Abstract
More than any other neuron, olfactory sensory neurons are exposed to environmental insults. Surprisingly, their only documented response to damaging stress is apoptosis and subsequent replacement by new neurons. However, they expressed unfolded protein response genes, a transcriptionally regulated defense mechanism activated by many types of insults. The unfolded protein response transcripts Xbp1, spliced Xbp1, Chop (Ddit3), and BiP (Hspa5) were decreased when external access of stressors was reduced by blocking a nostril (naris occlusion). They, and Nrf2 (Nfe2l2), were increased by systemic application of tunicamycin or the selective olfactotoxic chemical, methimazole. Methimazole’s effects overcame naris occlusion and the unfolded protein response was independent of odor-evoked neuronal activity. Chemical stress is therefore a major and chronic activator of the unfolded protein response in olfactory sensory neurons. Stress-dependent repression of the anti-apoptotic gene Bcl2 was absent, however, suggesting a mechanism for disconnecting the UPR from apoptosis and tolerating a chronic unfolded protein response. Environmental stressors also affect both the sustentacular cells that support the neurons and respiratory epithelia because naris occlusion decreased expression of the xenobiotic chemical transformation enzyme Cyp2a5 in sustentacular cells and both naris occlusion and methimazole altered the abundance of the antibacterial lectin Reg3g in respiratory epithelia.
Keywords: Smell, ER stress, apoptosis, methimazole, tunicamycin, naris occlusion
INTRODUCTION
Of all mammalian neurons, the most directly exposed to environmental insults are the olfactory sensory neurons (OSNs). The only documented reaction of OSNs to environmental stressors is to accumulate damage, apoptose, and thereby stimulate their own replacement (Beites et al., 2005; Cowan and Roskams, 2002; Hinds et al., 1984; Schwob, 2002). Instead, the known protective mechanisms are all provided by neighboring cells: a mucus layer, a barrier of supporting sustentacular cells, and the robust expression of xenobiotic chemical transformation enzymes by sustentacular cells (Farbman, 1992; Kaliner, 1992; Ling et al., 2004; Piras et al., 2003; Whitby-Logan et al., 2004). Reasoning that OSNs must respond to chemical stress, we searched for evidence of stress response gene expression in mouse OSNs. While previous studies have detected Hmox1, Nqo1, and Nrf2 (Nfe2l2) in OSNs (Chen et al., 2003; Gussing and Bohm, 2004), a more complete view of gene expression in mouse OSNs identified genes fundamental to the unfolded protein response (UPR) as being abundantly expressed in OSNs (Sammeta et al., 2007).
The UPR is a mechanism by which cells respond to an increase in misfolded or unfolded proteins in the endoplasmic reticulum (ER) (Schroder and Kaufman, 2005). Stressors that evoke the UPR are numerous and include chemicals and pathogens, environmental factors that result in hypoxia and nutrient depletion, and even physiological conditions such as secretion of large amounts of protein. The activation of the UPR begins with three ER-resident transmembrane proteins, IRE1a (ERN1), PERK (EIF2AK3), and ATF6, whose lumenal domains act as sensors for misfolded and unfolded proteins by competing with them for binding to the molecular chaperone, BiP (HSPA5) (Fig. 1). Activation of IRE1a permits its cytosolic endoribonuclease to act upon Xbp1 mRNA, removing an intron and converting the encoded protein from inactive Xbp1 into the transcriptionally active spliced Xbp1. Activation of PERK via autophosphorylation transiently attenuates translation of most mRNAs and increases transcription of UPR target genes via phosphorylation of eIF2a, ATF4, and NRF2. Activation of ATF6 is accomplished by proteolytic release of an active bZIP transcription factor domain. All three sensors lead to altered transcription of target genes, including the sensors themselves, ER chaperones such as BiP (Hspa5), ER-associated degradation factors, and proteins involved in protein trafficking, small molecule transport, lipid synthesis, carbohydrate synthesis, amino acid synthesis, energy production, control of redox state, and apoptosis (Cullinan and Diehl, 2006; Rutkowski and Kaufman, 2007; Zhang and Kaufman, 2008a; b). The UPR can mediate both acute and chronic stress responses. Acute responses are either resolved quickly or the cell undergoes apoptosis via the UPR target CHOP, which represses the anti-apoptotic gene Bcl2 and activates pro-apoptotic genes (McCullough et al., 2001; Oyadomari and Mori, 2004; Zhang and Kaufman, 2008b). How cells manage a chronic UPR without undergoing apoptosis is poorly understood (Rutkowski and Kaufman, 2007; Zhang and Kaufman, 2008b). Understanding how cells survive the UPR is a particularly compelling issue for neurobiology given evidence that the UPR is associated with neural aging and myelination disorders, and is evoked in neurons by ischemia, neurotrauma, and neurodegenerative disorders (Banhegyi et al., 2008; DeGracia and Montie, 2004; Lin and Popko, 2009; Naidoo, 2009; Paschen and Mengesdorf, 2005; Uehara, 2007).
Fig. 1.
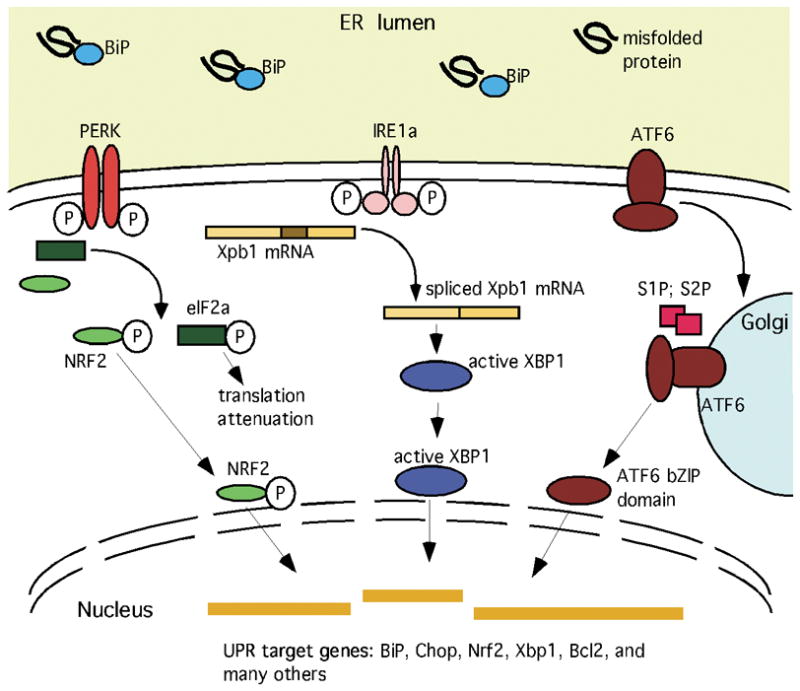
Schematic depiction of selected core elements of the unfolded protein response (UPR), featuring the components measured herein (Schaefer et al., 2001; Schroder and Kaufman, 2005; Zhang and Kaufman, 2008a; b). Accumulation of unfolded proteins in the ER lumen competitively reduces BiP binding to PERK, IRE1a, and ATF6, releasing them to carry out their cytoplasmic and nuclear activities. PERK phosphorylates several substrates, including NRF2 and EIF2a. The latter event decreases translation of most mRNAs, but also increases translation of a few others, such as the transcription factor Atf4 (not depicted). IRE1a dimerizes, activating its endoribonuclease activity and thereby reducing Xbp1 mRNA into a transcript that encodes an active bZIP transcription factor. ATF6 translocates to the Golgi apparatus where the action of two proteases (S1P and S2P) release a bZIP transcription factor domain. The increased availability of these transcriptional regulators alters expression of many genes, including the core elements of the URP pathway itself, molecular chaperones, proteins involved in the degradation of ER proteins, antioxidant proteins, metabolic enzymes, regulators of the cell cycle, and survival/apoptotic factors.
