 3.22 (dd, 1 H, 5′-Ha, J = 6.6, 9.9 Hz), 3.11 (dd, 1 H, 5′-Hb, 3.3, 9.9 Hz), 2.85 (dt, 1 H, 2′-Ha, J = 6.6, 13.2 Hz), 2.35 (ddd, 1 H, 2′-Hb, J = 4.6, 6.6, 13.2 Hz), 1.62∼1.43 (m, 4 H, (CH2)2), 1.37∼1.17 (m, 4 H, (CH2)2), 0.92 (t, 3 H, Me, J = 7.4 Hz), 0.85 (t, 3 H, Me, J = 7.2 Hz). 13C NMR (67.8 MHz, DMSO-d6) δ: 159.52 (C), 157.59 (C), 157.54 (C), 154.42 (CH), 151.05 (C), 149.42 (C), 146.77 (C), 144.54 (C), 141.79 (C), 140.69 (CH), 135.20 (C), 135.14 (C), 129.31 (CH), 129.21 (CH), 128.88 (CH), 127.38 (CH), 127.27 (CH), 126.72 (CH), 126.48 (CH), 126.23 (CH), 123.68 (C), 112.76 (CH), 105.90 (CH), 85.37 (CH), 85.12 (C), 82.53 (CH), 70.54 (CH), 64.13 (CH2), 54.73 (CH3), 54.68 (CH3), 50.43 (CH2), 44.13 (CH2), 38.79 (CH2), 30.51 (CH2), 28.57 (CH2), 19.57 (CH2), 19.05 (CH2), 13.67 (CH3), 13.46 (CH3).
3-[2′-Deoxy-5′-O-(4,4′-dimethoxytrityl)-β-d-ribofuranosyl- 3′-O-(2-cyanoethyl-N,N-diisopropylphosphoramidite)]-5- (N,N-di-t-butylformamidino)amino-7-(N,N-diphenylcarb amoyl)oxyimidazo[4,5-b]pyridine (14). To a mixture of 13 (440 mg, 0.49 mmol) and N,N-diisopropylethylamine (260 µl, 1.50 mmol) in CH2Cl2 (15 ml) was added 2-cyanoethyl N,N-diisopropylchlorophosphoramidite (223 µl, 1.00 mmol) at room temperature. After the mixture was stirred at the same temperature for 20 min, the mixture was diluted with CHCl3 (60 ml). The mixture was washed with aqueous NaHCO3 (saturated, 25 ml), H2O (25 ml), brine (25 ml), then dried (Na2SO4) and concentrated in vacuo. The residue was purified by silica gel column chromatography (neutral silica gel, 2.5 × 15 cm, 50% AcOEt in hexane) to give 14 (487 mg, 90%) as a white foam. FAB-LRMS m/z 1103 (MH+); FAB-HRMS calculated for C63H76N8O8P (MH+) 1103.5524, found 1103.5550. 1H NMR (270 MHz, DMSO-d6) δ: 8.36, 8.33, 8.31, 8.29, 6.65 and 6.65 (each s, each 0.5 H, 2-H, 6-H and N=CH-N), 7.50∼7.39 (m, 8 H, Ph2), 7.32∼7.26 (m, 4 H, DMTr and Ph2), 7.23∼7.15 (m, 7 H, DMTr), 6.80∼6.70 (m, 4 H, DMTr), 6.45 and 6.44 (each t, each 0.5 H, 1′-H), 4.75 (m, 1 H, 3′-H), 4.08 (m, 1 H, 4′-H), 3.68, 3.67, 3.66 and 3.65 (each s, each 1.5 H, OMe2), 3.73∼3.20 (m, 10 H, 5′-Ha,b and (CH2)4), 3.03 (m, 1 H, 2′-Ha), 2.74, 2.63 (each t, each 1 H, (CH)2, J = 5.9 Hz), 2.49 (m, 1 H, 2′-Hb), 1.58∼1.45 (m, 4 H, (CH2)2), 1.34∼1.18 (m, 4 H, (CH2)2), 1.20∼1.08 (m, 9 H, Me3), 1.01∼0.99 (m, 3 H, Me), 0.91 (t, 3 H, Me, J = 7.3 Hz), 0.86 and 0.85 (each t, each 1.5 H, Me, J = 7.3 Hz). 31P NMR (109 MHz, DMSO-d6) δ: 149.01, 148.83.
Oligodeoxynucleotide synthesis
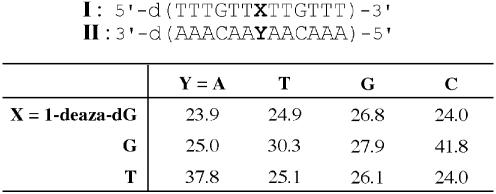
Sequences of ODNs were as follows: ODN I, 5′-d(TTT GTT XTT GTT T)-3′, X = 1-deaza-dG, G or T; ODN II, 5′-d(AAA CAA YAA CAA A)-3′, Y = A, T, G or C; ODN III, 5′-d(CGT AXC CAC CAG CTT ATA TTC CGT C)-3′, X = 1-deaza-dG, A, T, G or C; ODN IV, 5′-fluorecein-d(GAC GGA ATA TAA GCT GGT GG)-3′.
Oligodeoxynucleotides containing 1-deaza-dG (ODNs I and III, X = 1-deaza-dG) were synthesized on an Expedite 8900 Nucleic Acid Synthesis System (PerSeptive Biosystems) using standard phosphoramidite chemistry. The longer coupling time (300 s) for the 1-deaza-dG phosphoramidite (14) was used to ensure the high yield coupling. The 5′-terminal DMTr group of synthesized ODNs was removed prior to its cleavage from the CPG support. Each ODN was then cleaved and deprotected by treating the CPG-bound ODNs with concentrated NH4OH (28%) containing 10% (w/v) AcONH4 at 55°C for 16–20 h. The ODNs were desalted by RP silica gel column (YMC dispo SPE) and then purified by RP-HPLC (Shiseido Capcell Pak C18, 10 × 250 mm, 0.1 M TEAB buffer and CH3CN solvent system, pH 8.0). The yields of the ODNs were 98.1 nmol (10.9 OD260 nm units) for I and 88.2 nmol (20.2 OD260 nm units) for III, respectively, starting from the 1 µmol scale synthesis. MALDI-TOF/MS analysis of ODN I and III: for I, calculated mass 3969.6 (monoanion), observed mass 3969.1; for III, calculated mass 3776.4 (dianion), observed mass 3776.5. The molar extinction coefficients of ODNs containing 1-deaza-dG (1) were approximated by calculating the extinction coefficients of ODNs where the analog was replaced by thymidine, since the molar extinction coefficients of these two nucleosides were similar: 1 ε260 = 8400 (pH 7.0); thymidine ε260 = 8700 (pH 7.0). Unmodified ODNs and 5′-fluorescein-labeled primer were purchased from Sigma Genosys Japan (Japan).
Complete enzymatic digestion of synthesized ODNs
Each synthesized ODN (0.2 OD) was incubated with snake venom phosphodiesterase (0.2 U) and alkaline phosphatase (10 U) in a reaction buffer containing 50 mM Tris–HCl, 1 mM MgCl2 (pH 8.0, 100 µl) at 37°C for 6 h. Then the reaction tube was heated at 95°C for 5 min. Cold EtOH (300 µl) was added and the mixture was kept at –20°C for 1 h. After the tube was centrifuged at 12 000 r.p.m. at 0°C for 20 min, supernatant was collected and concentrated. The nucleoside components were analyzed by RP-HPLC (Tosoh TSK-Gel ODS-100S, 4.6 × 250 mm, 0.1 M TEAA buffer and CH3CN solvent system, pH 7.0). 1-Deaza-dG (1) was eluted just prior to 2′-deoxyguanosine. The nucleoside components calculated from peak area and molar extinction coefficients were as follows: for ODN I, T:G:1 = 9.8:2.0:1.0 (10:2:1); for ODN III, A:T:G:C:1 = 4.8:7.1:3.1:9.0:1.0 (5:7:3:9:1). The theoretical values are indicated in parentheses.
