Abstract
IGPS is a 51 kDa heterodimeric enzyme comprised of two proteins, HisH and HisF, that catalyze the hydrolysis of glutamine to produce NH3 in the HisH active site and the cyclization of ammonia with N’-[(5’-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide-ribonucleotide (PRFAR) in HisF to produce imidazole glycerol phosphate (IGP) and 5-aminoimidazole-4-carboxamide ribotide (AICAR). Binding of PRFAR and IGP stimulates glutaminase activity in the HisH enzyme over 5000 and 100-fold, respectively, despite the active sites being > 25 Å apart. The details of this long-range protein communication process were investigated by solution NMR spectroscopy and CPMG relaxation dispersion experiments. Formation of the heterodimer enzyme results in a reduction in millisecond motions in HisF that extend throughout the protein. Binding of lGP results in an increase in protein-wide millisecond dynamics evidenced as severe NMR line broadening. Together, these data demonstrate a grouping of flexible residues that link the HisF active site with the protein interface to which HisH binds and provide a model for the path of communication between the IGPS active sites.
Keywords: allostery, protein dynamics, millisecond motions, IGP synthase
Introduction
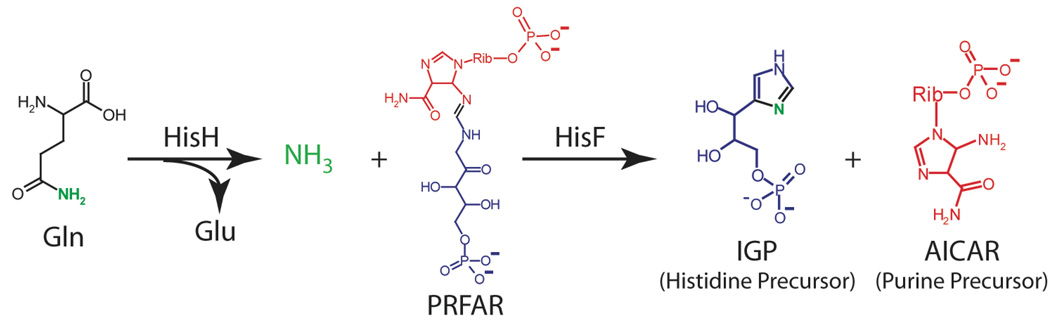
The source of nitrogen for use in biosynthetic pathways is commonly supplied in the form of ammonia produced by the hydrolysis of glutamine by a class of enzymes known as glutamine amidotransferases (Massiere et al. 1998; Zalkin et al. 1998). Imidazole glycerol phosphate synthase (IGPS) is one such enzyme located at the metabolic branch point of histidine and nucleotide biosynthesis (Scheme 1). IGPS is found only in bacteria, some plants and fungi, making it a potential therapeutic target (Chaudhuri et al. 2001; Sinha et al. 2004). IGPS catalyzes two reactions in two distinct and spatially separated active sites (Figure 1) (Chaudhuri et al. 2001; Douangamath et al. 2002; Omi et al. 2002). The first reaction is the hydrolysis of glutamine to yield glutamate and NH3 by the glutaminase domain of IGPS (also known as HisH) (Klem et al. 1993; Klem et al. 2001). The second reaction occurs in the cyclase domain (also known as HisF) and involves the incorporation of NH3 produced by HisH into the nucleotide N’-[(5’-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide-ribonucleotide (PRFAR) to form imidazole glycerol phosphate (IGP) and 5-aminoimidazole-4-carboxamide ribotide (AICAR) (Klem et al. 1993; Klem et al. 2001). After production, IGP continues along the histidine biosynthetic pathway and AICAR is recycled in the de novo synthesis of purines. In some species, such as S. cerevisiae, the cyclase and glutaminase activities reside on a single 60 kDa polypeptide chain (Kuenzler et al. 1993; Chittur et al. 2000); whereas in bacteria, including the thermophile T. maritima, IGPS is a heterodimer composed of two separate proteins, HisH (23 kDa) and HisF (28 kDa) (Figure 1).
Scheme 1.
Enzymatic reaction catalyzed by the HisH and HisF subunits of Imidazole glycerol phosphate synthase (IGPS).
Figure 1.
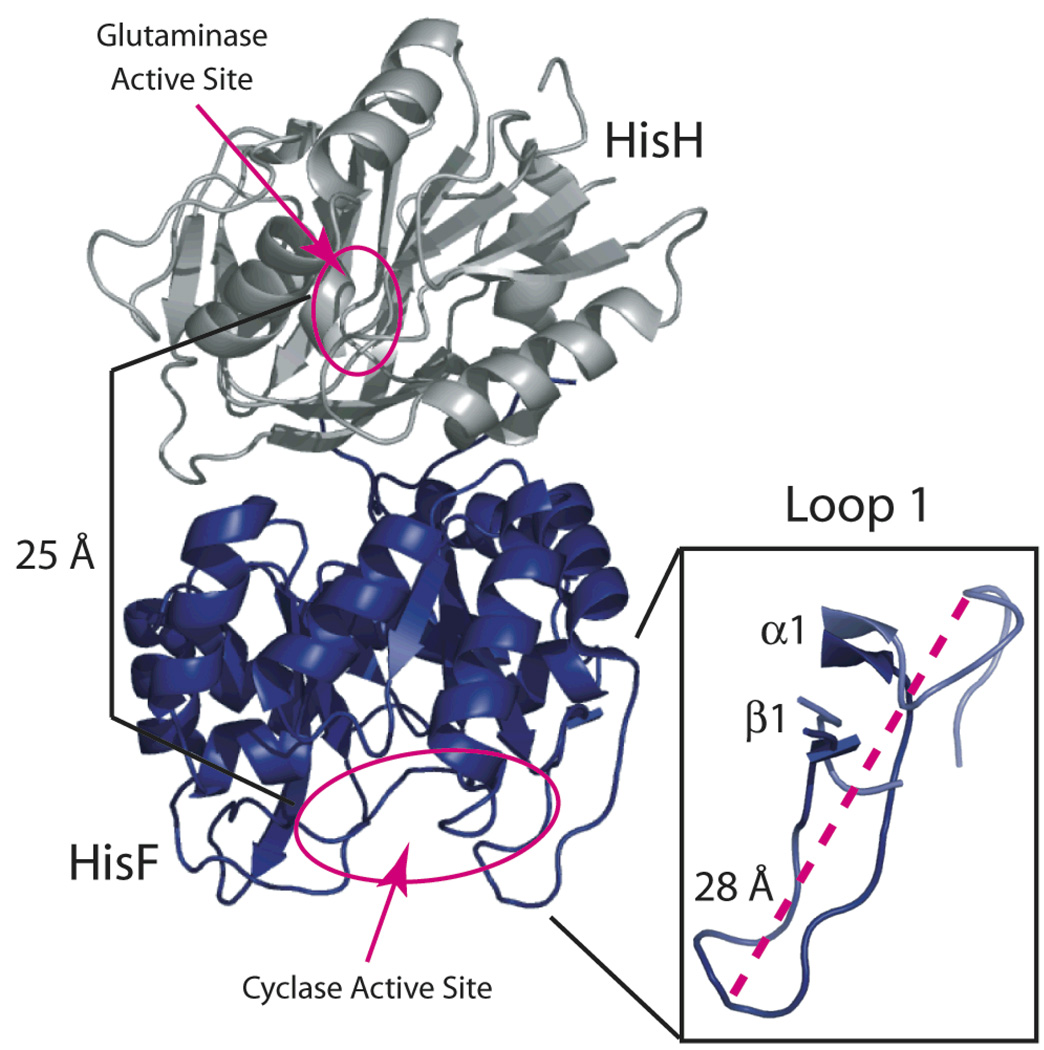
Structure of IGPS. Ribbon representation of the X-ray structure of (Left) the HisF subunit of IGPS shown in blue and HisH shown in gray. The two active sites are circled in magenta. On the right a close-up view of the loop 1 conformation with (blue) and without (light blue) inorganic phosphate bound to the HisF active site; the dashed magenta line shows the 28 Å movement of the Cα carbon of Asn25. The coordinates were from the 2.40 Å X-ray structure (1GPW) (Douangamath et al. 2002) and depicted in MacPymol (DeLano 2005).