We found that unilateral naris occlusion caused an ipsilateral reduction in UPR mRNAs in OSNs that is independent of neuronal activity, indicating that OSNs experience continuous stress caused by external factors. Given these findings, the necessary capacity of the olfactory epithelium to absorb chemicals and our observation that systemic application of chemical stressors increased UPR mRNAs, we conclude that chemical stress is a major activator of the UPR in OSNs.
EXPERIMENTAL PROCEDURES
Mice, naris occlusion, and chemical injection
C57Bl/6 mice were purchased from Harlan Laboratories (Indianapolis, IN) and housed in the animal facility at the University of Kentucky. Mice carrying a targeted deletion of Cnga2 were obtained from Dr. Peter Mombaerts, Max Planck Institute of Biophysics, Frankfurt, Germany (Zheng et al., 2000). In these mice, the Cnga2 locus lacks codons for amino acids 178–678, thereby encoding a truncated protein that cannot function as an ion channel and generating a mouse whose OSNs have little or no response to odorants or forskolin. Cnga2 mice are a mixed C57Bl/6-129 genetic background. All procedures with mice were done according to approved Institutional Animal Care and Use Committee protocols and conformed to NIH guidelines.
For the end point of all major experiments (naris occlusion, reduced OSN activity, methimazole effects), this project targeted mice at age postnatal day 11 (P11). This age was chosen to balance our success rate for naris occlusion, which decreases with age, with the need to avoid survival longer than a week after naris occlusion, which leads to potentially confounding changes in the olfactory system, such as increased survival of OSNs and reduced neurogenesis in both the epithelium and olfactory bulb (Brunjes et al., 1991; Cummings and Brunjes, 1994; Maruniak et al., 1989; Zou et al., 2004). Some proof-of-principle experiments (e.g., tunicamycin) and confirmatory experiments were also done at age P21 – P22. Naris occlusion was done on anesthetized mice on postnatal day 5 (P5) using a surgical cautery device (Perfectemp Cautery, Bovie Medical Corp), followed by sealing with cyanoacrylate glue (Coppola et al., 1994). Verification of sealing of the naris was done just prior to euthanasia by inspection under a dissecting microscope for bubbles emanating through saline applied to the closed naris. Naris occluded mice were sacrificed at P11.
Methimazole (in saline) was given as a single intraperitoneal injection at a concentration of 50 mg/kg body weight. Mice were sacrificed 24h after the injection. Methimazole is metabolized in Bowman’s gland duct cells and sustentacular cells by a cytochrome P450-dependent pathway leading to the production of reactive intermediates that cause dose- and time-dependent apoptosis of OSNs (Bergman and Brittebo, 1999; Bergstrom et al., 2003). The methimazole experiment was done at age P10–P11 and again at age P21–P22.
Mice received a single intraperitoneal injection of tunicamycin in saline at a concentration of 500 μg/kg body weight at age P21 and were sacrificed 24h after the injection (Iwawaki et al., 2004).
Genes
Mouse genes and mRNAs are displayed according to the gene symbol conventions of Entrez Gene (National Center for Biotechnology Information), typically as a single capital followed by lower case, while references to the protein products are in all capital letters. To avoid ambiguity, the Entrez gene IDs, gene names, gene symbols, and common synonyms for all mouse genes mentioned herein are listed in Supplemental Table S1.
In situ hybridization and immunohistochemistry
In situ hybridization was done on 10 μm coronal cryosections of the anterior head as described previously (Sammeta et al., 2007). Briefly, fixation was accomplished by placing freshly dissected tissue of young mice (age P11) into ice-cold 4% paraformaldehyde in phosphate buffered saline (PBS, pH 7.4) overnight. Older mice (age P22) were first perfused transcardially with ice-cold 4% paraformaldehyde. The fixed tissue was then decalcified in 0.5M EDTA in PBS (pH 8.0) for 24h at 4°C and cryoprotected in 10%, 20% and 30% sucrose successively for at least 1h each at 4°C before being embedded in OCT compound (Sakura Finetech USA, Inc., Torrance, CA) and stored at −80° C. For each mRNA species, cDNA fragments (400–500 bp) were amplified by PCR from olfactory epithelium cDNA and cloned into pBluescript. The primer positions are listed in Supplemental Table S1. The fragments chosen were selected to have less than 80% identity to any other mRNA. This was not possible for Cyp2a5, however, so the probe used hybridizes to both Cyp2a5 and Cyp2a4. The Xbp1 probe detected both spliced and unspliced Xbp1 mRNAs. Sense and antisense recombinant RNA probes (approximately 500 bp in length) labeled with digoxygenin were prepared. Briefly, tissue sections on slides were prepared at room temperature by incubation in 4% PFA for 15 min, washed in PBS (pH 7.4) for 3 min, treated with proteinase K for 12 min (37° C), post-fixed again in 4% PFA for 10 min, washed in PBS for 3 min, treated with 0.2M HCl for 10 min, washed in PBS for 3 min, incubated for 1 min in 500 ml of 0.1M triethalonamine-HCl (pH 8.0) under stirring before mixing in 1.25 ml of acetic anhydrate, stirred for 10 min, washed in PBS for 3 min, dehydrated in 60%, 80%, 95%, and 100% ethanol for 1.5 min each, and allowed to dry completely. Hybridization was done overnight in a 50% formamide, 0.6M NaCl, 1mM EDTA, 10mM Tris-HCl (pH 8.0) containing 10% dextran sulfate, 0.25% sodium dodecyl sulfate, 200 μg/ml yeast tRNA, and 1X Denhardt’s solution with 1ng/μl of riboprobe at 65° C. Washing was 2X standard saline citrate (SSC), 50% formamide at 37° C for 30 min, TNE (10 mM Tris-Cl (pH 7.5), 0.5 M NaCl, 1 mM EDTA) at 37° C for 10 min, TNE containing 20 μg/ml RNase A at 37° C for 30 min, TNE at 37° C for 10 min, 2X SSC at 37° C for 20 min, and 0.2X SSC at 37° C for 20 min, 2X SSC at 37° C for 20 min, and 0.1X SSC at 37° C for 20 min. Detection was done using a 1,000-fold dilution of an alkaline phosphatase-conjugated antibody to digoxygenin (#1093274, Roche Applied Science, Indianapolis, IN) with NBT/BCIP (Nitro-Blue Tetrazolium Chloride/5-Bromo-4-Chloro-3′-Indolyphosphate p-Toluidine) as the substrate. Detection reactions, which were stopped when reaction product became visible, ran 16 – 48 hours. Yoked sense controls were invariably negative. A detailed protocol, which follows the methods of Ishii and colleagues (Ishii et al., 2003; Ishii et al., 2004), is available from the authors.