Thermal denaturation study
A pair of complementary ODNs, 5′-d(TTTGTTXTTGTTT)-3′ (ODN I, 3.0 µM) and 3′-d(AAACAAYAACAAA)-5′ (ODN II, 3.0 µM), where X = 1-deaza-dG, G or T and Y = A, T, G or C, were heated at 70°C for 3 min in a buffer solution containing 100 mM NaCl and 10 mM sodium cacodylate (pH 7.0), then cooled gradually to 10°C. Each duplex was heated from 10 to 60°C at a rate of 0.5°C/min and the thermally induced transition of the duplex at 260 nm was recorded on a Shimadzu UV-2500PC equipped with a Shimadzu TMSPC-8 temperature control system.
Synthesis of 1-deaza-2′-deoxyguanosine-5′-triphosphate (16)
Compound 12 (60 mg, 0.10 mmol) and 1,8-bis(dimethylamino)naphthalene (21 mg, 0.10 mmol) were dissolved in trimethylphosphate (0.5 ml) under an inert atmosphere. The mixture was cooled to 0°C, phosphorus oxychloride (14 µl, 0.15 mmol) was added dropwise and the mixture was stirred at 0°C for 5 min. Tributylamine (200 µl, 0.84 mmol) and a 0.5 M solution of tris(tetrabutylammonium)hydrogen pyrophosphate in DMF (2 ml) were mixed by vortex and then added to the above reaction mixture. After stirring for another 5 min at 0°C, 0.1 M triethylammonium bicarbonate (TEAB) solution (pH 8.0, 10 ml) was poured into the reaction mixture. The mixture was diluted with methanol (15 ml). To this mixture was added concentrated NH4OH (28%) containing 10% (w/v) AcONH4 (35 ml), and the whole was heated at 60°C for 3 h in a barotolerant tube. The solvent was removed and the residue was diluted with 20% aqueous CH3CN solution (120 ml). The mixture was purified by anion exchange chromatography (DEAE-Toyopearl 650S, 2.5 × 25 cm, buffer A = 20% CH3CN and 0.2 M TEAB; buffer B = 20% CH3CN and 0.7 M TEAB; gradient of 20–100% B in A over 3 h). The fractions (eluted at 0.4 M TEAB) were collected and concentrated in vacuo, then lyophilized to give 16 (25 µmol, 25% as determined from UV absorbance; ε260 = 8400) as a yellow foam. FAB-LRMS m/z 507 (MH+), 608 (M+Et3NH)+, 709 [M–H++2(Et3NH)]+; FAB-HRMS calculated for C11H17N4O13P3 (MH+) 507.0083, found 507.0079. 1H NMR (270 MHz, D2O) δ: 8.25 (s, 1 H, 2-H), 6.29 (dd, 1 H, 1′-H, J = 6.3, 7.2 Hz), 5.83 (s, 1 H, 6-H), 4.70 (m, 1 H, 3′-H), 4.17 (m, 1 H, 4′-H), 4.13∼4.09 (m, 2 H, 5′-Ha and b), 2.60 (ddd, 1 H, 2′-Ha, J = 6.3, 7.2, 14.0 Hz), 2.43 (ddd, 1 H, 2′-Hb, J = 3.3, 6.3, 14.0 Hz). 31P NMR (109 MHz, D2O and 0.1 M TEAB buffer, pH 8.0) δ: –7.23 (d, J = 19 Hz), –10.70 (d, J = 19 Hz), –22.33 (t, J = 19 Hz).
Single nucleotide insertion reactions by Klenow fragment (exo–)
5′-Fluorescein-labeled primer (ODN IV) and template (ODN III) were mixed in a 1:2 ratio and were incubated at 37°C for 1 h prior to the polymerase reactions. Annealed primer (IV, 0.4 µM) and template (III, 0.8 µM) mixture, dNTP (20 µM) and Kenow fragment (exo–) (0.5 U/5 µl) in a reaction buffer (25 µl) containing 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2 and 1 mM DTT was incubated at 37°C. At indicated time points, 5 µl aliquots of the reaction mixture were sampled and added into the loading solution (5 µl) containing 10 M urea, 5 mM EDTA, 0.05% (w/v) bromophenol blue and 0.05% (w/v) xylene cyanol. The samples were analyzed by electrophoresis on a 20% denaturing polyacrylamide gel, which was visualized and analyzed with a Molecular Imager FX (Bio-Rad).
Steady-state kinetics
Steady-state kinetics for single nucleotide insertions were carried out under the conditions described above. The duplex concentration was 0.4 µM, and 0.5 U/5 µl of Klenow fragment (exo–) was used. The amount of dNTP (2 µM–2 mM) and reaction time (60 s–10 min) were adjusted for each combination to give extents of reaction of 20% or less. The extents were analyzed by electrophoresis on a 20% denaturing polyacrylamide gel, which was quantitated by a Molecular Imager FX (Bio-Rad).
Single nucleotide insertion reactions
Therminator™ DNA polymerase. Annealed primer (IV, 0.4 µM) and template (III, 0.8 µM) mixture, dNTP (10 µM) and Therminator DNA polymerase (0.05 U) in a reaction buffer (5 µl) containing 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4 and 0.1% Triton X-100 were incubated at 74°C for 10 min. To the reaction mixture was added loading solution (5 µl) and the samples were analyzed by electrophoresis on a 20% denaturing polyacrylamide gel.
AMV reverse transcriptase. Annealed primer (IV, 0.4 µM) and template (III, 0.8 µM) mixture, dNTP (200 µM) and AMV reverse transcriptase (1.0 U) in a reaction buffer (5 µl) containing 50 mM Tris–HCl (pH 8.3), 100 mM KCl, 10 mM MgCl2 and 4 mM DTT was incubated at 37°C for 30 min. To the reaction mixture was added loading solution (5 µl) and the samples were analyzed by electrophoresis on a 20% denaturing polyacrylamide gel.
M-MLV reverse transcriptase. Annealed primer (IV, 0.4 µM) and template (III, 0.8 µM) mixture, dNTP (200 µM) and M-MLV reverse transcriptase (1.0 U) in a reaction buffer (5 µl) containing 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2 and 10 mM DTT was incubated at 37°C for 30 min. To the reaction mixture was added loading solution (5 µl) and the samples were analyzed by electrophoresis on a 20% denaturing polyacrylamide gel.
Extension reactions beyond 1-deaza-2′-deoxyguanosine residue in the template
Extension reactions were carried out under the conditions described above for each polymerase, except that dNTP mixture (each dNTP at 200 µM) was used.
Extension reactions subsequent to 1-deaza-2′-deoxyguanosine in the primer
Extension reactions were carried out under the conditions described above for each polymerase, except that the reactions were carried out in the presence of 1-deaza-dGTP (20 µM for Therminator DNA polymerase and 200 µM for AMV reverse transcriptase) and dNTPs (2 µM for Therminator DNA polymerase and 20 µM for AMV reverse transcriptase).