The HisH glutaminase enzyme is a member of the type I glutamine amidotransferase (GAT) family and possesses a flavodoxin-like tertiary structure (Zalkin et al. 1998). The active site of HisH is comprised of three residues that form a conserved catalytic triad of Cys84-His178-Glu180. The mechanism of ammonia production from Gln involves (1) thioester bond formation between the Gln substrate and Cys84 and (2) hydrolysis to release NH3 and glutamate (Chittur et al. 2001). In order to be an efficient nucleophile in the cyclase reaction, NH3 must be sequestered from solvent to prevent rapid conversion to NH4+ in aqueous solution. To facilitate the necessary transfer of NH3 to the cyclase active site, the sequential HisH and HisF reactions are tightly coupled, in part by placing the HisH active site in close proximity to the HisF/HisH interface (Figure 1). The ammonia molecule produced by HisH enters the interface between HisF and HisH where it encounters a gate formed by four universally conserved charged side chains from residues within HisF. The four residues (Arg5, Glu46, Lys99, and Glu167) are linked by salt bridges, which are believed aid in the exclusion of water from the β-barrel core (Amaro et al. 2005). It is not clear whether these gate residues move to allow NH3 to pass or if NH3 can simply go around the gate (Douangamath et al. 2002; Amaro et al. 2005). There is no experimental data regarding the role of motions in the access of NH3 to the ammonia tunnel. Nevertheless, computational and structural studies (Chaudhuri et al. 2001; Douangamath et al. 2002; Amaro et al. 2007) as well as homology to other amidotransferases with established NH3 tunneling (Rudolph et al. 1995; Krahn et al. 1997; Thoden et al. 1997) support a HisF/H model in which NH3 passes the gate and enters a hydrophobic tunnel formed by the β-strands of the HisF (β/α)8 barrel (Figure 1).
The hydrolysis of glutamine and its subsequent reaction with PRFAR are tightly coupled chemical reactions despite the two active sites being separated by >25Å and residing on different polypeptide chains (Figure 1). The kinetic mechanism is random sequential, indicating no preferred order for the binding of Gln or PRFAR (Myers et al. 2003). The tightly coupled nature of the two active sites was demonstrated by kinetic studies that showed negligible hydrolysis of Gln by HisH in the absence of bound ligand at the HisF active site (Klem et al. 1993; Myers et al. 2003). In the presence of the substrate, PRFAR, a 5300-fold increase in basal Gln hydrolysis is observed (Myers et al. 2003). This active site synchrony ensures that Gln is not wasted by needless hydrolysis in the absence of PRFAR and results in a 1:1 stoichiometry for the HisH:HisF reactions. Additionally, it has been found that the product IGP has a reduced, but significant, effect on the basal levels of Gln hydrolysis, resulting in a 110-fold increase in kcat/Km (Myers et al. 2003). Together these data suggest a model in which conformational changes propagate from one active site to the other upon substrate binding in HisF to enable catalysis. One such necessary rearrangement identified in HisH is a proposed flipping of the backbone carbonyl oxygen of Gly50 to stabilize the oxyanion tetrahedral intermediate that forms after nucleophilic attack of Cys84 and prior to hydrolysis (Chaudhuri et al. 2001; Sinha et al. 2004). Several mutations in HisF have also been identified that alter active site coupling and yield reaction stoichiometries ranging from 2:1 to greater than 100:1 ratios for Gln hydrolysis:IGP/AICAR production. A conformational change that could be involved in the allosteric coupling between active sites is hinted at from x-ray crystallography experiments in the presence and absence of PO43−, a competitive inhibitor that mimics the effects of PRFAR binding due to the two phosphate moieties present in the substrate (Scheme 1). In the presence of phosphate, loop1 in HisF (residues 19–31) occupies a closed conformation (Figure 1b); however, in the absence of active-site ligand the Cα of Asn25 moves 27 Å in a direction away from the PRFAR binding site creating an open loop conformation. Flexibility in this region is further supported by a lack of electron density for regions of loop 1. Additionally, mutation of a universally conserved residue located in loop 1, Lys19, to an arginine or alanine results in an increase in the reaction stoichiometry to 3:1 and 43:1, respectively (Myers et al. 2003). The details of how ligand binding at the HisF active site activates the chemical reactivity of the HisH active site, which is over 25 Å away, are unknown.
