Abstract
Although gene expression profiling by microarray analysis is a useful tool for assessing global levels of transcriptional activity, variability associated with the data sets usually requires that observed differences be validated by some other method, such as real-time quantitative polymerase chain reaction (real-time PCR). However, non-specific amplification of non-target genes is frequently observed in the latter, confounding the analysis in ∼40% of real-time PCR attempts when primer-specific labels are not used. Here we present an experimentally validated algorithm for the identification of transcript-specific PCR primers on a genomic scale that can be applied to real-time PCR with sequence-independent detection methods. An online database, PrimerBank, has been created for researchers to retrieve primer information for their genes of interest. PrimerBank currently contains 147 404 primers encompassing most known human and mouse genes. The primer design algorithm has been tested by conventional and real-time PCR for a subset of 112 primer pairs with a success rate of 98.2%.
INTRODUCTION
Quantitative transcript abundance analysis by real-time PCR has become widely applied in recent years (1,2). Typical applications of this method monitor the amplicon production after each thermocycle by the appearance of a fluorescent signal that is dependent on dye binding to the DNA product of the reaction or generated from a fluorophore engineered in the primer sequences. Because of its simplicity and sensitivity, real-time PCR is now widely used for precise evaluation of gene expression.
However the sensitivity of the method is also a liability. PCR can generate unintended products, especially when an RNA sample containing thousands of genes is used as the template. The unexpected amplicons are usually the result of primer mispriming to non-target sites. The presence of extraneous amplicons complicates data analysis and can lead to incorrect inferences about message abundance. Extraneous amplicons are a particularly serious problem for the cheapest and most widely practiced form of real-time PCR, which relies on fluorescent detection of amplified DNA by sequence non-selective dyes, such as SYBR Green I.
Most existing primer design programs are predicated on a single template of limited genetic complexity (3). Primer failures resulting from flawed design tool predictions are sufficiently widespread that several online databases have been established as repositories for empirically validated primer sequences submitted by researchers (4). Unfortunately these databases contain primers for only a few hundred genes at present. Thus, in most cases, an investigator will need to identify primers for genes of interest by trial and error.
With the advent of microarray technology (5), gene-specific oligo probe design has become the subject of multiple studies (6–10). A few genome-wide primer design programs have been developed for the production of amplicons with minimal potential for cross-hybridization, which are then spotted as probes in cDNA microarrays (11–13). Most of these programs have used BLAST (14) to identify gene-specific regions from which the PCR primers are designed. This strategy is appropriate for cDNA microarrays, since amplicon probe cross-hybridization is the main concern for microarray specificity. However, these programs are not designed for real-time PCR studies.
Here we present an algorithm and its implementation to identify specific primers for real-time PCR. An online primer database has been created to allow any investigator to freely retrieve primer information for genes of interest. The algorithm has been tested by conventional and real-time PCR experiments for a subset of 112 primer pairs and has been shown to be highly reliable.
MATERIALS AND METHODS
Mouse total RNA
C57BL6 mouse liver total RNA was either purchased from Stratagene or prepared with Trizol (protocol available at http://pga.mgh.harvard.edu/Parabiosys/resources/microarrays.php). Total RNAs from other mouse tissues were from the Mouse Total RNA Master Panel (Clontech). DNA contamination of RNA samples was not assessed prior to use, but has been evaluated for the above protocol in prior microarray analyses.
PCR primer preparation
PCR primer sequences were retrieved from the online PrimerBank database. These primers were synthesized at the Molecular Biology Core Facility, Massachusetts General Hospital. Both UV absorbance and capillary electrophoresis were used to assess the quality of primer synthesis.
RT–PCR
Reverse transcription (RT) was carried out with the SuperSript First-Strand Synthesis System using the manufacturer’s protocol (Invitrogen). A 20 µl RT reaction included 5 µg of total RNA, 150 ng of random hexamers, 2 µl of 10× RT buffer, 4 µl of 25 mM MgCl2, 2 µl of 0.1 M dithiothreitol, 1 µl of RNaseOUT, 1 µl of 50 U/µl SuperScript II and DEPC-treated water. The RNA template was then removed by adding 1 µl of RNase H and incubating at 37°C for 20 min.