RESULTS AND DISCUSSION
Synthesis of 1-deaza-2′-deoxyguanosine
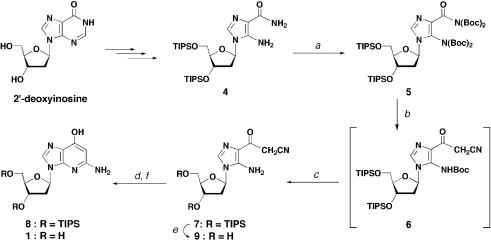
Synthesis of the target compound, 1-deaza-dG (1), was based on the synthetic strategy of 1-deazaguanosine with some modifications (24), as shown in Scheme 1. The starting synthon was an AICA 2′-deoxyriboside derivative 4 in which the hydroxyl groups on the deoxyribose moiety were protected with triisopropylsilyl groups. Compound 4 was synthesized from 2′-deoxyinosine in three steps by a previously reported method (25,26). t-Butyloxycarbonyl (Boc) groups were introduced to both the carboxamide group and amino group on the imidazole ring of compound 4 to give tetra-Boc derivative 5. Nucleophilic substitution of the Boc-protected carboxamide moiety with acetonitrile anion gave cyanoacetyl derivative 6, with one Boc group on the 5-amino group eliminated concomitantly. Cyclization of the amino moiety with a cyano group would give the desired 1-deazaguanine aromatic system. However, removal of the remaining Boc group on the amino moiety was found to be troublesome. Subjecting compound 6 to acidic conditions, such as TFA in CH2Cl2 or aqueous acetic acid, resulted in cleavage of the glycosidic bond. Lewis acids, which are known to remove various carbamate groups, were also tested, but only gave a mixture of decomposed compounds. After several trials, it was found that heating compound 6 on dried silica gel resulted in efficient removal of the Boc group on the 5-amino group. Practically, a slurry mixture of compound 6 and silica gel in CHCl3 was dried using a rotary evaporator and was then heated at 80°C under reduced pressure to give 4-cyanoacetyl-5-aminoimidazole nucleoside (7). This reaction seemed to proceed with the help of silica gel since no conversion was observed even at an elevated temperature in the organic solvent. Intramolecular cyclization of compound 7 advanced smoothly under basic conditions to give the ribose-protected target 1-deaza-dG (8). Deprotected target compound 1 was also synthesized for physical and structural analyses. The triisopropyl silyl groups of compound 7 were first removed to give compound 9, and this was followed by a ring closure reaction, giving 1-deaza-dG (1).
Scheme 1. Conditions: (a) di-t-butyl dicarbonate, Et3N, DMAP, 1,2-dichloroethane, 75°C, 1 h, 78%; (b) CH3CN, n-BuLi, THF, –78°C, 1.5 h; (c) silica gel, 80°C, 2.5 h, 41% (two steps); (d) aqueous Na2CO3–EtOH, 90°C, 3.5 h, 86% (from 7 to 8); (e) TBAF, THF, 0°C–room temperature, 1 h, 75%; (f) aqueous Na2CO3–EtOH, 90°C, 3 h, 58% (from 9 to 1).
Conformational analysis of 1-deaza-2′-deoxyguanosine
Since the base moiety of 1-deaza-dG is comprised of 4-hydroxypyridine and an imidazole ring, a tautomeric equilibrium between hydroxypyridine and pyridone has been predicted (Fig. 1, 1 and 2). In a pyridone tautomer (2), the keto oxygen atom is a potent hydrogen bond acceptor, while the nitrogen atom at position-4 (N3 as purine numbering) is a potent hydrogen bond donor. On the other hand, this nitrogen atom acts as a proton acceptor in the hydroxypyridine tautomer (1). In this tautomer, the hydroxyl group can act as both a proton acceptor and a proton donor depending on the orientation of hydroxyl group. Clarification of the most stable structure and the difference between energies of the tautomeric structures is important in determining the structure–activity correlations (27).
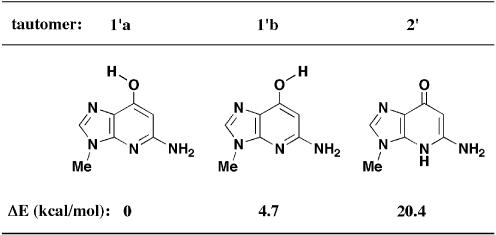
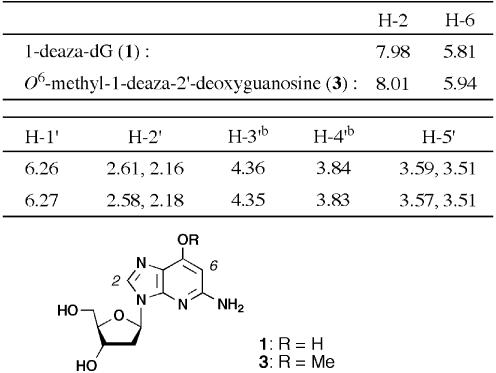
To search for a stable tautomeric structure, the energy differences of these 1-deaza-dG tautomers were estimated by ab initio calculation (27,28). In the calculation, deoxyribose moieties were substituted by a methyl group. Calculated differences in energies of three conformations (1′a, 1′b and 2′) are shown in Figure 2. The most stable conformation was tautomer 1′a, which was 4.7 kcal/mol more stable than 1′b and 20.4 kcal/mol more stable than tautomer 2′. A clear advantage was found for hydroxypyridine tautomers (1′a and 1′b), and internal hydrogen bond formation with a hydroxyl group and N1-nitrogen atom (N7 as purine numbering) had a further stabilizing effect on the syn-rotamer 1′a. From these results, it was anticipated that 1-deaza-dG was as the syn-hydroxypyridine tautomer (1′a). Furthermore, base pair formation between 1′a and cytidine with a geometry similar to that of a canonical G:C base pair but with two hydrogen bonds was expected. The 1H NMR data for 1-deaza-dG (1), compared with that of O6-methyl-1-deaza-dG (3), also indicated the stable structure to be the hydroxypyridine tautomers (1′a and 1′b) (Table 1). Proton signals on the base moiety of O6-methyl-1-deaza-dG (3), in which the tautomeric structure was fixed in the hydroxyl form, were observed at 8.01 (H-2) and 5.94 (H-6) p.p.m. (21). Similarly, the same proton signals of 1-deaza-dG (1) were observed at 7.98 (H-2) and 5.81 (H-6) p.p.m., indicating that the two analogs have the same hydroxypyridine structures.
Figure 2.
Energy differences (ΔE kcal/mol) between three conformations, calculated at the [MP2/6-31G(d,p)//MP2/6–31G+(2d′,p′)] level.
Table 1. 1H NMR spectra in DMSO-d6a.
aChemical shifts are indicated in p.p.m.
bIn the original paper (21) assignment for 3′ and 4′ protons of compound 3 were inverted, which we have corrected based on our assignment.