Immunohistochemistry was done on the same 10 μm coronal cryosections described above. The frozen sections were thawed and permeabilized in PBS with 1% Triton X-100. Blocking buffer, 2% BSA and 0.4% Triton X100 in PBS, was applied for 1h at room temperature followed by incubation in primary antibody diluted in the blocking buffer overnight at 4° C. Sections were washed with 0.05% Tween-20 in PBS for 30 min and then incubated with Cy3 conjugated secondary antibody diluted in PBS. Nuclei were counterstained using Hoechst 33342 (# H1399, Invitrogen, Carlsbad, CA). The anti-caspase-3 rabbit polyclonal antibody (Table 1) detects the 17kDa active fragment of caspase-3. It does so on western blots only when apoptosis-initiating proteases such as caspase-8 and caspase-9 are active, and it does not detect the 17kDa fragment if these enzymes are inhibited (Cheong et al., 2003). Its ability to label cells in tissue sections is directly correlated with both the detection of the active 17kDa fragment in western blots and the presence of conditions known to cause apoptosis (Cheong et al., 2003; Hu et al., 2000; Kaiser et al., 2008). To confirm the ability of this antiserum to bind its antigen, we used a 10-fold molar excess of the antigen to block immunoreactivity (#1050, Cell Signaling Technology, Inc.), and as a control used a 10-fold molar excess of an irrelevant peptide antigen (KNNLKECGLY, #sc-262 P, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Cy3-conjugated donkey anti-rabbit (# 711-165–152, Jackson Immunoresearch Laboratories Inc., West Grove, PA) was used at a dilution of 1:1000.
Table 1.
Primary antibodies used for immunohistochemistry.
| Antigen | Immunogen | Manufacturer | Dilution |
|---|---|---|---|
| Active Caspase-3 | residues 165 – 175 in human caspase-3 | # 9661S, Cell Signaling Technology Inc., Danvers, MA | 1:200 |
For counts of immunoreactive cells, three tissue sections per mouse were selected, matched for anterior-posterior position between treatment and control, and used to generate an average count for each mouse. Immunoreactive cells were visually identified and included in the count if their nuclei were contained in the section. Counts were made at three positions along the olfactory epithelium (the dorsal recess, the dorsal septum, and the ventral septum) per section and normalized by dividing by the total length of epithelium viewed in that tissue section. Abercrombie’s correction for over-counting of profiles in sections was applied using a mean nuclear diameter of 3.23 ± 0.3 μm to obtain a correction factor of 0.76 (Abercombie, 1946).
Wide-field images were obtained on a Nikon Diaphot 300 inverted microscope using a Spot 2e camera and Spot software version 4.0.6 through a 40×/0.75 numerical aperture Plan Fluor objective or a 4×/0.13 numerical aperture Plan objective. Images were processed in Adobe Photoshop (version 7.0) by adjusting size and brightness. Images were combined and labeled in Deneba Canvas (version 8.0).
Quantitative RT-PCR
Reverse transcription was performed on 1 μg of total RNA using SuperScript II and random hexamers (Life Technologies, Rockville, MD) in a 50 μl reaction as we have published previously (Shetty et al., 2005; Yu et al., 2005). Primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) and purchased from Integrated DNA Technologies (Coralville, IA). The Cyp2a5 primers do not distinguish between Cyp2a5 and Cyp2a4. Specificity for spliced Xbp1 was achieved with a primer that spanned the splice junction formed by removal of an intron by IRE1a. Amplification was performed on triplicate samples in an ABI Prism 7700 Sequence Detection System using 1 μl of the cDNA reaction and the Syber Green Core Reagent Kit under conditions prescribed by the manufacturer (Applied Biosystems). Thermal cycler conditions were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Whole olfactory epithelium cDNA was used as a template for standard curves, which were required to exceed a criterion correlation coefficient of 0.98 before being accepted for analysis. Melt curves were performed on each sample to verify that each reaction produced a single product. Results were normalized to the geometric mean of four stable mRNAs in each sample: Gapdh, Actb, Ubc, and Hprt1 (Vandesompele et al., 2002). Statistical analyses were done with Student’s t-tests, paired tests for the naris occlusion experiments and unpaired tests for all others. Because each experiment requires testing the same samples for nine independent transcripts, correction for multiple testing within each experiment was done using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). All data are reported as means ± standard errors of the mean. For graphical display, quantitative RT-PCR data were converted to fold change from the mean control value.
RESULTS
Olfactory sensory neurons expressed UPR genes
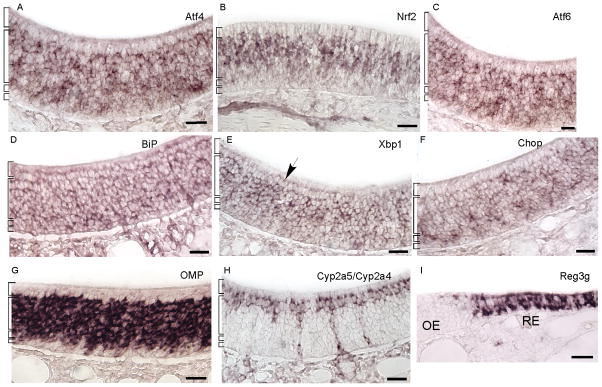
We discovered that unfolded protein response (UPR) mRNAs were abundant among the 10,000 genes expressed by olfactory sensory neurons (OSNs) (Sammeta et al., 2007), leading to the hypothesis that chemical stress evokes the UPR in OSNs, thereby increasing the capacity of OSNs to tolerate, reconstitute, and degrade unfolded and misfolded proteins. We first confirmed that several key UPR mRNAs did indeed localize to the OSNs of C57Bl/6 mice (Fig. 2). These UPR transcripts, with the exception of BiP, were not detected in neighboring respiratory epithelium. Transcripts used as controls in this study, Omp, Cyp2a5/Cyp2a4, and Reg3g, are also expressed only in certain cell types. Omp is specific to mature OSNs, Cyp2a5/Cyp2a4 are expressed in sustentacular cells and Bowman’s gland duct cells, and Reg3g is specific to an as yet unidentified cell type in the respiratory epithelium (Fig. 2A; Supplemental Fig. S1) (Monti-Graziadei et al., 1977; Piras et al., 2003; Yu et al., 2005).
Fig. 2.
UPR transcripts were detected primarily in the OSN layers of the olfactory epithelium. (A) A map of the cell body layering of the pseudostratified olfactory epithelium using mRNAs specific to each cell type. Omp is specific to mature OSNs (mOSN), Gap43 to immature OSNs (iOSN), Ngn1 (Neurog1) to basal progenitor cells (basal), and Cyp2a5/Cyp2a4 to sustentacular cells (Sus) and Bowman’s gland duct cells (BG). The lines mark the basal lamina separating the olfactory epithelium from the lamina propria (LP). (B – G) Atf4, Nrf2, Atf6, BiP, Xbp1, and Chop were detected in the OSN layers central to the epithelium. BiP (panel E) was also detected in the sustentacular cells, basal cells, and cells of the underlying lamina propria. (H) An example of the lack of signal in sense strand hybridizations. Scale bars, 25μm.
To confirm that the UPR in the olfactory epithelium was responsive to chemically-induced stress, we applied tunicamycin, which directly evokes the UPR by inhibiting N-glycosylation of proteins in the ER, thereby increasing the misfolding of membrane proteins in the ER (Iwawaki et al., 2004). Because the UPR depends on changes in the level of transcription of certain critical genes, we used mRNA abundance as a measure of activation of the UPR. As expected, tunicamycin increased the abundance of UPR transcripts Xbp1, spliced Xbp1, BiP, Nrf2, and Chop in olfactory epithelium samples (Fig. 3A; Supplemental Table S2). Not affected were the controls Cyp2a5, Omp, and Reg3g.
Fig. 3.