Conventional and real-time PCRs were carried out on an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Conventional PCRs were sometimes also carried out on a PTC-200 cycler (MJ Research). In both cases, hot-start PCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems). In brief, the PCR mixtures were pre-heated at 50°C for 2 min and then at 95°C for 10 min to activate the AmpliTaq Gold DNA polymerase, followed by 40 cycles of amplification (95°C for 15 s; 60°C for 30 s; 68°C for 40 s). A final extension step was performed at 60°C for 10 min. The PCR products were checked on 3.5% NuSieve 3:1 Agarose gel (Cambrex Bio Science Rockland). Real-time PCR results were also analyzed using the ABI Prism 7000 SDS software (Applied Biosystems).
PrimerBank website
The PrimerBank database is freely accessible at http://pga.mgh.harvard.edu/primerbank/index.html. Detailed information for the primers in Supplementary Material Table S1 can be obtained from this website.
RESULTS
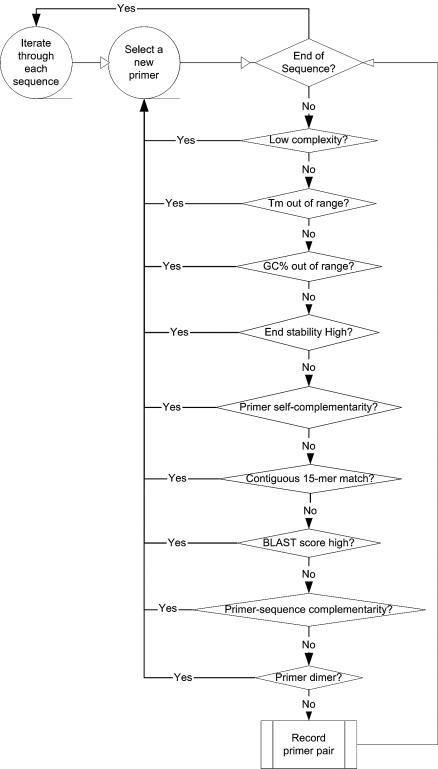
Figure 1 shows a simplified flow chart describing the primer selection algorithm. The algorithm was implemented in Perl as a program called uPrimer. uPrimer requires ∼2 days on a 1.5 GHz Linux system to design primers for human or mouse genes.
Figure 1.
A simplified flow chart describing the primer design algorithm.
Gene sequences
The principle source of gene sequence information for this project is the NCBI protein database GenPept (http://www.ncbi.nlm.nih.gov/Entrez/). The corresponding DNA coding sequences were retrieved and redundant sequences were clustered using a program called DeRedund (8). Low complexity regions may contribute to primer cross-reactivity (15) and thus are excluded by the DUST program (16). To further enhance sequence complexity, a primer sequence is rejected if it contains six or more contiguous identical residues and no primer candidate is considered from sequence regions with ambiguous residues.
Two kinds of priming reactions are commonly used in RT reactions: random priming and oligo(dT) priming. Oligo(dT) priming usually results in cDNA libraries enriched for mRNA and tends to over-represent the 3′ ends of transcripts. As the detection of different splice isoforms is one major goal in gene expression analysis, we expect to perform random priming in RT reactions. In general, maximum sensitivity in random priming lies close to the 5′ end of a coding sequence (8). Therefore coding regions were scanned from the 5′ end to the 3′ end until three qualified primer pairs had been picked.
Primer uniformity
To facilitate the conduction of multiple PCRs, all human and mouse primers are designed to have similar properties. All primers are 19–23 nt long, with a preferred length of 21 residues. This is long enough to permit generation of gene-specific primers, while reducing the potential for cross-reactivity and allowing cost-effective generation of large primer sets. The GC contents are also similar (35–65%) to ensure uniform priming. Because 3′ end residues contribute most to non-specific primer extension, especially if the binding of these residues is relatively stable (17), the algorithm evaluates the ΔG value for the last five residues at the 3′ end and a threshold value of –9 kcal/mol is adopted for primer rejection.