Stability of the N-glycosidic bond
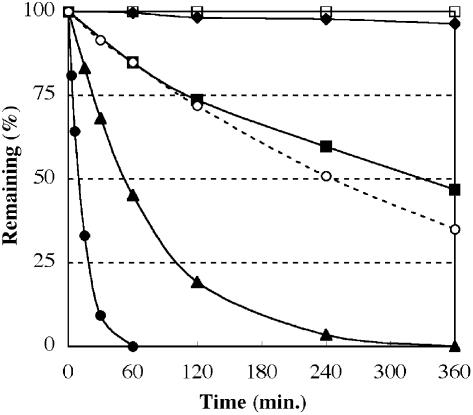
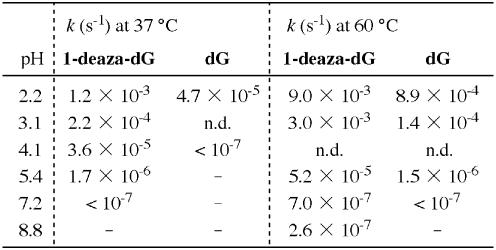
During the synthesis and structural analysis of 1-deaza-dG (1) we found that this nucleoside was rather sensitive to acid and that the nucleoside underwent acid-catalyzed hydrolysis of the N-glycosidic bond. To elucidate the susceptibility of 1-deaza-dG to deglycosidation, nucleosides (1 and deoxyguanosine) in various pH solutions were incubated at 37 and 60°C and hydrolysis of the N-glycosidic bond was monitored by HPLC (Fig. 3, at 37°C). All of the deglycosidation reactions showed first order kinetics. The rate constants (k) for hydrolysis of the N-glycosidic bond were calculated by a first order reaction equation (29,30), and the results are shown in Table 2. At pH 2.2 and at 37°C, 1-deaza-dG (1) underwent rapid cleavage of the N-glycosidic bond to release 1-deazaguanine and was completely decomposed within 60 min. Its half-life at pH 2.2 and 37°C was calculated to be 9.6 min with a rate constant (k) of 1.2 × 10–3 s–1. This rate is 25 times faster than the hydrolysis of natural deoxyguanosine, for which the rate constant (k) is 4.7 × 10–5 s–1 (t1/2 = 250 min) under the same conditions. No hydrolysis of the N-glycosidic bond was observed for deoxyguanosine at pH 4.1, whereas 1-deaza-dG was still hydrolyzed with a rate constant of 3.6 × 10–5 s–1 (t1/2 = 320 min). An increase in temperature increased the rate constant of N-glycosidic bond cleavage. 1-Deaza-dG completely decomposed within 10 min at 60°C and pH 2.2, with k = 9.0 × 10–3 s–1 (t1/2 = 77 s). Only a little decomposition of 1-deaza-dG was observed under physiological and alkaline conditions, even at elevated temperature (60°C). The high susceptibility to acidic conditions was thought to be mainly due to the change in electron density of the aromatic ring system. Since, in 1-deaza-dG (1) the electronegative nitrogen atom was replaced with an electropositive carbon atom, which resulted in increased basicity of the remaining nitrogen atoms at the N1 and N4 positions (N7 and N3 positions as purine numbering).
Figure 3.
Hydrolysis of the N-glycosidic bond at 37°C. 1-Deaza-dG (1) at pH 2.2 (closed circles), pH 3.1 (closed triangles), pH 4.1 (closed squares) and pH 5.4 (closed diamonds). Deoxyguanosine at pH 2.2 (open circles) and 4.1 (open squares). Conditions used are described in Materials and Methods. Deglycosylation was monitored by RP-HPLC and the remaining percentages were calculated by comparing the ratio of compounds with thymine.
Table 2. Rate constants (k s–1) for the cleavage of an N-glycosidic bonda.
aConditions were 0.1 mM 1-deaza-dG (1) or deoxyguanosine and 0.04 mM thymine in 0.1 M sodium phosphate (pH 2.2–7.2), incubated at 37 and 60°C.
With regard to the susceptibility of 1-deaza-dG (1) to acidic conditions, we were concerned that the 1-deaza-dG monomer could not tolerate the conditions used in DNA synthesis. However, by introducing protecting groups on the amino and hydroxyl groups of the base moiety (see Scheme 2), 1-deaza-dG could be made tolerant to the acidic conditions used in DNA synthesis, such as 3% trichloroacetic acid in dichloromethane or 80% aqueous acetic acid solution. Less than 3% of the compound was decomposed after 24 h incubation of compound 12 in 80% acetic acid solution at 37°C.
Scheme 2. Conditions: (a) N,N-di-t-butylformamide dimethyl acetal, DMF, room temperature, 3 h; (b) diphenylcarbamoyl chloride, i-Pr2NEt, room temperature, 2 h, 97% (two steps); (c) TBAF, THF, 0°C–room temperature, 1 h, 87%; (d) DMTrCl, pyridine, room temperature, 1 h, 82%; (e) 2-cyanoethyl N,N-diisopropylchlorophosphoramidite, i-Pr2NEt, CH2Cl2, room temperature, 20 min, 90%.
Synthesis of oligodeoxynucleotides
The 1-deaza-dG derivative (8) was then converted into a phosphoramidite unit for incorporation into synthetic ODNs (Scheme 2). Both the N5 amino group and O7 hydroxyl group of 1-deaza-dG were protected with suitable protecting groups. At first, compound 8 was treated with di-t-butylformamide dimethylacetal (31) to introduce an amidine group on the amino group to give compound 10. A dimethylformamidine group was also tested, but di-t-butylamidine gave a better yield and was stable under the following reaction conditions. The hydroxyl group was then protected with a diphenyl carbamoyl group by treating compound 10 with diphenyl carbamoyl chloride and diisopropylethylamine in pyridine to give compound 11 in 97% yield in two steps. After removing the silyl groups on the deoxyribose moiety, compound 12 was converted into a phosphoramidite unit (14) in two steps by a standard procedure.
ODNs containing 1-deaza-dG (ODNs I and III) were synthesized using a phosphoramidite unit (14) and standard deoxynucleoside phosphoramidite monomers. The average coupling efficiency for amidite monomer 14 was 95% as determined from the released trityl cation. The dimethoxytrityl group on the 5′-terminus of the completed ODNs was removed prior to its cleavage from the CPG support. Concentrated ammonia solution containing 10% (w/v) ammonium acetate (31) was used for the cleavage and deprotection steps. Each ODN was purified by RP-HPLC. The purities of ODNs were analyzed on both reverse phase and ion exchange columns and showed a single peak. The nucleoside components were checked by complete enzymatic digestion. Also, ODNs were analyzed by MALDI-TOF mass spectrometry and the observed molecular weights supported their structures. The molar extinction coefficients (ε260) of ODNs were calculated according to the nearest neighbor method (see Materials and Methods for details).
Thermal denaturation studies
Because of the lack of a single nitrogen atom, 1-deaza-dG (1) was expected to show distinct base pairing ability compared to that of its deoxyguanosine counterpart. Initial conformational analysis suggested the feasibility of a Watson–Crick type base pair with cytidine with two hydrogen bonds. On the other hand, a possible rotamer (1′b in Fig. 2) could show distinctive base pairing selectivity when incorporated into duplex DNA. In order to study hydrogen bonding selectivity and base pair stabilities of 1-deaza-dG (1) in the context of duplex DNA, thermal denaturing experiments were carried out.
ODNs containing 1-deaza-dG at the middle of a 13 nt sequence (I, Table 3) and the complementary sequence (II) were used in the study. In these sequences, 1-deaza-dG (1) was paired with each of four canonical nucleobases. Melting temperatures were obtained from the midpoints of the thermal denaturation curves, and the results are shown in Table 3. The Tm of a natural G:C base pair was 41.8°C and that of a T:A base pair was 37.8°C. The mismatched base pairs showed lower Tm values, in the range 24.0–30.3°C. For the duplex containing 1-deaza-dG (1) paired with natural bases, Tm values were in the range 23.9–26.8°C. Despite the base pair with two hydrogen bonds being expected, the 1-deaza-dG:C base pair exhibited a Tm of 24.0°C. This value is significantly lower not only than the Tm of the corresponding G:C base pair but also than that of the two hydrogen bond A:T base pair and is similar to the Tm values of mismatched base pairs. The most stable combination formed by 1-deaza-dG was the base pair with guanosine, showing a Tm of 26.8°C. This value was still lower than that of the matched base pairs. Hydrogen bonds are not the only factor responsible for stable duplex formation; other factors, such as base stacking abilities, also contribute to duplex stability. As judged from these results, however, it was presumed that 1-deaza-dG had little ability to form hydrogen bonded base pairs with natural nucleosides, including Watson–Crick type and wobble type base pairs, in the context of duplex DNA.
Table 3. Melting temperatures (°C) of ODN I:IIa.
aConditions were 100 mM NaCl, 10 mM sodium cacodylate, pH 7.0, 3.0 µM of each strand.
Single nucleotide insertion studies by the Klenow fragment (exo–)
Since 1-deaza-dG (1) possesses characteristic hydrogen bonding patterns with a geometry similar to those of natural purine nucleosides, we were interested in determining whether the analog is accepted by nucleoside/nucleotide-utilizing enzymes. Accordingly, we focused on polymerase extension reactions.