Summary of quantitative RT-PCR experiments displaying the fold-differences for treatment versus control for mRNAs in olfactory epithelium samples. (A) Tunicamycin increased all UPR transcripts compared to saline injection. N = 3 mice. (B) Naris occlusion decreased BiP, Xbp1, spliced Xbp1, and Chop, as well as Bcl2, Cyp2a5/Cyp2a4, and Reg3g. N = 6 mice. (C) Methimazole increased all UPR transcripts and Reg3g, but decreased Cyp2a5/Cyp2a4, compared to saline. N = 5 mice. (D) Naris occlusion did not impede the ability of methimazole to increase UPR mRNA abundance. Olfactory epithelium samples ipsilateral to naris occlusion (closed side) of mice injected with methimazole were compared to olfactory epithelia contralateral to naris occlusion (open side) from an independent group of mice injected with saline. N = 5 mice. (E) No differences were observed in methimazole effects between samples ipsilateral and contralateral to naris occlusion. N = 5 mice. (F) None of the transcripts differed in abundance between Cnga2−/− and Cnga2+/+ mice. N = 6 mice. Error bars, standard error of the mean; Black, significant increase; grey, significant decrease; *, corrected p < 0.05; **, corrected p < 0.01.
Naris occlusion reduced the abundance of UPR transcripts
If UPR mRNAs were detectable in the OSNs of untreated mice because OSNs are in an exposed position where exogenous chemicals, pathogenic microorganisms, and particulate matter can wreak damage on them, then reducing access of these agents should reduce the UPR. We used unilateral naris occlusion to block orthonasal entry of chemicals and other agents into the nasal cavity. Naris occlusion caused reduced abundance of UPR transcripts Xbp1, spliced Xbp1, BiP, and Chop in the olfactory epithelium (Fig. 3B), indicating that naris occlusion reduced the level of unfolded protein stress experienced by OSNs.
Naris occlusion also decreased the amount of Bcl2 mRNA (Fig. 3B), consistent with a decrease in orthonasal odor stimulation because odor stimulation increases expression of Bcl2 (Watt et al., 2004).
The mRNAs encoding the xenobiotic transformation enzymes Cyp2a5/Cyp2a4 were also reduced. These enzymes are believed to be part of the powerful odorant and chemical clearance function performed by sustentacular cells and Bowman’s gland duct cells (Adams et al., 1991; Chen et al., 1992). Also decreased was Reg3g, an antibacterial lectin that is specific to neighboring respiratory epithelium (Yu et al., 2005). These reductions are consistent with the ability of naris occlusion to reduce access of most environmental stressors to the nasal cavity.
Reduced OSN activity did not reduce UPR transcript abundance
Naris occlusion reduces odor stimulation of the olfactory epithelium. To test whether the reduction in UPR transcripts caused by naris occlusion was simply due to reduced OSN activity rather than reduced chemical stress, we turned to Cnga2 knockout mice in which the ability of odors to evoke electrical activity in OSNs is nearly gone (Brunet et al., 1996; Lin et al., 2004; Zhao and Reed, 2001; Zheng et al., 2000). Suspecting that chemical stress is the primary regulator of UPR gene transcription in OSNs, we predicted that Cnga2−/− mice would not differ from wild-type mice. Indeed, we observed no difference in abundance for any of the UPR transcripts tested: Nrf2, Xbp1, BiP, Chop, and spliced Xbp1 (Fig. 3F). As a positive control for decreased OSN activity, we determined by quantitative RT-PCR that the Cnga2−/− mice used in this experiment had significantly less Fos mRNA in the olfactory bulb than did the wild-type mice used (n = 5; means and standard errors of 2.5±0.5 versus 0.6±0.15 units; p = 0.01; Student’s t-test). Odor stimulation of OSNs is known to elevate Fos expression in the olfactory bulb (Guthrie et al., 1993; Onoda, 1992; Sallaz and Jourdan, 1993). These data indicate that UPR gene expression in OSNs is not regulated by OSN activity.
An olfactoxic chemical increased the abundance of UPR transcripts
Chemical stress is the most likely reason OSNs express UPR genes. Odorants and other chemicals accumulating in the olfactory epithelium presumably either react with cellular proteins directly or induce other conditions that damage cellular proteins, such as oxidative stress and reactive oxygen species. While naris occlusion readily evoked decreases in UPR transcripts, we reasoned that the extraordinary capacity of the olfactory epithelium to detoxify and clear odorant chemicals would limit the ability of most odorants to evoke stress. We therefore turned to an alternative approach that is known to chemically stress the olfactory epithelium, the systemic injection of the olfactotoxic chemical methimazole. Methimazole appears to be converted by cytochrome P450 enzymes located in the sustentacular cells and Bowman’s glands of the olfactory epithelium into toxic intermediates that are difficult to clear from the tissue (Bergman and Brittebo, 1999; Bergstrom et al., 2003). Systemic methimazole therefore mimics one mechanism by which odorants that are difficult to clear might damage OSNs. We used a combination of dose, duration, mouse age, and mouse strain that produced a strong chemical challenge without inducing OSN apoptosis. Indeed, in none of the mice used in this experiment did we observe disruption of the pseudostratification of the olfactory epithelium or changes in the appearance of OSN nuclei (Supplemental Fig. S2) observed in older mice, primarily of other strains (Bergman and Brittebo, 1999; Bergman et al., 2002; Brittebo, 1995; Piras et al., 2003). Our observations were further supported by quantifying apoptosis via counts of cells immunoreactive for activated caspase-3 (n = 2 mice; means of 3.6±0.3 versus 3.5±0.2 cells per 0.1mm of epithelial length; p = 0.3; Student’s t-test). In contrast, methimazole injection increased the abundance of the UPR transcripts Xbp1, spliced Xbp1, BiP, Nrf2, and Chop (Fig. 3C). Repeating this experiment with mice at age P21, we obtained similar effects.
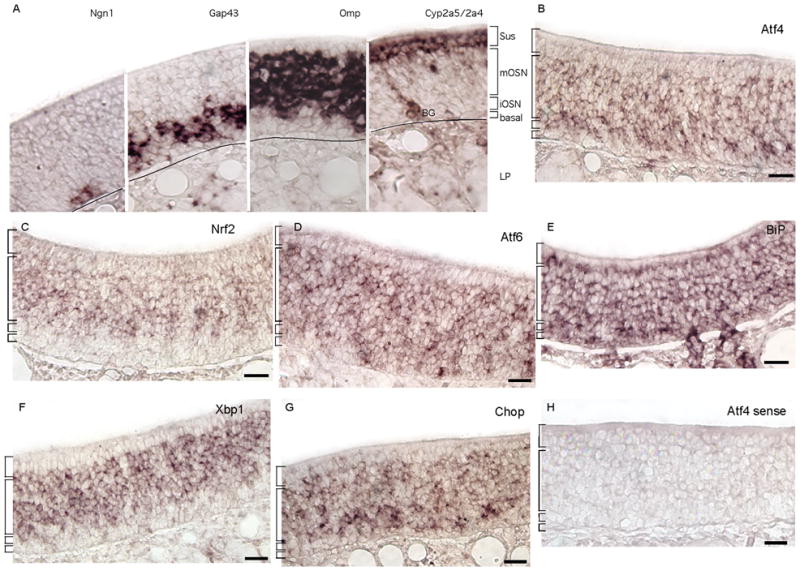
If these many-fold increases occurred primarily in cells other than OSNs, then in situ hybridization after methimazole treatment would detect stronger signals in these other cell types. However, if these differences were instead mostly due to changes within the OSNs, in situ hybridization would detect little expansion of the expression pattern of UPR genes shown in Fig. 2. The latter result was what we observed (Fig. 4). The only easily observable difference was that Xbp1 became detectable in sustentacular cells (Fig. 4E; Fig. 2F). The intensity of Xbp1 labeling in sustentacular cells remained less than in OSNs, however, arguing that much of the more than 2-fold increase in Xbp1 and spliced Xbp1 caused by methimazole (Fig. 3C) must have occurred in OSNs. This conclusion is even more certain for Nrf2 and Chop, which were detected only in the OSN layers after methimazole treatment (Fig. 4B, F). These data demonstrate that at least a large fraction of the increases observed for UPR mRNAs must have occurred in OSNs.