The melting temperature (Tm) determines the optimal annealing temperature. In recent years, significant progress has been made to accurately estimate the Tm of oligonucleotides (18–20). The nearest neighbor method is to date the most accurate approach and is implemented by the following formula (18):
Tm = ΔH°/[ΔS°–R ln(CT/4)]
where R is the gas constant (1.987 cal/Kmol), CT is the primer concentration, ΔH° is the enthalpy change and ΔS° is the entropy change. ΔH° and ΔS° are calculated by using the published thermodynamic parameters (18). The entropy change is dependent on salt concentration, so an entropy correction is performed:
ΔS° = ΔS° (1 M Na+) + 0.368 × (N – 1) × ln[Naeq+],
where N is the length of the primer and [Naeq+] is the Na+ equivalent concentration from all salts in a reaction. The default parameters for Tm calculation are 250 nM primer and 0.15 M Naeq+ (21). Variations in primer and salt concentrations in other typical PCR conditions affect the Tm values only slightly. All primer Tm values are in the narrow range 60–63°C.
Since PCR efficiency is decreased for very long amplicons, only short amplicons of 150–350 bp are considered during primer selection. Occasionally, if this requirement cannot be satisfied, a wider range of 100–800 bp is used. In general the larger amplicons are less attractive but they are included in the database because under some circumstances primer efficiency may not be the foremost consideration for the end user.
Primer cross-reactivity
Mismatches are known to significantly reduce priming stability (22,23) and at times even a single mismatch can destabilize a significant length of DNA duplex (24,25). Therefore we expect contiguous base pairing to be one of the most important factors in duplex stability. Our principal filter for cross-reactivity is the rejection of primers containing contiguous residues that are also found in other sequences. An analysis of the distribution of lengths of contiguous residues shared by two or more sequences in the design space of mammalian coding regions showed that a filter cut-off rejecting perfect 15mer matches was the most stringent feasible filter (8). Non-unique 15mers can be efficiently identified by a software ‘hashing’ technique with 10mers as the basic hash keys (8). Every possible 15mer in a primer sequence is compared to both strands of all known sequences in the design space. The presence of a repetitive 15mer excludes a primer from further consideration. To further reduce cross-reactivity, BLAST searches for primer sequence similarity were carried out against all known sequences in the design space and qualified primers were required to have BLAST scores of less than 30 [these threshold values were recommended from previous studies (8)].
Random priming in RT reactions results in a significant contribution of template from non-coding RNAs. To compensate for the abundance of these templates, more stringent filters were applied to minimize primer residues also found in non-coding RNAs.
The primer 3′ end residues are essential for controlling non-specific amplicons because DNA polymerase extension can be greatly reduced by mismatches (26,27). Therefore a more stringent filter should apply to cross-hybridization at the 3′ ends. In our algorithm the cross-hybridizing Tm for the 3′ end perfectly matched residues does not exceed 46°C; the Tm does not exceed 42°C when compared to non-coding RNA sequences.
Sequence self-complementarity
Secondary structure in the primer or target can retard primer annealing, leading to reduced PCR efficiency. Although the prediction of primer secondary structure is still challenging at present, secondary structure is most likely to occur in regions of self-complementarity (28). To reduce self-complementarity, no contiguous 5mer match is allowed anywhere between a primer and its complementary sequence. To avoid picking primers from a sequence region with high likelihood of secondary structure, no contiguous 9mer match is allowed when a primer sequence is compared to the complementary strand of its cognate gene sequence. A BLAST similarity search for the primer sequence is also carried out on the complementary strand and the score is required to be less than 18.
The formation of products arising from primers serving as template (primer dimers) can deplete free primers and result in poor PCR yield. Primer dimers are a common cause of real-time PCR quantitation failures when DNA intercalating dyes (e.g. SYBR Green I) are used. To prevent primer homodimer formation, candidate primers are rejected if the four residues at the 3′ end of a primer could be found in its complementary sequence. Complementarity of the forward and reverse primers in a primer pair is examined in the same way to prevent detrimental heterodimer formation.