Recent works have suggested that the fidelity of replication by DNA polymerase depends not only on the formation of a Watson–Crick base pair but also on the geometry of the base (32–35). A number of crystal structures of DNA polymerase bound to DNA have shown that most of the DNA–protein interactions occur between polymerase and the minor groove of DNA (32–34). The X-ray crystallography of polymerase β revealed the existence of hydrogen bonds between the N3 position of guanine in the template and residue Arg283 and between the O2 position of the incoming cytosine triphosphate and residue Asn279 (32). A minor groove interaction with an Arg residue has been found in many DNA–polymerase crystallographic studies and is thus thought to be a common mechanism for accurate polymerization (32–34). A study using the 3-deaza analog of deoxyguanosine has shown the importance of these interactions (14,15). The calculated stable conformation of 1-deaza-dG (1) has a minor groove geometry similar to that of deoxyguanosine, in which a hydrogen bond acceptor nitrogen atom is retained at the N4 position (N3 with purine numbering). On the other hand, the Watson–Crick hydrogen bond pattern of 1-deaza-dG is different from that of its natural guanosine counterpart. Because of this interesting feature of 1-deaza-dG, we used the compound to DNA polymerase reactions to evaluate the effects of modification on extension reactions. At first, a template containing 1-deaza-dG (1) was used in replication reactions. The triphosphate analog was also used in single nucleotide insertion reactions to determine the effect of modification of incoming nucleotides on polymerase reactions.
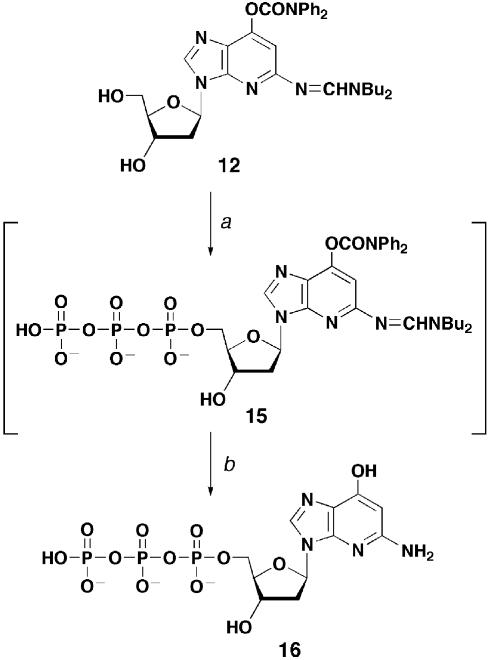
1-Deaza-dGTP (16) was synthesized according to the modified Yoshikawa method (Scheme 3) (36,37). Base-protected derivative 12 was used to prevent acid-catalyzed cleavage of the N-glycosidic bond. Compound 12 was treated with phosphorus oxychloride in trimethyl phosphate in the presence of a proton sponge (37) at 0°C. The resultant reaction mixture was allowed to react with pyrophosphate to give triphosphate derivative 15. After the protecting groups had been removed by treatment with concentrated ammonia containing 10% ammonium acetate at 55°C (31), the target 1-deaza-dGTP (16) was purified by ion exchange column chromatography. The use of the same method starting from unprotected 1-deaza-dG (1) failed to give the triphosphate analog 16, probably due to the sensitivity to acidic conditions. The 31P NMR spectrum of 1-deaza-dGTP (16) showed the expected signals.
Scheme 3. Conditions: (a) 1) POCl3, 1,8-bis(dimethylamino)naphthalene, (CH3O)3PO, 0°C, 5 min; 2) tri-n-butylamine, tris(tetrabutylammonium)hydrogen pyrophosphate, DMF, 0°C, 5 min; (b) NH4OH, aqueous AcONH4, MeOH, 60°C, 3 h, 25% (two steps).
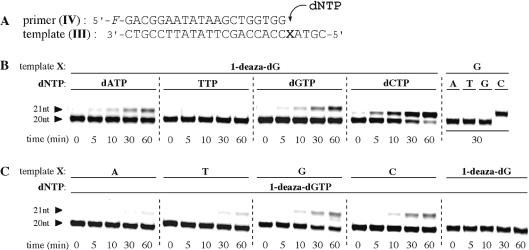
Initially, single nucleoside insertions by the Klenow fragment (exo–) [KF(exo–)] were carried out using a template containing 1-deaza-dG. KF(exo–) is a fragment of Escherichia coli DNA polymerase I enzyme that lacks both 3′- and 5′-exonuclease activities. The template and primer set are shown in Figure 4A. A synthesized 25 nt DNA template (III) in which 1-deaza-dG was introduced at position 21 from the 3′-terminus was annealed with a 5′-fluorescein-labeled 20 nt primer (IV) in the presence of KF(exo–) and dNTP. Single nucleotide incorporations were analyzed by polyacrylamide gel electrophoresis.
Figure 4.
Single nucleotide insertion studies with Klenow fragment (exo–). (A) Sequences of a 25 nt template (III) and 20 nt primer (IV). (B) PAGE of dNTP incorporation opposite the 1-deaza-dG residue. (C) PAGE of 1-deaza-dGTP incorporation opposite a natural nucleoside residue. Conditions used are described in Materials and Methods. Both X in the template sequence and added dNTP are indicated at the top of the gel.
Figure 4B shows the time courses of extension reactions. Among the four natural dNTPs, dCTP was found to be inserted most rapidly opposite 1-deaza-dG by KF(exo–). More than 70% and >90% of the primer (IV) were elongated by inserting dCTP after 30 min incubation and after 60 min incubation, respectively. dATP and dGTP were also found to be inserted opposite the 1-deaza-dG site, although with lower efficiencies. Under the same conditions, dCTP was selectively and efficiently inserted opposite the natural G residue. No mismatched base pair incorporations were observed. These data indicate that KF(exo–) incorporation of dCTP opposite the 1-deaza-dG residue was less efficient than was dCTP incorporation opposite the natural G residue. The steady-state efficiencies (Vmax/Km, % min–1 M–1) for the insertion of dNTPs opposite the 1-deaza-dG residue were calculated to be 1.2 × 104 (dATP), 1.4 × 103 (TTP), 1.7 × 104 (dGTP) and 6.6 × 104 (dCTP). dCTP incorporation opposite the 1-deaza-dG residue was somewhat more selective than the other nucleotide triphosphates by a factor of ∼4–50. However, the efficiency of dCTP incorporation opposite the 1-deaza-dG residue was ∼10–2–10–3 times lower than incorporation of a matched dNTP opposite an unmodified template (15,38,39).
Enzymatic incorporation of 1-deaza-dGTP (16) across a natural template was next performed using the same primer IV and template III containing a natural residue (X = A, T, G or C). Figure 4C shows the time courses of KF(exo–) elongations analyzed by PAGE. The results indicated that there were only subtle selectivities for the insertion of 1-deaza-dGTP across natural bases. In the template containing G and C residues, ∼25% of the primers were elongated by incorporation of 1-deaza-dGTP after 60 min incubation. At the same time, 8 and 14% insertions were observed for A- and T-containing templates, respectively. The calculated steady-state efficiencies (Vmax/Km, % min–1 M–1) for the insertion of 1-deaza-dGTP across natural nucleoside residues were 9.2 × 103 (for A), 1.6 × 104 (for T), 2.1 × 104 (for G) and 2.9 × 104 (for C). 1-Deaza-dGTP incorporation across the C residue was only slightly preferred by a factor of ∼1.4–3. Incorporation of 1-deaza-dGTP (16) across a template containing a 1-deaza-dG residue was not observed.