Here we use solution NMR to begin to address these questions. We focus on IGPS from T. maritima for four primary reasons. (1) This enzyme is highly homologous to those from bacteria and fungi. (2) Initial structural and kinetic analysis has been completed on the system and provides a framework for a more detailed study of the mechanism of long-range communication in proteins. (3) IGPS from T. maritima is particularly stable at higher temperatures resulting in narrower NMR resonances. (4) The proteins HisF and HisH can be expressed, purified, and isotopically labeled individually to produce simplified NMR spectra compared to their single-polypeptide chain counterparts. Here we investigate the details of allosterism in IGPS by NMR methods on monomeric HisF and heterodimeric HisF (HisF-IGPS) in both the apo and IGP bound forms
Materials and methods
Protein expression and purification
HisF from T. maritima was expressed, purified and prepared for NMR study as previously described (Lipchock et al. 2008). The full length gene for HisH was cloned from T. maritima genomic DNA into the pET43.1b(+) vector between the NdeI and XhoI restriction sites to yield a C-terminally histidine tagged HisH construct. For routine preparations, the C-His-HisH plasmid was transformed into Rosetta(DE3) cells (Novagen) and 1.0 mL of a 20 mL LB overnight culture was used to inoculate 1.0 L of LB with 100 µg/mL ampicillin. Cells were allowed to grow at 37°C until the OD600 reached 0.9, at which point protein expression was induced with 1.0 mM IPTG. Cells were harvested by centrifugation after four hours and lysed by sonication in 5 mL lysis buffer (10 mM Tris pH 7.5, 10 mM CAPS, 0.3 M NaCl, 1 mM β-mercaptoethanol) per gram of wet cells. The cell debris was removed by centrifugation at 10,500 × g for 30 minutes. The supernatant was applied to a Ni/NTA (Qiagen) gravity column and was washed with 10 mM Tris pH 9.5, 10 mM CAPS, 0.3 M NaCl, 1 mM β-mercaptoethanol and 10 mM imidazole. Purified HisH was eluted with 10 mM Tris pH 9.5, 10 mM CAPS, 1 mM β-mercaptoethanol and 250 mM imidazole and dialyzed into 10 mM HEPES pH 9.5 and 10 mM glycine. The isolated HisH protein was complexed with HisF immediately after dialysis. Complex formation was achieved by binding an excess of purified HisH to a Ni gravity column and then adding purified labeled HisF. The column was then sealed and allowed to rotate at 4°C for 30 minutes. Bound protein was washed and eluted as above. NMR samples were dialyzed into 10 mM HEPES pH 9.5, 10 mM KCl, 0.5 mM EDTA, 1 mM DTT, 5% D2O and concentrated to ~1 mM and carefully adjusted to pH 7.3 with 1 M HCl to precipitate excess HisH, which was subsequently removed by centrifugation.
NMR Assignments
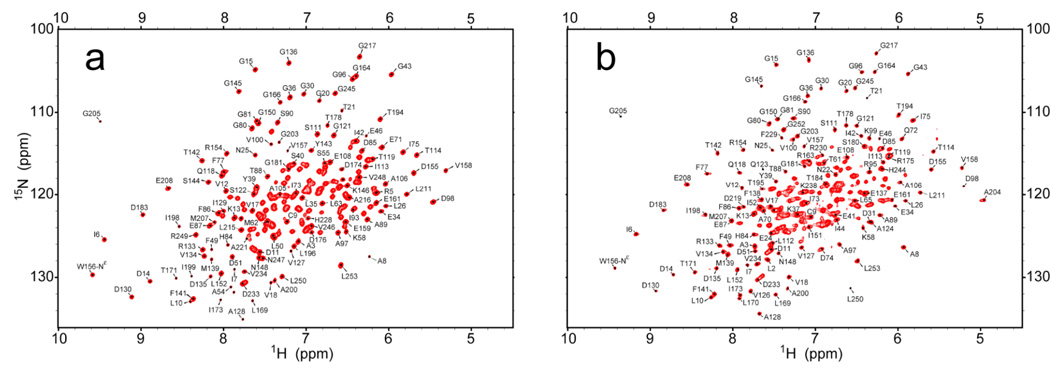
All NMR data were collected on Varian 600 and 800 MHz spectrometers at 30°C equipped with pulsed field gradients and triple resonance probes. Spectra were processed using NMRPipe (Delaglio et al. 1995) and analyzed with Sparky (Kneller et al. 1993). Backbone resonances for perdeuterated 15N HisF complexed with unlabeled HisH (HisF-IGPS) were assigned by comparing 1H-15N two-dimensional TROSY spectra for the assigned HisF enzyme with IGPS (HisF/HisH complex). Ambiguities were resolved by collecting a TROSY-based HN(CA)CB experiment (Pervushin et al. 1997) on a perdeuterated 15N, 13C HisF sample complexed with unlabeled HisH. 97% and 87% of the backbone resonances were assigned in the HisF and HisF-IGPS forms, respectively.
R2 measurements
Transverse relaxation rates (R2) were measured using the TROSY-detected R2 experiment (Zhu et al. 2000). Experiments for perdeuterated 15N HisF were collected at 14.1 T at 30°C in 10 mM MES pH 6.7 with 10 mM KCl, 1.0 mM EDTA, 1.0 mM DTT and 5% D2O. The 1H spectral width was set to 10,000 Hz and the 1H carrier frequency centered on the water resonance. The 15N carrier frequency was centered at 120 ppm with a 15N spectral width of 2,400 Hz. 24 transients were collected for each of 128 t1 points. The recycle delay was set to 2.0 s. R2 values were measured from the mono-exponential decay of peak heights collected with eight different relaxation delays: 0.0, 9.5, 19.0, 38.0, 57.0, 76.0, 114.0 and 190.0 ms. Error analysis was achieved by collecting duplicate spectra with relaxation delays of 9.5 and 76.0 ms. Peak heights were measured using a modified Sparky ‘rh’ command which utilizes a 3×3 grid placed on the peak center.
Experiments for perdeuterated 15N HisF-IGPS were performed at 14.1 T at 30°C in 10 mM KH2PO4 H 7.2 with 1.0 mM EDTA, 1.0 mM DTT and 5% D2O. The 1H frequency was centered at the water resonance with a spectral width of 7,500 Hz. The 15N carrier frequency was centered at 120 ppm with a 15N spectral width of 2,600 Hz. 32 transients were collected for each of 128 t1 points. The recycle delay was set to 2.0 s. R2 values were measured from peak heights measured in spectra collected with two relaxation delay times: 0.0 and 18.9 ms. Experiments were collected in an interleaved fashion for a total of 10 spectra (3 × 0.0 ms, 7 × 18.9 ms). R2’s were calculated using the constant time approximation
| (1) |
in which T is the total relaxation time, 19.8 ms in this case and IT/0 are the NMR peak intensities, measured as above with a modified Sparky ‘rh’ command, for relaxation times T ms and 0 ms (reference spectrum), respectively.