Fig. 4.
After methimazole treatment UPR transcripts were still detected primarily in the OSN layers of the olfactory epithelium. (A – C, F) Atf4, Nrf2, Atf6, and Chop were detected only in the OSN layer. (D) BiP was detected in all cell layers, its normal pattern. (E) Xbp1 was detected in the OSN layers, and also faintly in the sustentacular cell layer (arrow). (G – I) Expression patterns of the control mRNAs Omp, Cyp2a5/Cyp2a4, and Reg3g were normal. OE, olfactory epithelium; RE, respiratory epithelium; Scale bars, 25μm.
The control mRNAs tested in this experiment also gave interesting results. Omp mRNA was unchanged, but Cyp2a5/Cyp2a4 and Reg3g were altered (Fig. 3C). The stability of Omp is further evidence that OSNs survived the methimazole treatment. The reduction in Cyp2a5/Cyp2a4 is consistent with previous evidence that methimazole, albeit at longer durations and greater quantities, damages sustentacular cells and reduces expression of Cyp2a5 in other mouse strains (Piras et al., 2003). The increase in Reg3g parallels the induction of this gene in gut epithelia by gram-positive bacterial colonization and associated inflammatory and immune responses (Cash et al., 2006; Ogawa et al., 2003).
Methimazole overcame the effect of naris occlusion
Given that methimazole applied systemically increased the abundance of UPR mRNAs, it should be able to substitute for the stressors whose access is blocked by naris occlusion. Methimazole appeared to have a much stronger effect than naris occlusion. We therefore predicted that when applied simultaneously with naris occlusion, methimazole would again cause large increases in UPR mRNA abundance. Indeed, this is what we observed (Fig. 3D). In addition, methimazole’s effects were not reduced when naris occlusion was applied simultaneously (Fig. 3E), arguing that the scale of the effects of methimazole and naris occlusion applied independently were similar and did indeed reflect large differences in response magnitude.
DISCUSSION
Though olfactory sensory neurons (OSNs) do ultimately respond to stress by undergoing apoptosis and thereby helping to stimulate their own replacement, they are not passive victims that lack defense mechanisms. Instead, OSNs responded to chemical stress with a robust unfolded protein response (UPR). The possibility remains that misfolding of endogenous proteins such as the odorant receptors, which may be difficult to fold (Gimelbrant et al., 2001; Gimelbrant et al., 1999; McClintock and Sammeta, 2003), or other factors might contribute to the UPR in OSNs, but we found that much of the UPR in OSNs was due to external stressors, especially chemicals. The ability of chemical stress to increase Chop expression, the critical element linking the UPR to apoptosis, suggests a potential mechanism whereby chemical stress induces apoptosis of OSNs. That OSNs are almost continuously exposed to chemical stress, yet appear to be slow to apoptose, suggests that they have an ability to survive chronic UPR. Like other cells with chronic UPRs, such as brain neurons impacted by neurodegenerative diseases, they must have a mechanism of uncoupling CHOP from the intrinsic pathway of apoptosis (Rutkowski and Kaufman, 2007). More broadly, the UPR of OSNs appears to be only one component of a multicellular response to damaging agents that enter the nasal cavity. Other components include increased capacity to metabolize xenobiotic compounds by sustentacular cells and increased antibacterial protein secretion by the neighboring respiratory epithelium.
OSNs have a chronic UPR disconnected from apoptotic mechanisms
Even in mice housed in our animal facility and otherwise untreated, we observed significant expression of UPR genes in OSNs (Sammeta et al., 2007). That naris occlusion reduced the amount of these mRNAs in samples of nasal epithelium can only mean that these decreases occurred primarily in OSNs rather than in neighboring cell types. A large fraction of the increases in UPR transcripts observed after tunicamycin and methimazole treatments must also have occurred in OSNs. These treatments caused increases in mRNA abundance of up to 15-fold yet in situ hybridization after the treatments continued to detect signals primarily in OSNs (Fig. 4, Supplemental Fig. S3). If the response had occurred mostly in cell types other than OSNs, then the in situ hybridization data would have shown stronger signals in these other cells than in OSNs. These findings cannot be interpreted to mean the UPR occurred only in OSNs, however, but rather that much of each increase was in OSNs. Other cells certainly contributed, as evidenced by the behavior of Xbp1, which became detectable in sustentacular cells only after chemical stress. Why OSNs were so highly responsive is unknown, though a contributing factor may be the tendency for certain toxic chemicals such as toluene and methimazole (or their metabolites) to accumulate in or near OSNs (Bergman and Brittebo, 1999; Ghantous et al., 1990).
The extent of the UPR response in OSNs appeared to be complete in that we detected expression of mRNAs that are downstream of each of the three sensor proteins. PERK activation was indicated by increased expression of Nrf2, which when phosphorylated by PERK induces its own expression (Kwak et al., 2002). IRE1a activity was demonstrated by the presence of spliced Xbp1. ATF6 activity was indicated by the behavior of unspliced Xbp1 mRNA, for which ATF6 is the primary activator (Yoshida et al., 2001). Many other downstream UPR genes, such as Chop and BiP, are regulated coordinately by the three sensor pathways (Schroder and Kaufman, 2005).
The activation of all three UPR pathways typically results in increased expression of Chop, consequent repression of Bcl2, activation of pro-apoptotic genes and pathways, and apoptosis (Rutkowski and Kaufman, 2007; Schroder and Kaufman, 2005; Szegezdi et al., 2009). Several types of neurons are protected from stress-induced apoptosis if CHOP is lacking (Chen et al., 2007; Silva et al., 2005; Tajiri et al., 2004; Tajiri et al., 2006). However, this consensus mechanism may not be true for all cells, and differences in the dynamics of the UPR signaling pathways can result in differences in cell survival (Lin et al., 2007; Southwood et al., 2002). For example, sustained IRE1a pathway activation appears to be associated with increased cell survival (Lin et al., 2007). Our data suggest that OSNs survive at least a low level of chronic UPR for weeks or months, but don’t yet indicate how. Perhaps the IRE1a pathway predominates and persists in OSNs during chronic stress. Alternatively, perhaps the increase in Chop mRNA we observed in response to chemical stress is disconnected from the intrinsic apoptotic pathway in OSNs. Bcl2, a potent regulator of OSN apoptosis that decreases in severely damaged OSNs, is expressed in OSNs and is modulated by odor-stimulated cyclic AMP and mitogen activated protein kinase signaling (Jourdan et al., 1998; Sammeta et al., 2007; Shetty et al., 2005; Watt et al., 2004; Yagi et al., 2007). We observed that Bcl2 mRNA decreased along with Chop after naris occlusion and failed to decrease when Chop increased after methimazole or tunicamycin treatment, leading to the hypothesis that OSNs survive chronic UPR in part by disconnecting CHOP from Bcl2 repression until such time that damage accumulates sufficiently or is acutely severe, as when OSN axons are severed (Carson et al., 2005; Shetty et al., 2005). The interactions between the UPR and regulators of apoptosis, especially Bcl2 family proteins, is multifaceted and bidirectional (Szegezdi et al., 2009) so the regulation of Bcl2 by the UPR presumably represents only one component of the OSN’s ability to resist apoptosis driven by ER stress.