Single nucleotide insertion studies using other polymerases
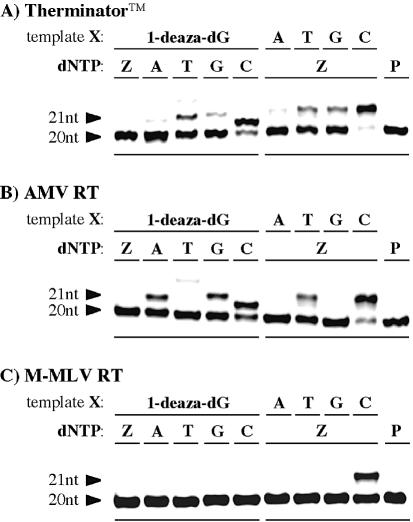
Other commercially available polymerases were also tested for their ability to recognize 1-deaza-dG in elongation reactions. Therminator™ DNA polymerase, which has been reported to have an enhanced ability to incorporate a modified substrate (40), as well as two reverse transcriptases, Avian myeloblastosis virus reverse transcriptase (AMV-RT) and Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (39), were used in the single nucleotide insertion studies. These enzymes lack exonuclease and endonuclease activities, as does KF(exo–), and therefore have no proofreading activities. The same template (III) and primer (IV) set was used, and the resultant PAGE images are shown in Figure 5.
Figure 5.
PAGE of single nucleotide insertion studies (A) with Therminator™ DNA polymerase (reaction mixtures being incubated at 74°C for 10 min), (B) with AMV reverse transcriptase (reaction mixtures being incubated at 37°C for 30 min) and (C) with M-MLV reverse transcriptase (reactions mixture being incubated at 37°C for 30 min). Conditions used are described in Materials and Methods. Both X in the template sequence and added dNTP are indicated at the top of each gel. Z indicates 1-deaza-dGTP; P indicates reaction without dNTP.
Figure 5A shows the single nucleotide insertions by Therminator DNA polymerase. With this thermal polymerase, dCTP was incorporated most rapidly across the template containing 1-deaza-dG residue, followed by a small incorporation of TTP. Under the same conditions, 1-deaza-dGTP (16) was incorporated across the C residue with good selectivity and almost complete elongation was observed.
AMV reverse transcriptase incorporations of natural nucleotide triphosphate across the 1-deaza-dG residue were similar to those in the case of reactions with KF(exo–) (Fig. 5B). dCTP was most efficiently incorporated, and some incorporation of dATP and dGTP was also observed. However, with AMV reverse transcriptase quite good selectively was found for incorporation of 1-deaza-dGTP (16) opposite the C residue. This was not the case with KF(exo–).
Most of the single nucleotide insertion reactions with M-MLV reverse transcriptase were less effective than those with other polymerases (Fig. 5C). Only a single combination, 1-deaza-dGTP (16) incorporation opposite the C residue, gave an extended 21 nt product. No other combinations incorporated dNTPs across the 1-deaza-dG residue or incorporated 1-deaza-dGTP across natural nucleoside residues. Incorporation of 1-deaza-dGTP (16) across the template containing the 1-deaza-dG residue was not observed with any of the enzymes tested.
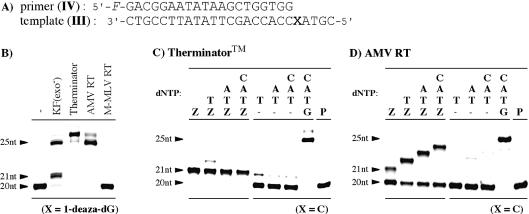
Extension reactions beyond a 1-deaza-2′-deoxyguanosine residue in the template
Polymerase extension reactions beyond the analog were studied using a template containing 1-deaza-dG in the presence of all four natural triphosphates (Fig. 6B). In the case of KF(exo–), a fully elongated product along with an aborted product at the position of the analog were observed. This result indicates that KF(exo–) accepted 1-deaza-dG, presented in the template, to some extent and continued to elongate the primer DNA. Therminator polymerase and AMV reverse transcriptase both accepted the analog and showed fully elongated reactions. Also, as expected from the results of single nucleotide insertion studies, M-MLV reverse transcriptase showed no elongation at all. 1-Deaza-dG in the template sequence was not accepted by this enzyme.
Figure 6.
(A) Sequences of the 25 nt template (III) and 20 nt primer (IV). (B) PAGE of extension reactions beyond the 1-deaza-dG residue in the template. (C) PAGE of extension reactions subsequent to 1-deaza-dG in the primer with Therminator™ DNA polymerase and (D) with AMV reverse transcriptase. Conditions used are described in Materials and Methods. Added dNTPs are indicated at the top of each gel. Z indicates 1-deaza-dGTP; P indicates reaction without dNTP.
Extension reactions subsequent to 1-deaza-2′-deoxyguanosine in the primer
Finally, extension reactions subsequent to 1-deaza-dG, incorporated opposite the C residue, were studied using Therminator™ DNA polymerase and AMV reverse transcriptase. These two enzymes incorporated 1-deaza-dGTP (16) opposite the C residue with good selectivity. A template (III) containing cytidine at position 21 (X = C) was used. In the case of Therminator DNA polymerase, the reactions were stopped after incorporation of 1-deaza-dGTP at position 21 and no additional elongation was observed (Fig. 6C). Clearly, 1-deaza-dG incorporated at the 3′-terminus of the primer inhibited incorporation of subsequent triphosphates and terminated the extension reactions. In contrast to Therminator DNA polymerase, AMV reverse transcriptase exhibited continuous elongations beyond the 1-deaza-dG residue upon addition of appropriate dNTPs (Fig. 6D). These further elongations occurred due to the incorporation of TTP subsequent to the incorporation of 1-deaza-dGTP, since the enzyme could not elongate the 20 nt primer in the absence of 1-deaza-dGTP. AMV reverse transcriptase accepted the 1-deaza-dG residue incorporated opposite a C residue and continued to elongate the primer, although the incorporation of 1-deaza-dGTP opposite a C residue was slower compared with incorporation of natural dGTP opposite a C residue. A second 1-deaza-dGTP incorporation at position 25, opposite a C residue, was not observed, presumably due to the low efficiency of 1-deaza-dGTP incorporation by this enzyme.
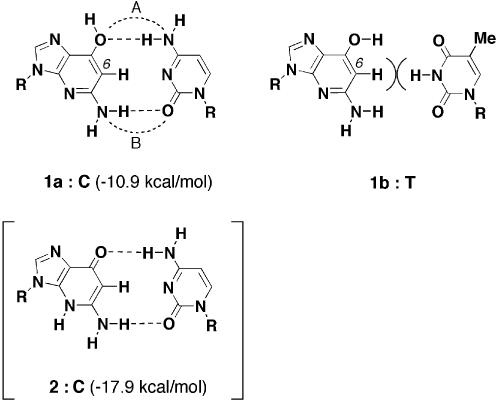
Putative base pair between 1-deaza-2′-deoxyguanosine and dC
In all cases, 1-deaza-dGTP (16) incorporation opposite the C residue or dCTP incorporation opposite the 1-deaza-dG residue had some advantages compared with other combinations. Although no stable base pair formed with 1-deaza-dG and natural nucleosides was estimated from the thermal denaturing experiments, it was likely that these selective incorporations occurred through the formation of a base pair between 1-deaza-dG and dC. In Figure 7, predicted base pairs having Watson–Crick geometries are depicted with anticipated hydrogen bonds. A syn tautomer of 1-deaza-dG (1a) was able to form a Watson–Crick type base pair with cytidine, which has a geometry similar to that of a canonical G:C base pair (1a:C). In this base pair, two hydrogen bonds, one between the 4-amino of dC and 7-hydroxyl oxygen of 1-deaza-dG and one between the 5-amino of 1-deaza-dG and 2-keto of dC, were formed. The N3-nitrogen atom of cytidine was unable to form a hydrogen bond because the imino proton of the complement guanosine was replaced with an aromatic proton in 1-deaza-dG. It might be because of formation of this two hydrogen bond base pair (1a:C) that the polymerase advantagously incorporated dCTP opposite a 1-deaza-dG residue and the reverse combination. Searls and McLaughlin reported a similar base pair formed between 3-deaza-2′-deoxycytidine and deoxyguanosine in their polymerase studies (41). They observed selective incorporation of dGTP opposite a 3-deaza-2′-deoxycytidine residue in the template sequence by Klenow fragment and pointed out the formation of a Watson–Crick type base pair with two hydrogen bonds. Another two hydrogen bond base pair formed between the anti-hydroxyl tautomer of 1-deaza-dG and thymidine has been suggested (1b:T). However, in this base pair, steric repulsion between the aromatic proton at the 6 position of 1-deaza-dG and the imino proton at N3 of thymidine forcedly prevents base pair formation.