Detection of µs – ms motions
Microsecond-millisecond protein dynamics were identified and characterized using an interleaved pseudo-four-dimensional TROSY relaxation-compensated CPMG experiment (Loria et al. 1999) with a heat compensation pulse train during the recycle delay. To quantitate the contribution of µs-ms motions to the of transverse relaxation rate, R2 was measured as a function of τcp, the interpulse delay in the CPMG pulse train. Because only a single static magnetic field strength was used and no attempt was made to quantitate chemical shift differences (Δω) or populations (pA/B) of conformationally exchanging sites the simplified, fast-limit expression was used to fit to the relaxation dispersion data (Luz et al. 1963).
| (2) |
In equation (2) kex is the sum of the forward and reverse exchange rate constants, , τcp is the delay between 180° refocusing pulses, and R2 (τcp → ∞) is the transverse relaxation rate in the limit of infinitely fast pulsing. Transverse spin-relaxation data using the rcCPMG experiment for the apo and IGP bound forms of monomeric HisF were acquired at 600 MHz with interpulse delays, τcp, of 0.0, 0.625, 0.714 (x2), 1.0, 1.25, 1.67, 2.0, 2.50 (x2), 3.33, 5.0, and 10.0 ms during the nitrogen relaxation period for a constant, total relaxation time of 40 ms (Mulder et al. 2001). Data for the apo and IGP-bound forms of HisF-IGPS were acquired at 800 MHz with the relaxation period as described above for monomeric HisF. Due to reduced signal-to-noise, additional relaxation data for IGP-bound HisF-IGPS were collected at 800 MHz with interpulse delays of 0.625 and 6.25 ms for a total relaxation delay of 25 ms. Dispersion analysis was completed for the monomeric and apo-complexed forms of HisF by fitting Equation (2) to the relaxation data using in-house processing scripts as described previously (Beach et al. 2005).
IGP Titrations
IGP (Toronto Research Chemicals) was dissolved in the appropriate NMR buffer and the pH adjusted to 6.7 for titration to monomeric HisF and 7.3 for HisF-IGPS. For HisF, 1H-15N two-dimensional TROSY spectra were collected at the following concentrations: 0.1, 0.2, 0.5, 1.0, 2.0, 5.0 and 10.0 mM IGP. For HisF-IGPS, spectra were collected at 0.6, 1.4, 2.8, 4.1, 6.8, 9.3 and 11.8 mM IGP. IGP titrations for both HisF and HisF-IGPS were preformed at 30° C at 14.1 T. For 2H,15N HisF, 32 transients were collected for each titration point. For 2H,15N HisF-IGPS, 48 transients were acquired due to the reduced signal-to-noise of the larger HisF-IGPS molecule. In both HisF and HisF-IGPS, saturation by IGP was noted by monitoring the shift of NMR resonances upon increasing [IGP] until no further shift was noted. Dissociation constants for IGP were estimated from the NMR titration series with IGP. This value was determined for HisF and HisF-IGPS by a global fitting of data from multiple amino acid residues that experience chemical shifts greater than 1σ from the protein-wide average value.
Results
NMR Assignments and HisF-IGPS formation
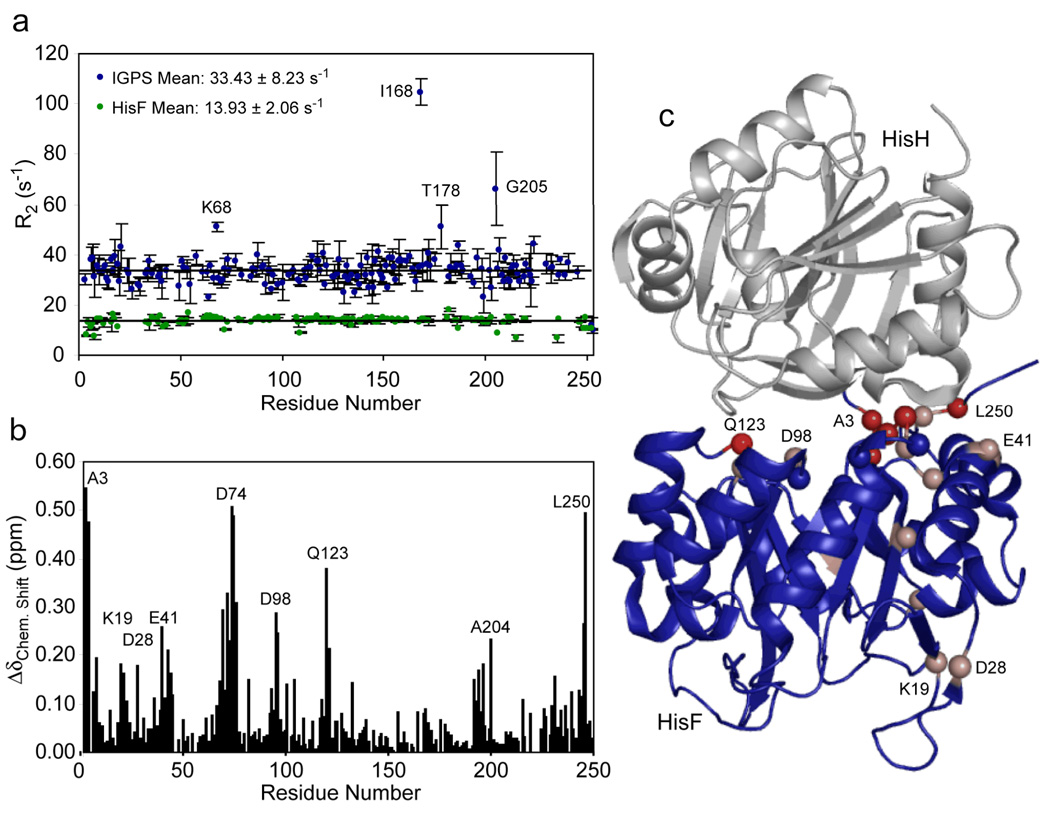
The amino acid assignments for 97% of the backbone 1H-15N resonances of HisF haven been determined as previously published (Lipchock et al. 2008) (Figure 2). The heterodimer HisF-IGPS complex was formed as described in the methods section and was confirmed by tryptophan fluorescence spectroscopy and gel filtration (not shown). As shown in Figure 3a, the 90% trimmed mean R2 for 15N-HisF increases from 13.93 ± 2.06 s−1 to 33.43 ± 8.23 s−1 in the presence of stoichiometric amounts of HisH, indicative of an increased rotational correlation time and formation of HisF-IGPS. The resonances for 87% of the backbone 1H-15N resonances of HisF-IGPS have also been assigned as described in the methods section (Figure 2). Mapping of chemical shift perturbations upon formation of IGPS yields a patch of residues with ΔδC.S. > 0.4 ppm localized at the proposed HisF/HisH interface (Figure 3b–c). However, residues as far as 25 Å from the HisH/HisF interface experience ΔδC.S. greater than one standard deviation from the average. Of particular interest is residue Lys19, which has a Δδc.s. = 0.18 ppm and has been demonstrated by mutagenesis and x-ray crystallography to be involved in a ligand binding-induced conformational change. Chemical shift data suggests that the HisH binding platform is comprised primarily of residues in the loop connecting α2/β3 and the N- and C-terminal tails of HisF. Interestingly, five of the twelve residues at the interface for which significant chemical shift perturbations are measures are charged (Lys4, Glu41, Asp74, Asp98, Arg249). Additionally, Gln123, which has been implicated in a hinging mechanism for the glutaminase activation in HisH, experiences a Δδc.s. = 0.38 ppm, despite being over 15 Å from the region where the residues with the largest chemical shift perturbations are concentrated.