Chemical stress activates the UPR in OSNs
The UPR can be activated by any stress that increases the load of unfolded proteins in the ER, either via increased expression of difficult-to-fold proteins or more commonly by insults that cause increased frequency of protein misfolding. Naris occlusion reduces access of several types of external stressors to the olfactory epithelium, including chemicals, microorganisms, and particulates. That the olfactory epithelium must absorb volatile chemicals in order to detect them favors the interpretation that chemical stress causes UPR transcripts to be abundant in OSNs. By measuring UPR transcripts in Cnga2−/− mice, we were able to rule out the potentially confounding factor of OSN activity, which also decreases after naris occlusion. More directly, we found that the olfactotoxic chemical methimazole increased expression of fundamental UPR genes in OSNs. The powerful effect of methimazole on UPR transcript abundance compared to the intermediate level of abundance evoked by ambient odorant chemical exposure in our mouse colony correlates with methimazole’s unusually high toxicity to the olfactory epithelium. While not ruling out the possibility that other stressors would also activate the UPR in OSNs, we conclude that chemical stress is sufficient to do so in OSNs. That systemic chemical stress, the route we used for methimazole and tunicamycin, has indirect effects on the UPR via hormonal signals seems unlikely. For example, tunicamycin does not induce a corticosterone response in rats (Gonzales et al., 2008). In addition, the evidence is strong that systemic methimazole acts locally in the olfactory epithelium (Bergman and Brittebo, 1999; Bergman et al., 2002; Bergstrom et al., 2003; Brittebo, 1995). Overall, our data implicate the absorption of chemicals as the major driving force behind the expression of UPR genes in mouse OSNs, at least under standard laboratory housing conditions.
Does the level of the UPR in OSNs directly correlate with the amount of odorant recently experienced? We suspect that this correlation is very nonlinear. The powerful capacity of the sustentacular cells to metabolize and presumably clear odorant chemicals may be sufficient to prevent significant accumulation of odorants and their metabolites, keeping chemical stress within the epithelium at low to moderate levels. Consistent with this idea, in pilot experiments with increased odor stimulation we detected no significant increase in UPR mRNAs. We hypothesize that only in cases of extreme odorant concentrations that overcome clearance mechanisms, or the rare instances of chemicals that are unusually difficult to metabolize and clear (e.g., methimazole), would the full UPR become apparent.
The results of our methimazole treatments should not be interpreted as contradictory to previous demonstrations of the ability of methimazole to cause lesions of the olfactory epithelium (Bergman and Brittebo, 1999; Bergstrom et al., 2003; Brittebo, 1995). Indeed, we have used two 50 mg/kg doses of methimazole to lesion the sustentacular cell and OSN layers of the olfactory epithelium of adult C57Bl/6 mice (unpublished data), just as others have done in other mouse strains (Bergman et al., 2002; Piras et al., 2003). While strain differences in sensitivity to methimazole are as yet unproven, the extent of lesions at 24 hrs after a single methimazole dose in adult NMRI mice did appear to be more extensive than in adult C57Bl/6 mice (Brittebo, 1995). Age, however, may have been the greatest factor contributing to the lower susceptibility we observed. Though the effect of methimazole on juvenile mice has not been documented, aging increases the response of humans to methimazole (Yamada et al., 1994). In addition, the UPR, which appears to help protect OSNs against methimazole toxicity, also decreases with age (Naidoo, 2009).
A multifaceted defense response in the nasal cavity
Naris occlusion evoked decreases in expression of components of three distinct defense mechanisms in nasal epithelia: the UPR in OSNs, xenobiotic chemical transformation in sustentacular cells, and expression of the antibacterial protein Reg3g in an as yet unidentified cell type in the respiratory epithelium. The chemical transformation capacity of sustentacular cells is well known (Banger et al., 1994; Chen et al., 1992; Dahl et al., 1982; Heydel et al., 2001; Krishna et al., 1994; Matsui et al., 1998; Miyawaki et al., 1996; Piras et al., 2003; Thornton-Manning et al., 1997; Yu et al., 2005). This function would appear to be critical for a tissue whose purpose is to concentrate and detect exogenous chemicals, consistent with evidence that cytochrome P450 activity is even higher in the olfactory epithelium than in the liver (Reed et al., 1986). In contrast, environmental stressors had not previously been linked to either the UPR in OSNs or to antibacterial gene expression in the respiratory epithelium. The signals and pathways that regulate the expression of Cyp2a5/Cyp2a4 in sustentacular cells and Reg3g in respiratory epithelium remain unknown. While a mechanism to coordinately regulate expression of all three of these defense responses is possible given that all responded to a single manipulation (naris occlusion), a simpler explanation is that naris occlusion reduced access of distinct triggers for each.
In summary, nasal epithelia experience environmental insults ranging from chemicals to pathogenic microorganisms to particulate matter. They monitor and respond to these insults. To the view that mucus secretion and flow helps carry away all three types of insults, and that the sustentacular cells have a powerful ability to transform and presumably clear xenobiotic chemicals, must now be added two additional components. First, OSNs resist chemical damage by increasing their ability to protect and reconstitute misfolded proteins. Second, respiratory epithelial cells express, and presumably secrete, the gram-positive antibacterial lectin Reg3g in a manner that may be regulated by the level of environmental insult experienced.
Supplementary Material
Acknowledgments
Acknowledgements of support
This work was supported by NIH award R01 DC002736.
Abbreviations
- ER
endoplasmic reticulum
- OSN
olfactory sensory neuron
- P5
postnatal day 5
- UPR
unfolded protein response
References
- Abercombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- Adams DR, Jones AM, Plopper CG, Serabjit-Singh CJ, Philpot RM. Distribution of cytochrome P-450 monoxygenase enzymes in the nasal mucosa of hamster and rat. Am J Anat. 1991;190(3):291–298. doi: 10.1002/aja.1001900309. [DOI] [PubMed] [Google Scholar]
- Banger KK, Foster JR, Lock EA, Reed CJ. Immunohistochemical localisation of six glutathione S-transferases within the nasal cavity of the rat. Arch Toxicol. 1994;69(2):91–98. doi: 10.1007/s002040050143. [DOI] [PubMed] [Google Scholar]
- Banhegyi G, Mandl J, Csala M. Redox-based endoplasmic reticulum dysfunction in neurological diseases. J Neurochem. 2008;107(1):20–34. doi: 10.1111/j.1471-4159.2008.05571.x. [DOI] [PubMed] [Google Scholar]
- Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306(2):309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Bergman U, Brittebo EB. Methimazole toxicity in rodents: covalent binding in the olfactory mucosa and detection of glial fibrillary acidic protein in the olfactory bulb. Toxicol Appl Pharmacol. 1999;155(2):190–200. doi: 10.1006/taap.1998.8590. [DOI] [PubMed] [Google Scholar]
- Bergman U, Ostergren A, Gustafson AL, Brittebo B. Differential effects of olfactory toxicants on olfactory regeneration. Arch Toxicol. 2002;76(2):104–112. doi: 10.1007/s00204-002-0321-2. [DOI] [PubMed] [Google Scholar]