Distribution of the rejected primers
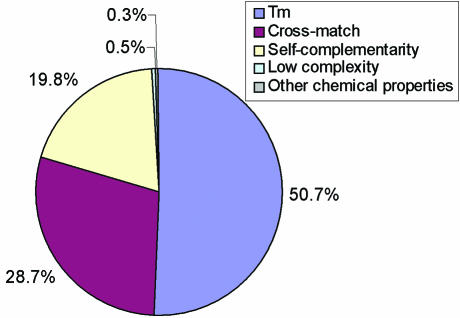
15 562 332 primers were evaluated before 37 277 primer pairs were picked to cover 15 697 mouse genes. The very high rejection rate, 99.5%, reflects filter stringency. The distribution of the rejected mouse primers is shown in Figure 2. Among the rejected primers, 50.7% had too high or too low Tm values, 28.7% cross-hybridized to non-target genes, 19.8% were rejected because of sequence self-complementarity, 0.5% were from low complexity regions and 0.3% were rejected because of other properties (GC content and end stability).
Figure 2.
Distribution of the rejected mouse primers. 15 562 332 primers were rejected during primer selection. They were rejected because they could not meet the primer selection criteria for melting temperature (Tm), cross-match to other sequences, sequence self-complementarity, sequence low complexity or other properties of the primers (GC content and end stability).
The online primer database
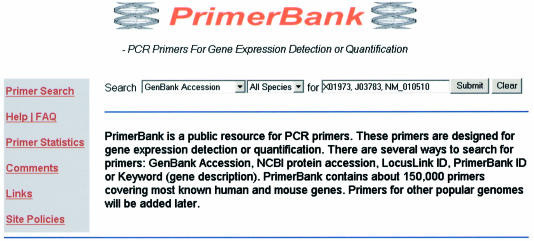
Successfully designed human and mouse primers were imported in a MySQL database installed on a Linux server. A web-based interface was established to allow users to query the primer database, PrimerBank. Figure 3 shows the search page of the website. 147 404 primers were picked and included in PrimerBank to cover 16 293 human and 15 697 mouse genes. There are several ways to search for primers: by GenBank accession no., NCBI protein accession no., LocusLink ID, PrimerBank ID or Keyword (gene description). Batch primer retrieval is also available by entering multiple IDs at the same time. Detailed instructions are included in the Help page of the website. Because of the sequence redundancy in public sequence databases, PrimerBank uses LocusLink index files (29,30), updated weekly from ftp://ftp.ncbi.nih.gov/, to map gene accessions to gene loci and associate the gene information with the primers.
Figure 3.
A screenshot of the web interface for PrimerBank. There are several ways to search for primers: GenBank accession no., NCBI protein accession no., LocusLink ID, PrimerBank ID or Keyword (gene description). PrimerBank currently contains 147 404 primers designed for human and mouse genes.
Experimental evaluation of the primers
To evaluate the quality of the primers identified by the algorithm, 112 primer pairs representing 108 genes were tested in conventional RT–PCR and real-time PCR experiments. The primer information was retrieved from PrimerBank and is summarized in Supplementary Material Table S1. The genes were chosen because they had been shown to be expressed in mouse liver by microarray experiments and were of interest to local investigators (unpublished data). Some genes were from closely related gene families. Among them, 16 genes were from the cytochrome P450 family and five genes were from the Dok family.