Figure 2.
1H-15N TROSY NMR spectrum of HisF in (a) as a monomer and (b) in 1:1 complex with HisH to form HisF-IGPS. Selected amino acid assignments are shown on each. The spectra were acquired at 14.1 T, 303 K at pH = 7.3. Each spectrum is the result of 128 t1 and 512 t2 points with spectral widths of 2500 and 10,000 Hz respectively.
Figure 3.
Formation of the IGPS heterodimer. (a) Transverse relaxation rates (R2) as a function of amino acid sequence for HisF (green) and HisF in 1:1 with HisH (HisF-IGPS) (blue). In (b) chemical shift changes in HisF upon interaction with 1 molar equivalent of HisH calculated using (Grzesiek et al. 1996). Values for Δδ between 1 and 4 standard deviations above the average are shown as a gradient from light to dark red spheres.
IGP Titration
The effects of ligand binding on glutaminase activity in IGPS have previously been studied and revealed increased glutamine hydrolysis in the presence of PRFAR, as well as the two reaction products AICAR and IGP (Myers et al. 2003). While the increase in glutaminase activity for HisH is greatest for PRFAR binding (kcat/KM increases 5300-fold relative to basal levels), AICAR and IGP increase kcat/KM for glutamine hydrolysis relative to basal levels by 2.6 and 110-fold, respectively. To probe the structural and dynamic effects that ligand binding has on HisF-IGPS, IGP was titrated into perdeuterated 15N-labeled HisF and HisF-IGPS. For monomeric HisF, a global fit of 25 residues with Δδ values greater than 1σ from the average yielded an estimate of the IGP dissociation constant of 2.8 ± 0.3 mM. For HisF-IGPS, 19 residues were fit globally to obtain a Kd estimate = 0.5 ± 0.1 mM.
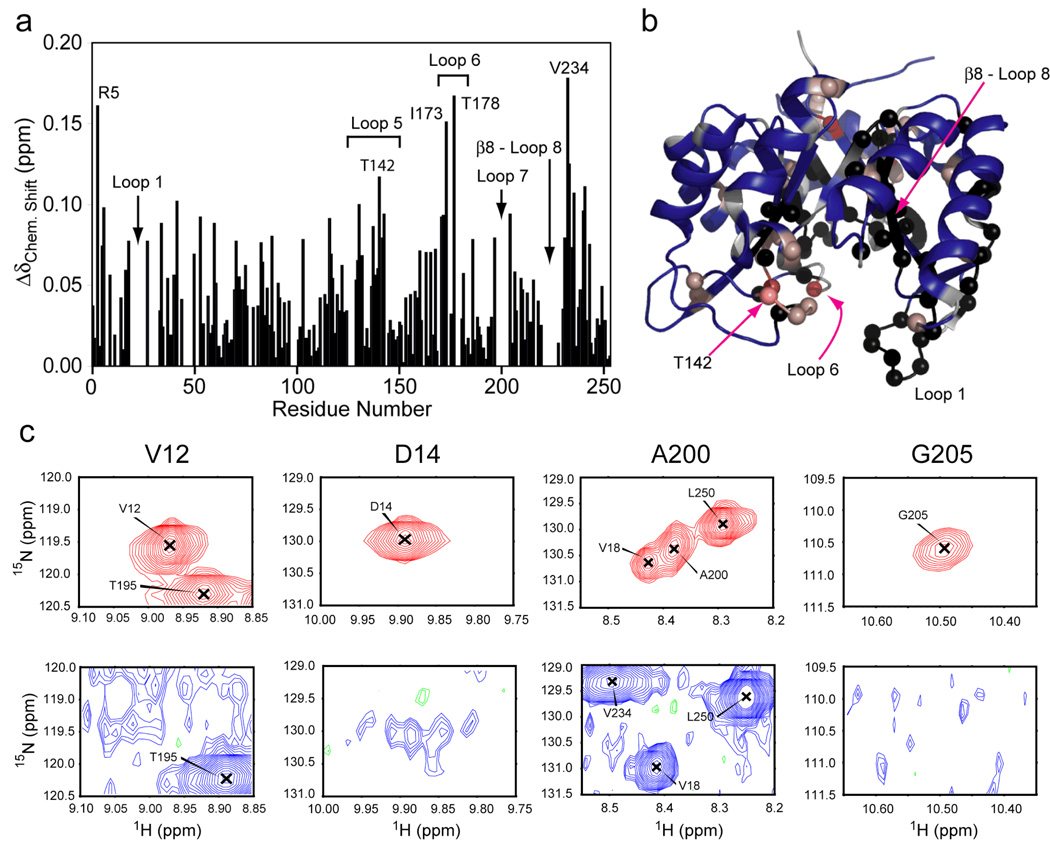
As shown in Figure 4, significant line broadening occurs in HisF upon titration with IGP, preventing the observation of 55 backbone resonances previously present in the apo form (Figure 4). These residues are not solely localized to the IGP binding site (loops 7 and 8), but rather spread throughout the cyclase active site with nearly all of loop 1 experiencing severe broadening. Additionally, in rather striking fashion resonance broadening extends from the cyclase active site into the β-sheets that comprise the core of the protein with significant broadening in β1–4, β7 and β8. This path of broadened residues provides a direct network that connects the cyclase active site of HisF with HisH. Figure 4 also shows that, like the case for complex formation, chemical shift perturbations upon binding of IGP are not localized to the expected binding site, but rather extend to opposite ends of the protein with two of the four largest shifts occurring for residues Arg5 and Val234 located at the HisF/HisH interface. Of additional significance is the fact that Arg5 is one of the four universally conserved residues that form salt bridges at the opening of the HisF β-barrel believed to function as a gate for ammonia tunneling.
Figure 4.
Interaction of imidazole glycerol phosphate (IGP) with HisF. (a) 1H/15N combination chemical shift changes in HisF upon saturation with IGP. Chemcial shift changes were determined as in Figure 3. (b) Composite chemical shift changes were mapped onto the structure of HisF. Residues with Δδ values between 1 and 3 standard deviations from the average are shown in a gradient from light to dark red spheres. In (c) selected examples of IGP binding-induced exchange broadening are shown for residues 12, 14, 200, and 205. Resonances from the apo enzyme are shown in red (positive peaks) and green (negative peaks). Resonances from the IGP form are shown in blue (positive) and green (negative) with a 50-fold decrease in the contour level.