- Bergstrom U, Giovanetti A, Piras E, Brittebo EB. Methimazole-induced damage in the olfactory mucosa: effects on ultrastructure and glutathione levels. Toxicol Pathol. 2003;31(4):379–387. doi: 10.1080/01926230390201101. [DOI] [PubMed] [Google Scholar]
- Brittebo EB. Metabolism-dependent toxicity of methimazole in the olfactory nasal mucosa. Pharmacol Toxicol. 1995;76(1):76–79. doi: 10.1111/j.1600-0773.1995.tb00107.x. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17(4):681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- Brunjes PC, Caggiano AO, Korol DL, Stewart JS. Unilateral olfactory deprivation: effects on succinate dehydrogenase histochemistry and [3H]leucine incorporation in the olfactory mucosa. Brain Res Dev Brain Res. 1991;62(2):239–244. doi: 10.1016/0165-3806(91)90171-e. [DOI] [PubMed] [Google Scholar]
- Carson C, Saleh M, Fung FW, Nicholson DW, Roskams AJ. Axonal dynactin p150Glued transports caspase-8 to drive retrograde olfactory receptor neuron apoptosis. J Neurosci. 2005;25(26):6092–6104. doi: 10.1523/JNEUROSCI.0707-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Fan Z, Wang X, Ma C, Bower KA, Shi X, Ke ZJ, Luo J. Brain-derived neurotrophic factor suppresses tunicamycin-induced upregulation of CHOP in neurons. J Neurosci Res. 2007;85(8):1674–1684. doi: 10.1002/jnr.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tu Y, Moon C, Nagata E, Ronnett GV. Heme oxygenase-1 and heme oxygenase-2 have distinct roles in the proliferation and survival of olfactory receptor neurons mediated by cGMP and bilirubin, respectively. J Neurochem. 2003;85(5):1247–1261. doi: 10.1046/j.1471-4159.2003.01776.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Getchell ML, Ding X, Getchell TV. Immunolocalization of two cytochrome P450 isozymes in rat nasal chemosensory tissue. Neuroreport. 1992;3(9):749–752. doi: 10.1097/00001756-199209000-00007. [DOI] [PubMed] [Google Scholar]
- Cheong JW, Chong SY, Kim JY, Eom JI, Jeung HK, Maeng HY, Lee ST, Min YH. Induction of apoptosis by apicidin, a histone deacetylase inhibitor, via the activation of mitochondria-dependent caspase cascades in human Bcr-Abl-positive leukemia cells. Clin Cancer Res. 2003;9(13):5018–5027. [PubMed] [Google Scholar]
- Coppola DM, Coltrane JA, Arsov I. Retronasal or internasal olfaction can mediate odor-guided behaviors in newborn mice. Physiol Behav. 1994;56(4):729–736. doi: 10.1016/0031-9384(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Roskams AJ. Apoptosis in the mature and developing olfactory neuroepithelium. Microsc Res Tech. 2002;58(3):204–215. doi: 10.1002/jemt.10150. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol. 2006;38(3):317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Brunjes PC. Changes in cell proliferation in the developing olfactory epithelium following neonatal unilateral naris occlusion. Exp Neurol. 1994;128(1):124–128. doi: 10.1006/exnr.1994.1119. [DOI] [PubMed] [Google Scholar]
- Dahl AR, Hadley WM, Hahn FF, Benson JM, McClellan RO. Cytochrome P-450-dependent monooxygenases in olfactory epithelium of dogs: possible role in tumorigenicity. Science. 1982;216(4541):57–59. doi: 10.1126/science.7063870. [DOI] [PubMed] [Google Scholar]
- DeGracia DJ, Montie HL. Cerebral ischemia and the unfolded protein response. J Neurochem. 2004;91(1):1–8. doi: 10.1111/j.1471-4159.2004.02703.x. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Cell Biology of Olfaction. Cambridge: Cambridge Unversity Press; 1992. p. 282. [Google Scholar]
- Ghantous H, Dencker L, Gabrielsson J, Danielsson BR, Bergman K. Accumulation and turnover of metabolites of toluene and xylene in nasal mucosa and olfactory bulb in the mouse. Pharmacol Toxicol. 1990;66(2):87–92. doi: 10.1111/j.1600-0773.1990.tb00711.x. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Haley SL, McClintock TS. Olfactory receptor trafficking involves conserved regulatory steps. J Biol Chem. 2001;276(10):7285–7290. doi: 10.1074/jbc.M005433200. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Stoss TD, Landers TM, McClintock TS. Truncation releases olfactory receptors from the endoplasmic reticulum of heterologous cells. Journal of Neurochemistry. 1999;72(6):2301–2311. doi: 10.1046/j.1471-4159.1999.0722301.x. [DOI] [PubMed] [Google Scholar]
- Gonzales JC, Gentile CL, Pfaffenbach KT, Wei Y, Wang D, Pagliassotti MJ. Chemical induction of the unfolded protein response in the liver increases glucose production and is activated during insulin-induced hypoglycaemia in rats. Diabetologia. 2008;51(10):1920–1929. doi: 10.1007/s00125-008-1094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussing F, Bohm S. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci. 2004;19(9):2511–2518. doi: 10.1111/j.0953-816X.2004.03331.x. [DOI] [PubMed] [Google Scholar]
- Guthrie KM, Anderson AJ, Leon M, Gall C. Odor-induced increases in c-fos mRNA expression reveal an anatomical “unit” for odor processing in olfactory bulb. Proc Natl Acad Sci U S A. 1993;90(8):3329–3333. doi: 10.1073/pnas.90.8.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydel J, Leclerc S, Bernard P, Pelczar H, Gradinaru D, Magdalou J, Minn A, Artur Y, Goudonnet H. Rat olfactory bulb and epithelium UDP-glucuronosyltransferase 2A1 (UGT2A1) expression: in situ mRNA localization and quantitative analysis. Brain Res Mol Brain Res. 2001;90(1):83–92. doi: 10.1016/s0169-328x(01)00080-8. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL, McNelly NA. An autoradiographic study of the mouse olfactory epithelium: evidence for long-lived receptors. Anat Rec. 1984;210(2):375–383. doi: 10.1002/ar.1092100213. [DOI] [PubMed] [Google Scholar]
- Hu BR, Liu CL, Ouyang Y, Blomgren K, Siesjo BK. Involvement of caspase-3 in cell death after hypoxia-ischemia declines during brain maturation. J Cereb Blood Flow Metab. 2000;20(9):1294–1300. doi: 10.1097/00004647-200009000-00003. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hirota J, Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr Biol. 2003;13(5):394–400. doi: 10.1016/s0960-9822(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol. 2004;33(6):657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10(1):98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Jourdan F, Moyse E, De Bilbao F, Dubois-Dauphin M. Olfactory neurons are protected from apoptosis in adult transgenic mice over-expressing the bcl-2 gene. Neuroreport. 1998;9(5):921–926. doi: 10.1097/00001756-199803300-00029. [DOI] [PubMed] [Google Scholar]
- Kaiser CL, Chapman BJ, Guidi JL, Terry CE, Mangiardi DA, Cotanche DA. Comparison of activated caspase detection methods in the gentamicin-treated chick cochlea. Hear Res. 2008;240(1–2):1–11. doi: 10.1016/j.heares.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliner MA. Human nasal host defense and sinusitis. J Allergy Clin Immunol. 1992;90(3 Pt 2):424–430. doi: 10.1016/0091-6749(92)90162-u. [DOI] [PubMed] [Google Scholar]