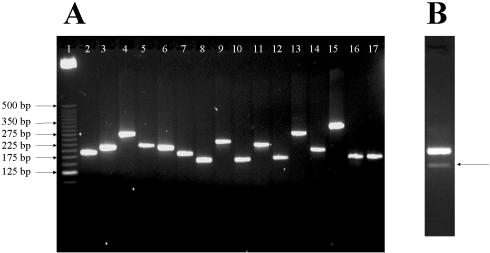
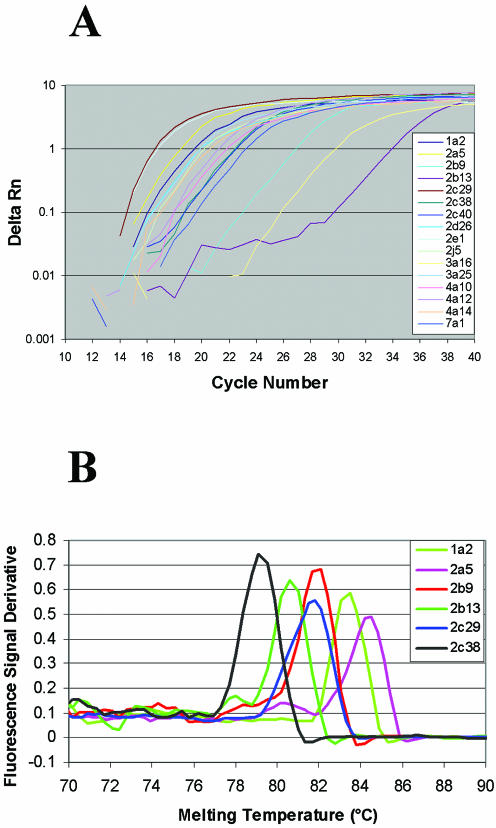
The results for the 16 cytochrome P450 genes are included here as examples and the relevant primer information is summarized in Table 1. The cytochrome P450 genes are closely related and the sequence similarity is ∼90% between some family members. Despite the high template homology, all 16 PCRs resulted in single specific amplicons, determined by gel electrophoresis (Fig. 4A). All 16 P450 genes were also efficiently amplified in real-time PCR and the amplification plots indicated no obvious correlation between amplicon length and PCR efficiency (Fig. 5A). The melting curve analysis indicated single amplicons for 15 of these P450 genes (six examples shown in Fig. 5B). PCR specificity was confirmed by sequencing the PCR products.
Table 1. Primer information for 16 cytochrome P450 genes.
| PrimerBank ID | Protein accession no. | Amplicon length | Forward primer | Reverse primer | Gene name |
|---|---|---|---|---|---|
| 6753566_1 | NP_034123 | 194 | ccaggtggtggaatcggtg | tcttaaacctcttgagggccg | P450, 1a2 |
| 6681103_1 | NP_031838 | 217 | atgctgacctcaggactcctc | ggtagatggtgaatacaggacca | P450, 2a5 |
| 6753578_1 | NP_034130 | 285 | gctcattctctggtcagatgttt | cgcttgtggtctcagttcca | P450, 2b9 |
| 6681105_1 | NP_031839 | 224 | cagatgaacagttcctgcgtt | gatgaagtctcgtggctcact | P450, 2b13 |
| 6681109_1 | NP_031841 | 218 | atctggtcgtgttcctagcg | agtaggctttgagcccaaatac | P450, 2c29 |
| 4249591_1 | AAD13720 | 193 | acaggcaaaccacatcgaaca | gctacggtgtctaccaaccac | P450, 2c38 |
| 6857779_1 | NP_034134 | 167 | gatccatttgtagtcttggtgct | aaattggtaaggcactgccca | P450, 2c40 |
| 13386414_1 | NP_083838 | 245 | ttggagatgacttatgggctgt | tccgttgaccacaaccacg | P450, 2d26 |
| 11276065_1 | NP_067257 | 168 | catcaccgttgccttgcttg | gccaacttggttaaagacttggg | P450, 2e1 |
| 6753586_1 | NP_034137 | 231 | tctgggaagcactccatctca | ccactggtgattggcccaa | P450, 2j5 |
| 6681117_1 | NP_031846 | 173 | atcctttgtccttgtcagtagca | cagataaataaagtccacgcggt | P450, 3a16 |
| 1914796_1 | CAA72720 | 297 | atatgggacctattctcatggct | tcctcagatatggtaatggcctt | P450, 3a25 |
| 3738263_1 | BAA33804 | 211 | ttccctgatggacgctcttta | ccttcagctcactcatagcaaa | P450, 4a10 |
| 21729747_1 | NP_031847 | 347 | atgagtgcctctgctctgag | ccattagcttttgggtctgatct | P450, 4a12 |
| 6681121_1 | NP_031848 | 183 | tttagccctacaaggtacttgga | gtccttcagatggtgcccc | P450, 4a14 |
| 6681125_1 | NP_031850 | 185 | agcatttctttgatctggggg | ccatgtttcctttgctttgctct | P450, 7a1 |
Figure 4.