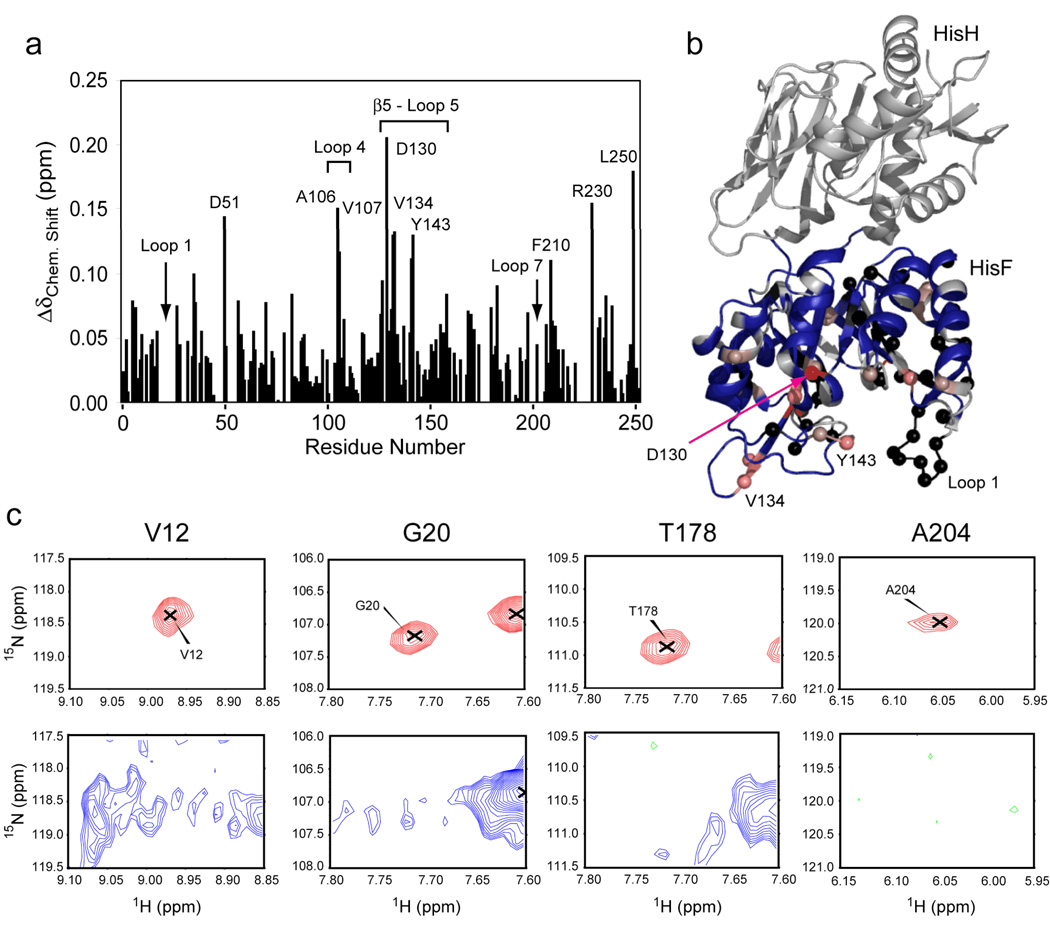
Titration of IGP with 15N-labeled HisF-IGPS yields a similar result with 38 residues becoming broadened beyond detection (Figure 5). Similarly, resonances in loops 1 and 7 are lost to exchange broadening, however, β1 and β4 experience reduced broadening effects. Broadening effects to β2 and β8 are largely unknown due to a lack of assignments for these amino acid sites in 15N-HisF-IGPS. Chemical shift perturbations are similar to HisF, however, and extend beyond the cyclase active site to the protein interface.
Figure 5.
Interaction of imidazole glycerol phosphate (IGP) with HisF-IGPS. (a) 1H/15N combination chemical shift changes in HisF upon saturation with IGP calculated as in figures 3 and 4. (b) Chemical shift changes were mapped onto the structure of HisF for residues with standard deviations of 1–4 from the average in a gradient of light to dark red. In (c) selected examples of IGP binding-induced exchange broadening is shown for residues 12, 20, 178, and 204. Resonances from the apo enzyme are shown in red (positive peaks) and green (negative peaks). Resonances from the IGP form are shown in blue (positive) and green (negative) with a 25-fold decrease in contour level.
Protein Dynamics
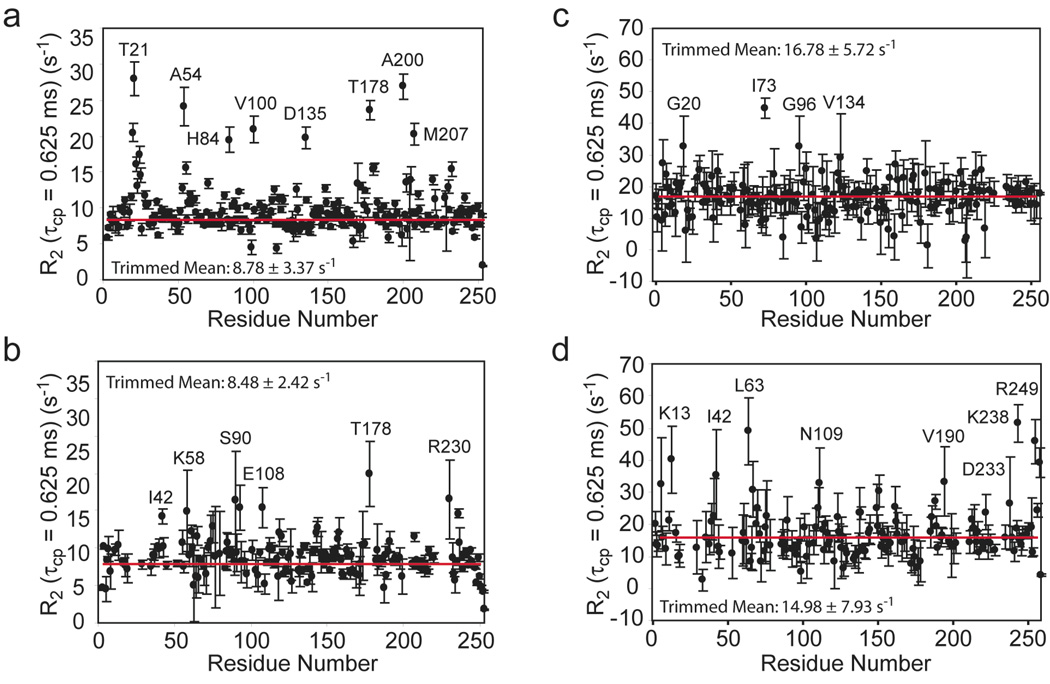
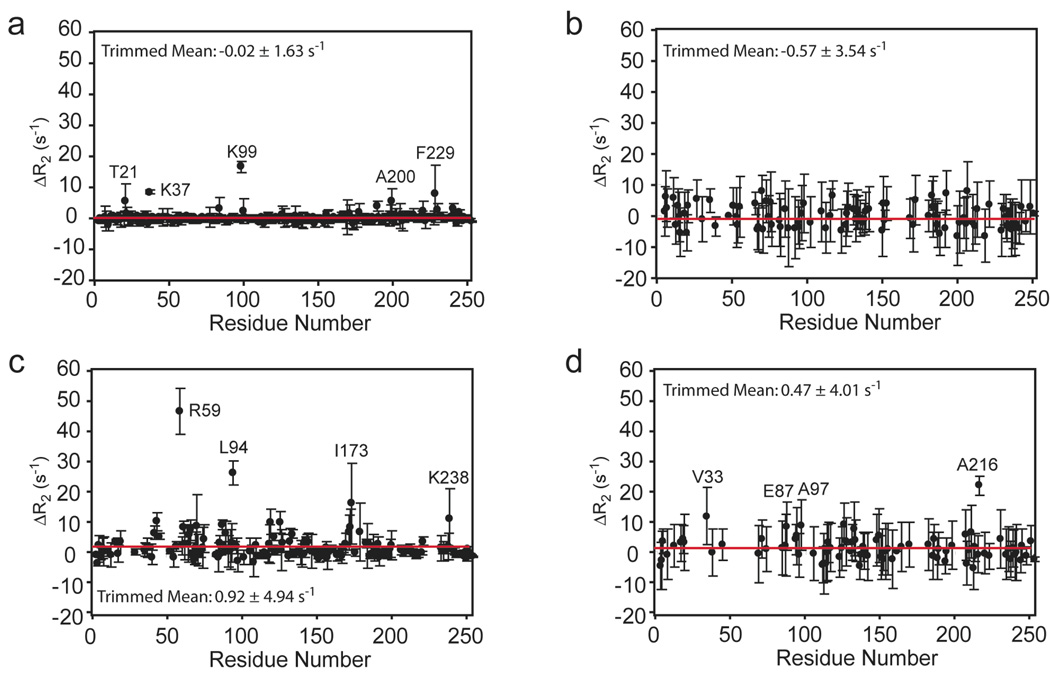
Transverse relaxation rates with a τcp value = 0.625 ms have been measured for HisF and HisF-IGPS in the presence and absence of IGP (Figure 6). In these measurements R2,in and R2,anti are the in-phase 15N and antiphase 1H/15N transverse relaxation rates and Rex is the additional contribution due to conformational exchange effects. Elevated values are observed for numerous residues suggesting the possible presence of µs-ms motions in these regions for apo HisF, IGP-bound HisF, apo HisF-IGPS and IGP-bound HisF-IGPS. To further investigate the possibility of µs – ms motions in IGPS, values were measured at short and long spin-echo pulse delays (Loria et al. 1999) to reveal any differences between