- Krishna NS, Getchell TV, Getchell ML. Differential expression of alpha, mu, and pi classes of glutathione S-transferases in chemosensory mucosae of rats during development. Cell Tissue Res. 1994;275(3):435–450. doi: 10.1007/BF00318813. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22(9):2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Arellano J, Slotnick B, Restrepo D. Odors detected by mice deficient in cyclic nucleotide-gated channel subunit A2 stimulate the main olfactory system. J Neurosci. 2004;24(14):3703–3710. doi: 10.1523/JNEUROSCI.0188-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Popko B. Endoplasmic reticulum stress in disorders of myelinating cells. Nat Neurosci. 2009;12(4):379–385. doi: 10.1038/nn.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling G, Gu J, Genter MB, Zhuo X, Ding X. Regulation of cytochrome P450 gene expression in the olfactory mucosa. Chem Biol Interact. 2004;147(3):247–258. doi: 10.1016/j.cbi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Maruniak JA, Lin PJ, Henegar JR. Effects of unilateral naris closure on the olfactory epithelia of adult mice. Brain Res. 1989;490(2):212–218. doi: 10.1016/0006-8993(89)90239-4. [DOI] [PubMed] [Google Scholar]
- Matsui M, Tamura H, Nagai F, Homma H, Miyawaki A, Mikoshiba K. On the nature of rat hepatic and mouse olfactory sulfotransferases. Chem Biol Interact. 1998;109(1–3):69–80. doi: 10.1016/s0009-2797(97)00121-x. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14(12):1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21(4):1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Homma H, Tamura H, Matsui M, Mikoshiba K. Zonal distribution of sulfotransferase for phenol in olfactory sustentacular cells. Embo J. 1996;15(9):2050–2055. [PMC free article] [PubMed] [Google Scholar]
- Monti-Graziadei GA, Margolis FL, Harding JW, Graziadei PP. Immunocytochemistry of the olfactory marker protein. J Histochem Cytochem. 1977;25(12):1311–1316. doi: 10.1177/25.12.336785. [DOI] [PubMed] [Google Scholar]
- Naidoo N. ER and aging-Protein folding and the ER stress response. Ageing Res Rev. 2009;8(3):150–159. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Fukushima K, Naito H, Funayama Y, Unno M, Takahashi K, Kitayama T, Matsuno S, Ohtani H, Takasawa S, Okamoto H, Sasaki I. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9(3):162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- Onoda N. Odor-induced fos-like immunoreactivity in the rat olfactory bulb. Neurosci Lett. 1992;137(2):157–160. doi: 10.1016/0304-3940(92)90393-l. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- Paschen W, Mengesdorf T. Endoplasmic reticulum stress response and neurodegeneration. Cell Calcium. 2005;38(3–4):409–415. doi: 10.1016/j.ceca.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Piras E, Franzen A, Fernandez EL, Bergstrom U, Raffalli-Mathieu F, Lang M, Brittebo EB. Cell-specific expression of CYP2A5 in the mouse respiratory tract: effects of olfactory toxicants. J Histochem Cytochem. 2003;51(11):1545–1555. doi: 10.1177/002215540305101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed CJ, Lock EA, De Matteis F. NADPH: cytochrome P-450 reductase in olfactory epithelium. Relevance to cytochrome P-450-dependent reactions. Biochem J. 1986;240(2):585–592. doi: 10.1042/bj2400585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32(10):469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Sallaz M, Jourdan F. C-fos expression and 2-deoxyglucose uptake in the olfactory bulb of odour-stimulated awake rats. Neuroreport. 1993;4(1):55–58. doi: 10.1097/00001756-199301000-00014. [DOI] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol. 2007;502:1138–1156. doi: 10.1002/cne.21365. [DOI] [PubMed] [Google Scholar]
- Schaefer ML, Finger TE, Restrepo D. Variability of position of the P2 glomerulus within a map of the mouse olfactory bulb. J Comp Neurol. 2001;436(3):351–362. [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269(1):33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- Shetty RS, Bose SC, Nickell MD, McIntyre JC, Hardin DH, Harris AM, McClintock TS. Transcriptional changes during neuronal death and replacement in the olfactory epithelium. Mol Cell Neurosci. 2005;30(1):90–107. doi: 10.1016/j.mcn.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Silva RM, Ries V, Oo TF, Yarygina O, Jackson-Lewis V, Ryu EJ, Lu PD, Marciniak SJ, Ron D, Przedborski S, Kholodilov N, Greene LA, Burke RE. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J Neurochem. 2005;95(4):974–986. doi: 10.1111/j.1471-4159.2005.03428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood CM, Garbern J, Jiang W, Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 2002;36(4):585–596. doi: 10.1016/s0896-6273(02)01045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E, Macdonald DC, Ni Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol. 2009;296(5):C941–953. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- Tajiri S, Oyadomari S, Yano S, Morioka M, Gotoh T, Hamada JI, Ushio Y, Mori M. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ. 2004;11(4):403–415. doi: 10.1038/sj.cdd.4401365. [DOI] [PubMed] [Google Scholar]
- Tajiri S, Yano S, Morioka M, Kuratsu J, Mori M, Gotoh T. CHOP is involved in neuronal apoptosis induced by neurotrophic factor deprivation. FEBS Lett. 2006;580(14):3462–3468. doi: 10.1016/j.febslet.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Thornton-Manning JR, Nikula KJ, Hotchkiss JA, Avila KJ, Rohrbacher KD, Ding X, Dahl AR. Nasal cytochrome P450 2A: identification, regional localization, and metabolic activity toward hexamethylphosphoramide, a known nasal carcinogen. Toxicol Appl Pharmacol. 1997;142(1):22–30. doi: 10.1006/taap.1996.7975. [DOI] [PubMed] [Google Scholar]
- Uehara T. Accumulation of misfolded protein through nitrosative stress linked to neurodegenerative disorders. Antioxid Redox Signal. 2007;9(5):597–601. doi: 10.1089/ars.2006.1517. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt WC, Sakano H, Lee ZY, Reusch JE, Trinh K, Storm DR. Odorant stimulation enhances survival of olfactory sensory neurons via MAPK and CREB. Neuron. 2004;41(6):955–967. doi: 10.1016/s0896-6273(04)00075-3. [DOI] [PubMed] [Google Scholar]
- Whitby-Logan GK, Weech M, Walters E. Zonal expression and activity of glutathione S-transferase enzymes in the mouse olfactory mucosa. Brain Res. 2004;995(2):151–157. doi: 10.1016/j.brainres.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Yagi S, Tsukatani T, Yata T, Tsukioka F, Miwa T, Furukawa M. Lipopolysaccharide-induced apoptosis of olfactory receptor neurons in rats. Acta Otolaryngol. 2007;127(7):748–753. doi: 10.1080/00016480601002062. [DOI] [PubMed] [Google Scholar]
- Yamada T, Aizawa T, Koizumi Y, Komiya I, Ichikawa K, Hashizume K. Age-related therapeutic response to antithyroid drug in patients with hyperthyroid Graves’ disease. J Am Geriatr Soc. 1994;42(5):513–516. doi: 10.1111/j.1532-5415.1994.tb04973.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yu TT, McIntyre JC, Bose SC, Hardin D, Owen MC, McClintock TS. Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J Comp Neurol. 2005;483(3):251–262. doi: 10.1002/cne.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008a;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. Identification and characterization of endoplasmic reticulum stress-induced apoptosis in vivo. Methods Enzymol. 2008b;442:395–419. doi: 10.1016/S0076-6879(08)01420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Reed RR. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 2001;104(5):651–660. doi: 10.1016/s0092-8674(01)00262-8. [DOI] [PubMed] [Google Scholar]
- Zheng C, Feinstein P, Bozza T, Rodriguez I, Mombaerts P. Peripheral olfactory projections are differentially affected in mice deficient in a cyclic nucleotide-gated channel subunit. Neuron. 2000;26(1):81–91. doi: 10.1016/s0896-6273(00)81140-x. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Feinstein P, Rivers AL, Mathews GA, Kim A, Greer CA, Mombaerts P, Firestein S. Postnatal refinement of peripheral olfactory projections. Science. 2004;304(5679):1976–1979. doi: 10.1126/science.1093468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.