Gel electrophoresis of PCR products. (A) PCR amplifications of 16 cytochrome P450 genes. Lane 1, 25 bp DNA ladder; lanes 2–17, 10 µl PCR products of P450 1a2, 2a5, 2b9, 2b13, 2c29, 2c38, 2c40, 2d26, 2e1, 2j5, 3a16, 3a25, 4a10, 4a12, 4a14 and 7a1. (B) PCR amplification using primer pair 7239366_1. The arrow indicates a non-specific amplicon.
Figure 5.
Real-time PCR of cytochrome P450 genes. (A) PCR amplification plots for 16 cytochrome P450 genes. (B) Melting curves of six genes from cytochrome P450 families 1 and 2 (plotted as the first derivative of the absorbance with respect to temperature).
An analysis of PCR efficiency was also conducted by measuring the slope of a standard curve created from serially diluted templates. Six primer pairs with a range in predicted amplicon length of 152–347 bp were analyzed and yielded an efficiency of 96 ± 4%.
Among the 112 primer pairs tested, 106 detected their target genes in liver total RNA. Literature searching indicated that five of the six undetected genes had been shown to be expressed in tissues other than liver (31–35). Thus total RNA from embryo, brain, kidney or testis was used to test primers designed for these genes (see Supplementary Material Table S1). In this case five primer pairs yielded single specific PCR products. Only one gene was not detected using the primer pairs we designed. Among the 106 genes detected in liver, all except one primer pair resulted in single specific amplicons on agarose gel (unpublished data). One primer pair yielded a minor band in addition to the desired major band (Fig. 4B). Sequencing indicated this is a novel splice isoform that was not identified in GenBank.
The 112 primer pairs were also tested in real-time PCR experiments. Melting curve analysis (plotted as the first derivative of the absorbance with respect to temperature) indicated the presence of single PCR products in 104 PCRs. Six reactions resulted in bimodal first derivative plots, although single bands were observed by agarose gel. Sequencing results confirmed that these PCR products were homogeneous and correct, indicating that the observed heterogeneity in melting temperature was due to internal sequence inhomogeneity (e.g. independently melting blocks of high and low GC content) rather than amplicon contamination. In summary, 110 out of 112 primer pairs led to single specific PCR products yielding a primer design success rate of 98.2%.
DISCUSSION
Primer specificity
Most approaches to primer design are based on the expectation of a single low complexity target sequence. With existing tools one can design a number of primer pairs and then individually check cross-matches of each primer with the entire genome by BLAST. However this is inefficient and inexhaustive and does not provide sensitivity to important design criteria. For example, cross-hybridization at the primer 3′ end is more likely to produce undesired amplicons and some templates, for example from non-coding RNAs, are more abundant than others and require more stringent filters to exclude them. In our experience, only about two-thirds of the primers designed in the conventional way are qualified for real-time PCR experiments (unpublished data).
The primer design algorithm described here is based on a successful approach to the prediction of oligonucleotides for interrogation of protein coding regions by microarrays (8), but differs from that design by the addition of filters thought to be relevant for PCR priming specificity. In the algorithm consideration is given to both contiguous residue matches and global sequence similarity. The highest filter stringency is applied to residues at the 3′ end and to adventitious matches with abundant non-coding RNAs. 74 544 mouse primers were identified after discarding 4 466 257 candidates for failure to meet specificity thresholds. The cross-match filters were the cause of 28.7% of primer rejections (Fig. 2). Among the 112 primer pairs we tested, only one pair yielded a minor non-specific amplicon. This represents a major advance in specificity over contemporary practice.