the two and thereby identify residues involved in a conformational exchange process (Figure 7). For apo HisF five residues exhibit significantly elevated ΔR2: Thr21, Lys37, Lys99, Ala200 and Phe229 (Figure 7a). Of particular note, are Thr21 and Lys99. Thr21 is located in loop 1, believed to be involved in a ligand induced conformational change. Lys99 is one of four universally conserved residues that form salt bridges at the entrance into the HisF β-barrel at the HisF/H interface. Biochemical studies reveal that mutation of Lys99 leads to a dramatic alteration in the PRFAR:Gln reaction stoichiometry. In IGP-bound HisF, four out of the five of these residues become broadened beyond detection, additionally, several residues near the HisF/H interface and in loop 6 acquire elevated ΔR2 (Figure 7c).
Figure 6.
Residue specific values. Data are shown for (a) HisF, (b) HisF + IGP, (c) HisF-IGPS, and (d) HisF-IGPS + IGP. Measurements were made as described in Materials and Methods. IGP was saturating (10 × Kd) in panels (b) and (d). The horizontal red line indicates the 10% trimmed mean of the data points. Amino acid residues with significantly elevated values are indicated.
Figure 7.
Exchange contribution to . Residue specific ΔR2 values are depicted for (a) HisF, (b) HisF + IGP, (c) HisF-IGPS, and (d) HisF-IGPS + IGP. The horizontal red line indicates the 10% trimmed mean of the data points. Amino acid residues with significantly elevated values are indicated. Missing data points are largely due to significant exchange broadening that precludes measurement.
In the IGPS heterodimer, the majority of apo HisF backbone amide sites do not exhibit Rex > 0 s−1 (Figure 7b). However upon binding of IGP the trimmed mean ΔR2 increases slightly from −0.57 ± 3.54 s−1 to 0.47 ± 4.01 s−1 and at least two residues (Val33 and Ala216) have values elevated above the average. Given that 18% of amide positions are exchange broadened by IGP binding, this slight overall increase in ΔR2 is a lower limit of the total effect (Figure 7d).