Figure 7.
Predicted base pairs with 1-deaza-dG (R = deoxyriboside). In the hydrogen bond energy calculations; R was replaced with a methyl group. The calculated energies are shown in parentheses.
Although the pyridone tautomer (2) was calculated to be less stable than the hydroxypyridine tautomers (1a and 1b), a two hydrogen bond base pair formed between tautomer 2 and cytidine was also indicated (2:C). To estimate the stabilities of the two predicted 1-deaza-dG:C base pairs (1a:C and 2:C), the hydrogen bond energies of these base pairs were calculated theoretically (Fig. 7) (28). The 1a:C base pair showed a hydrogen bond energy of –10.9 kcal/mol, while the 2:C pair showed a hydrogen bond energy of –17.9 kcal/mol. In base pair formation, an advantage was observed for the pyridone tautomer (2). However, because of the large energy difference between the two tautomeric conformations (Δ20.4 kcal/mol), the total energy advantage was for the 1a:C base pair by about –13 kcal/mol. By the same calculations, the hydrogen bond energies for canonical G:C and A:U base pairs were estimated to be –26.1 and –13.2 kcal/mol, respectively (28). From these data, the hydrogen bond energy of the 1a:C base pair, –10.9 kcal/mol, is thought to result in the formation of a base pair that is less stable than the canonical base pairs. The calculation also showed the hydrogen bond lengths of the base pair. The distances between the nitrogen atom and oxygen atom in the 1a:C base pair were 3.609 (bond A) and 3.259 Å (bond B), respectively. These values were ∼0.4–0.6 Å longer than the average distance in canonical base pairs (28). This is probably due to the small repulsive effect between the N3-nitrogen atom of cytidine and the aromatic proton at the 6 position of 1-deaza-dG. Because of these longer hydrogen bonds, the 1-deaza-dG:C base pair was assumed to be a sterically expanded base pair. This steric change affected not only the minor groove geometry of the base pair but also polymerase recognition through minor groove interactions. Because of this global structural change in the 1-deaza-dG:C base pair, it is thought that the polymerase incorporates this base pair with only low efficiency.
CONCLUSION
1-Deaza-dG (1), which lacks a nitrogen atom on position 1 of the guanosine ring, was successfully synthesized from the AICA-deoxyriboside. The tautomeric structure was suggested by theoretical calculation to be a hydroxyl form, in which the guanosine-like hydrogen bonding capability is retained. The compound was converted into a phosphoramidite derivative (14) for incorporation into synthetic DNAs. Melting temperatures (Tm) determined in thermal denaturing experiments showed no preference in base pairing selectivity and all base pairs formed with 1-deaza-dG had Tm values similar to those of normal mismatched base pairs. Recognition of 1-deaza-dG by polymerases was then investigated by single nucleotide insertion reactions. Klenow fragment (exo–) polymerase as well as Therminator™ DNA polymerase, AMV reverse transcriptase and M-MLV reverse transcriptase were used in the study. In these experiments, preferential incorporation of dCTP across the 1-deaza-dG residue and its reverse pair, 1-deaza-dGTP (16) incorporation across the C residue, was observed. However, both insertions were inefficient compared to the normal elongation reactions. Despite the Watson–Crick type base pair formed between 1-deaza-dG and cytidine suggested by theoretical calculation, the expanded global structure, due to the longer hydrogen bonds, seemed to inhibit effective elongation reactions.
Replacement of a single atom of an active molecule is a useful method for investigating the structure–function relationships of the molecule. In 1-deaza-dG (1) a single nitrogen atom of deoxyguanosine is replaced with a carbon atom. This small substitution can cause global changes in the heterocyclic ring, such as base stacking strength, hydrophobicity and potential hydrogen bonding abilities of the other nitrogen/oxygen atoms. The experiments described in this manuscript show that the behavior of the analog was different from the behavior of natural deoxyguanosine. Despite these altered properties, this analog could be a powerful tool for direct investigation of the motion of the nitrogen atom at the 1 position of guanosine in order to understand, for example, DNA–protein interactions (14,15,42) and structure–function relationships of guanine-related biological reactions (5,22,43).
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Dr Y. Komatsu (National Institute of Advanced Industrial Sciences and Technology, Hokkaido) for helpful discussions. The authors would also like to thank Drs A. Matsuda and N. Minakawa (Graduate School of Pharmaceutical Sciences, Hokkaido University) for valuable discussions and supports. We also thank Ms S. Oka (Center for Instrumental Analysis, Hokkaido University) for measurement of mass spectra.
REFERENCES
- 1.Sanghvi Y.S. (1993) Heterocyclic base modifications in nucleic acids and their applications in antisense oligonucleotides. In Crooke,S.T. and Lebleu,B. (eds), Antisense Research and Applications. CRC Press, Boca Raton, FL, pp. 273–288. [Google Scholar]
- 2.Luyten I. and Herdewijn,P. (1998) Hybridization properties of base-modified oligonucleotides within the double and triple helix motif. Eur. J. Med. Chem., 33, 515–576. [Google Scholar]
- 3.Mizuno Y., Itoh,T. and Nomura,A. (1982) Azanucleosides and deazanucleosides of biological interest. Heterocycles, 17, 615–644. [Google Scholar]
- 4.Kool E.T., Morales,J.C. and Guckian,K.M. (2000) Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed., 39, 990–1009. [DOI] [PubMed] [Google Scholar]
- 5.Seela F., Debelak,H., Usman,N., Burgin,A. and Beigelman,L. (1998) 1-Deazaadenosine: synthesis and activity of base-modified hammerhead ribozymes. Nucleic Acids Res., 26, 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piccirilli J.A., Krauch,T., Moroney,S.E. and Benner,S.A. (1990) Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature, 343, 33–37. [DOI] [PubMed] [Google Scholar]
- 7.Parkinson G.N., Lee,M.P.H. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 8.Bergholtz S., Andersen,T.Ø., Andersson,K.B., Borrebkæk,J., Lüscher,B. and Gabrielsen,O.S. (2001) The highly conserved DNA-binding domains of A-, B- and c-Myb differ with respect to DNA-binding, phosphorylation and redox properties. Nucleic Acids Res., 29, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanei-Ishii C., Sarai,A., Sawazaki,T., Nakagoshi,H., He,D.-N., Ogata,K., Nishimura,Y. and Ishii,S. (1990) The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem., 265, 19990–19995. [PubMed] [Google Scholar]