Successful primer design requires dependable genomic information and a comprehensive inventory of splice isoforms. Present estimates suggest there are many genes yet to be identified (36,37). It is also likely that among the known genes, there are still unidentified splice isoforms. The unrecognized genes or isoforms may contribute to primer cross-reactivity. For example, one primer pair, 7239366_1 in Supplementary Material Table S1, amplified both the target gene and a previously unidentified splice isoform (Fig. 4B). This design failure could have been avoided had the isoform been known. Since primer specificity could be improved with better genomic information, primers in PrimerBank will be regularly updated to reflect the latest progress from genomic research. As the PrimerBank version increases, superceded primers will not be deleted but will be retained and retrievable via their PrimerBank ID numbers. However only current version primers will be returned when other search terms are used.
Product specificity is very sensitive to the precise PCR conditions used for amplification. Since mispriming is most likely to occur at low temperatures, we have relied on hot-start PCR for gene quantitation studies, using chemically modified Taq polymerase (Applied Biosystems). Assembly of the reaction mixture can be carried out at room temperature because there is minimal enzyme activity before heat activation with this reagent. A high annealing temperature is also necessary to maintain PCR specificity. Previous studies indicated sufficient priming should occur at the primer Tm (21,38). Therefore, a 60°C annealing temperature was used in all of our experiments.
Primer secondary structures may reduce PCR efficiency. Our algorithm, like those in many existing primer design programs, screens for primer self-complementarity, which is the most important determinant for secondary structures. However, the global secondary structures of the gene sequences are largely ignored by other algorithms. If a primer is from a region of self-complementarity, the primer annealing process may be hindered, leading to a reduced PCR efficiency. Moreover, sequence self-complementarity can reveal multiple annealing sites for a primer and this introduces a serious problem for PCR specificity. To address this issue, our primers were not designed from gene sequence regions of self-complementarity (checked by both repetitive 9mer screening and global BLAST score).
The PrimerBank database
To help researchers easily retrieve primer information for their genes of interest, a web front end has been established for querying the primer database, PrimerBank. PrimerBank is tightly integrated with the information from the NCBI databases: NCBI database accession numbers may be used for primer query; NCBI sequences are attached to the primer information page and NCBI LocusLink indices are used internally for gene locus mapping.
PrimerBank currently contains 147 404 primers encompassing most known human and mouse genes. Additional primers will be included for genes from other organisms in the future. All primers in the database have uniform properties such as length, Tm and GC content, simplifying the process of analyzing multiple species from a single template preparation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our colleagues Yi Yang, Amy Stirman, Shukui Guan, Glenn Short and Najib El Messadi for their contributions. We also thank Mason Freeman and Harry Bjorkbacka for useful discussions and primer testing. This research was supported by PGA grant U01 HL66678 from the National Heart, Lung and Blood Institute.
REFERENCES
- 1.Walker N.J. (2002) Tech.Sight. A technique whose time has come. Science, 296, 557–559. [DOI] [PubMed] [Google Scholar]
- 2.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 3.Chen B.Y., Janes,H.W. and Chen,S. (2002) Computer programs for PCR primer design and analysis. Methods Mol. Biol., 192, 19–29. [DOI] [PubMed] [Google Scholar]
- 4.Pattyn F., Speleman,F., De Paepe,A. and Vandesompele,J. (2003) RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res., 31, 122–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerhold D., Rushmore,T. and Caskey,C.T. (1999) DNA chips: promising toys have become powerful tools. Trends Biochem. Sci., 24, 168–173. [DOI] [PubMed] [Google Scholar]
- 6.Li F. and Stormo,G.D. (2001) Selection of optimal DNA oligos for gene expression arrays. Bioinformatics, 17, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 7.Rouillard J.M., Herbert,C.J. and Zuker,M. (2002) OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 8.Wang X. and Seed,B. (2003) Selection of oligonucleotide probes for protein coding sequences. Bioinformatics, 19, 796–802. [DOI] [PubMed] [Google Scholar]
- 9.Wright M.A. and Church,G.M. (2002) An open-source oligomicroarray standard for human and mouse. Nat. Biotechnol., 20, 1082–1083. [DOI] [PubMed] [Google Scholar]
- 10.Chang P.C. and Peck,K. (2003) Design and assessment of a fast algorithm for identifying specific probes for human and mouse genes. Bioinformatics, 19, 1311–1317. [DOI] [PubMed] [Google Scholar]
- 11.Van Hijum S.A., De Jong,A., Buist,G., Kok,J. and Kuipers,O.P. (2003) UniFrag and GenomePrimer: selection of primers for genome-wide production of unique amplicons. Bioinformatics, 19, 1580–1582. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen H.B. and Knudsen,S. (2002) Avoiding cross hybridization by choosing nonredundant targets on cDNA arrays. Bioinformatics, 18, 321–322. [DOI] [PubMed] [Google Scholar]
- 13.Xu D., Li,G., Wu,L., Zhou,J. and Xu,Y. (2002) PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics, 18, 1432–1437. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 16.Hancock J.M. and Armstrong,J.S. (1994) SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput. Appl. Biosci., 10, 67–70. [DOI] [PubMed] [Google Scholar]
- 17.Rychlik W. (1995) Priming efficiency in PCR. Biotechniques, 18, 84–90. [PubMed] [Google Scholar]
- 18.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto N., Nakano,S., Yoneyama,M. and Honda,K. (1996) Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res., 24, 4501–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Ahsen N., Wittwer,C.T. and Schutz,E. (2001) Oligonucleotide melting temperatures under PCR conditions: nearest-neighbor corrections for Mg(2+), deoxynucleotide triphosphate and dimethyl sulfoxide concentrations with comparison to alternative empirical formulas. Clin. Chem., 47, 1956–1961. [PubMed] [Google Scholar]
- 22.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 23.Kane M.D., Jatkoe,T.A., Stumpf,C.R., Lu,J., Thomas,J.D. and Madore,S.J. (2000) Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res., 28, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 25.Willems V.D., Schroeder,H.W.J., Perlmutter,R.M. and Milner,E.C. (1989) Heterogeneity in the human Ig VH locus. J. Immunol., 142, 2547–2554. [PubMed] [Google Scholar]
- 26.Kwok S., Kellogg,D.E., McKinney,N., Spasic,D., Goda,L., Levenson,C. and Sninsky,J.J. (1990) Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res., 18, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang M.M., Arnheim,N. and Goodman,M.F. (1992) Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res., 20, 4567–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mount D.W. (2001) Bioinformatics: Sequence and Genome Analysis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 30.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ema M., Morita,M., Ikawa,S., Tanaka,M., Matsuda,Y., Gotoh,O., Saijoh,Y., Fujii,H., Hamada,H., Kikuchi,Y. et al. (1996) Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol., 16, 5865–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm J., Sachs,M., Britsch,S., Di Cesare,S., Schwarz-Romond,T., Alitalo,K. and Birchmeier,W. (2001) Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J. Cell Biol., 154, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori C., Welch,J.E., Fulcher,K.D., O’Brien,D.A. and Eddy,E.M. (1993) Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol. Reprod., 49, 191–203. [DOI] [PubMed] [Google Scholar]
- 34.Rendtorff N.D., Frodin,M., Attie-Bitach,T., Vekemans,M. and Tommerup,N. (2001) Identification and characterization of an inner ear-expressed human melanoma inhibitory activity (MIA)-like gene (MIAL) with a frequent polymorphism that abolishes translation. Genomics, 71, 40–52. [DOI] [PubMed] [Google Scholar]
- 35.Itoh S., Satoh,M., Abe,Y., Hashimoto,H., Yanagimoto,T. and Kamataki,T. (1994) A novel form of mouse cytochrome P450 3A (Cyp3a-16). Its cDNA cloning and expression in fetal liver. Eur. J. Biochem., 226, 877–882. [DOI] [PubMed] [Google Scholar]
- 36.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 37.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 38.Wu D.Y., Ugozzoli,L., Pal,B.K., Qian,J. and Wallace,R.B. (1991) The effect of temperature and oligonucleotide primer length on the specificity and efficiency of amplification by the polymerase chain reaction. DNA Cell Biol., 10, 233–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.