- 10.Mizusawa S., Nishimura,S. and Seela,F. (1986) Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res., 14, 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buhr C.A., Wagner,R.W., Grant,D. and Froehler,B.C. (1996) Oligodeoxynucleotides containing C-7 propyne analogs of 7-deaza-2′-deoxyguanosine and 7-deaza-2′-deoxyadenosine. Nucleic Acids Res., 24, 2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto A., Taiji,T., Tanaka,K. and Saito,I. (2000) Synthesis and duplex stability of oligonucleotides containing 7-vinyl-7-deazaguanine as a strong electron-donating nucleobase. Tetrahedron Lett., 41, 10035–10039. [Google Scholar]
- 13.Aubert Y., Perrouault,L., Hélène,C., Giovannangeli,C. and Asseline,U. (2001) Synthesis and properties of triple helix-forming oligodeoxyribonucleotides containing 7-chloro-7-deaza-2′-deoxyguanosine. Bioorg. Med. Chem., 9, 1617–1624. [DOI] [PubMed] [Google Scholar]
- 14.Washington M.T., Wolfle,E.T., Spratt,T.E., Prakash,L. and Prakash,S. (2003) Yeast DNA polymerase η makes functional contacts with the DNA minor groove only at the incoming nucleoside triphosphate. Proc. Natl Acad. Sci. USA, 100, 5113–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spratt T.E. (2001) Identification of hydrogen bonds between Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry, 40, 2647–2652. [DOI] [PubMed] [Google Scholar]
- 16.Spratt T.E. (1997) Klenow fragment–DNA interaction required for the incorporation of nucleotides opposite guanine and O6-methylguanine. Biochemistry, 36, 13292–13297. [DOI] [PubMed] [Google Scholar]
- 17.Versees W., Klaas,D., Von Holsbeke,E., Devroede,N. and Steyaer,J. (2002) Enzyme-substrate interactions in the purine-specific nucleoside hydrolase from Trypanosoma vivax. J. Biol. Chem., 277, 15938–15946. [DOI] [PubMed] [Google Scholar]
- 18.Minakawa N., Kojima,N. and Matsuda,A. (1999) Nucleosides and nucleotides 184. Synthesis and conformational investigation of anti-fixed 3-deaza-3-halopurine ribonucleosides. J. Org. Chem., 64, 7158–7172. [Google Scholar]
- 19.Cline B.L., Panzica,R.P. and Townsend,L.B. (1975) The synthesis of 1-deazaguanosine. J. Heterocyclic Chem., 12, 603–604. [Google Scholar]
- 20.Cline B.L., Panzica,R.P. and Townsend,L.B. (1978) Synthesis of 5-amino-3-(β-d-ribofuranosyl)imidazole[4,5-b]pyridin-7-one (1-deazaguanosine) and related nucleosides. J. Heterocyclic Chem., 15, 839–847. [Google Scholar]
- 21.Spratt T.E. and Campbell,C.R. (1994) Synthesis of oligodeoxynucleotides containing analogs of O6-methylguanine and reaction with O6-alkylguanine-DNA alkyltransferase. Biochemistry, 33, 11364–11371. [DOI] [PubMed] [Google Scholar]
- 22.Spratt T.E., Wu,J.D., Levy,D.E., Kanugula,S. and Pegg,A.E. (1999) Reaction and binding of oligodeoxynucleotides containing analogues of O6-methylguanine with wild-type and mutant human O6-alkylguanine-DNA alkyltransferase. Biochemistry, 38, 6801–6806. [DOI] [PubMed] [Google Scholar]
- 23.Spratt T.E. and Levy,D.E. (1997) Structure of the hydrogen bonding complex of O6-methylguanine with cytosine and thymine during DNA replication. Nucleic Acids Res., 25, 3354–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima N., Minakawa,N. and Matsuda,A. (2000) Nucleosides and nucleotides. Part 207: Studies in the chemical conversion of the 4-carboxamide group of 5-amino-1-β-d-ribofuranosylimidazole-4-carboxamide (AICA-riboside). Application for the synthesis of 1-deazaguanosine. Tetrahedron, 56, 7909–7914. [Google Scholar]
- 25.De Napoli L., Messere,A., Montesarchio,D., Piccialli,G. and Varra,M. (1997) 1-Substituted 2′-deoxyinosine analogues. J. Chem. Soc. Perkin Trans., 1, 14, 2079–2082. [Google Scholar]
- 26.Minakawa N., Kojima,N., Hikishima,S., Sasaki,T., Kiyosue,A., Atsumi,N., Ueno,Y. and Matsuda,A. (2003) New base pairing motifs. The synthesis and thermal stability of oligodeoxynucleotides containing imidazopyridopyrimidine nucleosides with the ability to form four hydrogen bonds. J. Am. Chem. Soc., 125, 9970–9982. [DOI] [PubMed] [Google Scholar]
- 27.Roberts C., Bandaru,R. and Switzer,C. (1997) Theoretical and experimental study of isoguanine and isocytosine: base pairing in an expanded genetic system. J. Am. Chem. Soc., 119, 4640–4649. [Google Scholar]
- 28.Kawahara S., Uchimaru,T., Taira,K. and Sekine,M. (2002) An ab initio study of the hydrogen bond energy of base pairs formed between substituted 9-methylguanine derivatives and 1-methylcytosine. J. Phys. Chem., 106A, 3207–3212. [Google Scholar]
- 29.Suzuki T., Matsumura,Y., Ide,H., Kanaori,K., Tajima,K. and Makino,K. (1997) Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligodeoxynucleotides. Biochemistry, 36, 8013–8019. [DOI] [PubMed] [Google Scholar]
- 30.Vongchampa V., Dong,M., Gingipalli,L. and Dedon,P. (2003) Stability of 2′-deoxyxanthosine in DNA. Nucleic Acids Res., 31, 1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Froehler B.C. and Matteucci,M.D. (1983) Dialkylformamidines: depurination resistant N6-protecting group for deoxyadenosine. Nucleic Acids Res., 11, 8031–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- 33.Eom S.H., Wang,J. and Steitz,T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature, 382, 278–281. [DOI] [PubMed] [Google Scholar]
- 34.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structure of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 35.Kool E.T. (1998) Replication of non-hydrogen bonded bases by DNA polymerase: a mechanism for steric matching. Biopolymers, 45, 3–17. [DOI] [PubMed] [Google Scholar]
- 36.Yoshikawa M., Kato,T. and Takenishi,T. (1967) A novel method for phosphorylation of nucleosides to 5′-nucleotides. Tetrahedron Lett., 8, 5065–5068. [DOI] [PubMed] [Google Scholar]
- 37.Kovács T. and Ötvös,L. (1988) Simple synthesis of 5-vinyl- and 5-ethynyl-2′-deoxyuridine-5′-triphosphate. Tetrahedron Lett., 29, 4525–4528. [Google Scholar]
- 38.Kamiya H., Maki,H. and Kasai,H. (2000) Two DNA polymerases of Escherichia coli display distinct misinsertion specificities for 2-hydroxy-dATP during DNA synthesis. Biochemistry, 39, 9508–9513. [DOI] [PubMed] [Google Scholar]
- 39.Morales J.C. and Kool,E.T. (2000) Varied molecular interactions at the active sites of several DNA polymerases: nonpolar nucleoside isosters as probes. J. Am. Chem. Soc., 122, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner A.F. and Jack,W.E. (2002) Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res., 30, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Searls T. and McLaughlin,L.W. (1999) Synthesis of the analogue nucleoside 3-deaza-2′-deoxycytidine and its template activity with DNA polymerase. Tetrahedron, 55, 11985–11996. [Google Scholar]
- 42.Xiang S., Short,S.A., Wolfenden,R. and Carter,C.W.,Jr (1996) Cytidine deaminase complexed to 3-deazacytidine: a “valence buffer” in zinc enzyme catalysis. Biochemistry, 35, 1335–1341. [DOI] [PubMed] [Google Scholar]
- 43.Cohen S.M., Mikata,Y., He,Q. and Lippard,S.J. (2000) HMG-domain protein recognition of cisplatin 1,2-intrastrand d(GpG) cross-links in purine-rich sequence contexts. Biochemistry, 39, 11771–11776. [DOI] [PubMed] [Google Scholar]