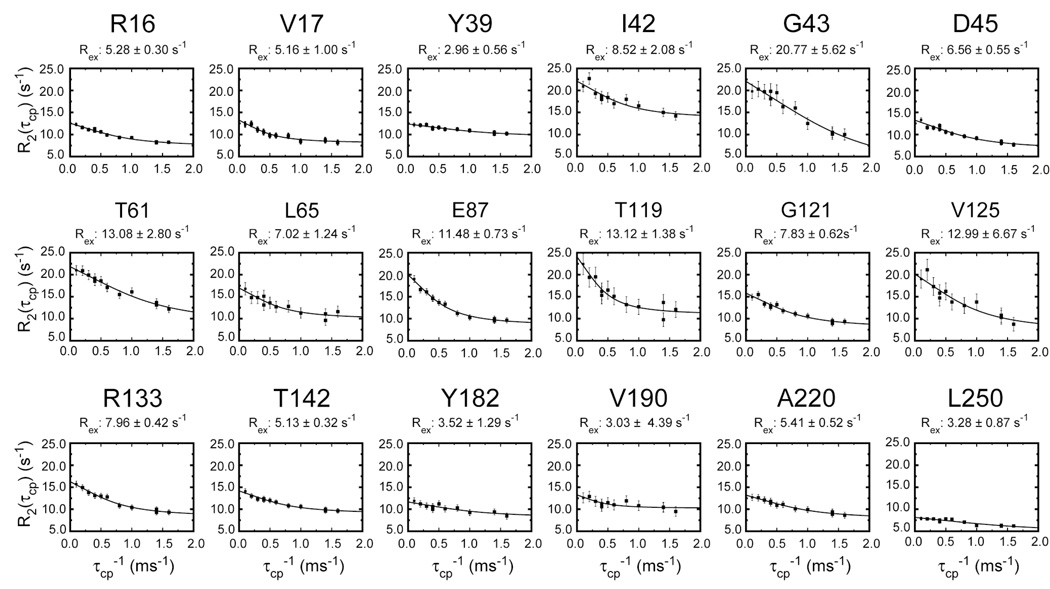
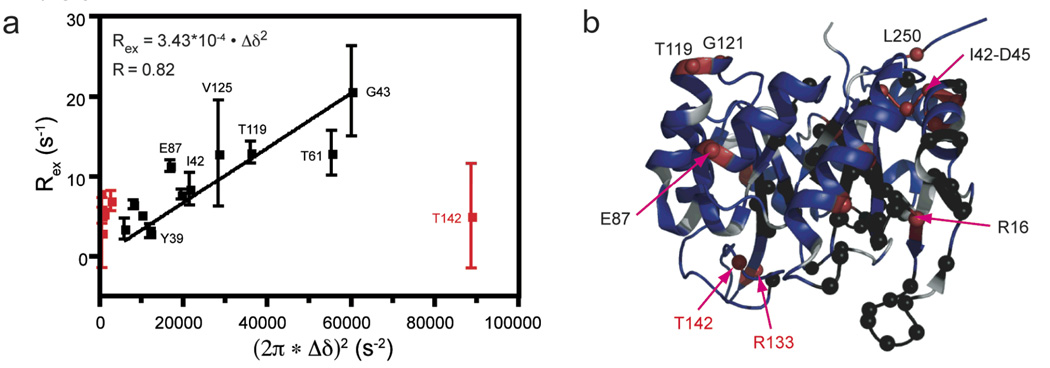
To provide a more complete view of µs-ms motions in IGPS, TROSY-CPMG based dispersion analysis was completed on the apo and IGP bound forms of HisF and HisF-IGPS as described in the methods (Figure 8). Data for IGP bound IGPS proved to have too low a signal-to-noise ratio and too severe exchange broadening for meaningful analysis. Measurements on both apo forms revealed no residues with measurable dispersion; however, the IGP saturated HisF enzyme yielded 18 residues amenable to dispersion analysis. These residues are located throughout the protein, but are concentrated in loop 1, the region connecting α1 and β2, α3 near the PRFAR binding site, and α4 at the HisF/H interface. Furthermore, when the Rex term measured from dispersion analysis using the fast-limit equation (2) is plotted versus the square of the 15N shifts ((2π × Δδ)2) between apo HisF and IGP bound HisF, a linear relationship with a obtained with a correlation coefficient of 0.82, excluding residues Thr142, Val17, Leu65, Val190, and Ala220 (Figure 9). T142 is located in the cyclase active site and has a significantly increased value for Δδ, relative to the other residues plotted in Figure 9. We hypothesize that Δδ is elevated for this value because of direct interaction with IGP, which precludes its inclusion in this analysis as the Δδ value measured is a combination of conformational and chemical effects. Val17, Leu65, Val190 and Ala220 were excluded because the Δδ values measured were considered too small (0.017–0.131 ppm) relative to the Rex measured to yield physically meaningful populations and exchange rates relevant to the IGP binding process. We believe these residues must be reporting on a different conformational exchange process for which Δω values are currently unknown. For the remaining 13 residues in Figure 9, given that this linear relationship indicates that papb/kex is constant and the exchange motion for this set of nuclear spins is a single, global process (Massi et al. 2006) between an IGP bound conformation and an apo-like conformation, even in the presence of IGP.
Figure 8.
CPMG dispersion curves for HisF. TROSY-based CPMG (Loria et al. 1999) dispersion curves were determined for 18 residues in apo HisF. Equation (2) was fit to each relaxation series to determine Rex, (pApBΔω2/kex), for each residue. The residue specific value of Rex is shown above each graph.
Figure 9.
Conformational exchange in HisF. Rex values determined for apo HisF from the data in Figure 8 were plotted against the square of the 15N HisF chemical shift changes (Δδ2) due to IGP binding. A linear fit was applied to the black data points, Thr142 being excluded from this fit. All data points with measurable dispersion curves are shown as red spheres in panel b with black spheres indicating exchange broadened amide sites due to IGP binding.
Discussion
The Monod-Wyman-Changeaux (MWC) (Monod et al. 1965) and Koshland-Nemethy-Filmer (KNF) (Koshland et al. 1966) models of protein allostery have proven useful for describing many experimental observations. Nonetheless the mechanistic details of allosterism have only been described for a limited number of proteins (Schachman 1988; Eaton et al. 1999; Kimmel et al. 2000). Recent advances in solution NMR spectroscopy have provided further insight into the mechanism of allosterism (Stevens et al. 2001; Volkman et al. 2001; Velyvis et al. 2007; Boyer et al. 2008; Bhattacharya et al. 2009; Bruschweiler et al. 2009), but much remains to be done. IGPS differs from these previous examples in that the allosteric activation by PRFAR or IGP essentially ‘turn-on’ an enzyme with negligible catalytic activity and the activation results in synchronization of the two active sites such that the net stoichiometry of Gln hydrolysis and AICAR/IGP synthesis is 1:1.
There are several observations that suggest a possible role for protein motions in the allosteric properties of IGPS. a) Upon formation of heterodimeric IGPS there is an overall decrease in residues experiencing ms motions and there are significant protein-wide changes in chemical shifts. These changes appear to be mainly of dynamic origin because the x-ray crystal structure for HisF and HisF-IGPS differ by only 0.67 Å (Lang et al. 2000; Douangamath et al. 2002). b) The binding of IGP, which stimulates glutaminase activity by two orders of magnitude, results in an increase in conformational exchange for both HisF and HisF-IGPS as evidenced by severe line broadening of 23% and 18% of assigned residues, respectively. Additionally, CPMG dispersion analysis reveals an increase in µs – ms motions throughout HisF upon IGP binding, including residues in loop 1 and in the loops comprising the interface with HisH between α1-β2 and α2-β3. These data establish a dynamic link between the active site of HisF and its interface with HisH. Plotting Rex for residues with measurable dispersion curves in IGP bound HisF with respect to the 15N (2π × Δδ)2 values for the IGP titration yields a linear slope, indicative of a single, global process. This relationship suggests that the Rex measured reflects a two-state conformational exchange between the apo and a conformation that resembles the IGP bound state that can work in a concerted fashion to relay binding in the cyclase active to the HisF/H interface, providing a framework for activation of the glutaminase active site in HisH. The time scale of these motions as estimated from a global analysis of the CPMG dispersion data in Figure 8 is 2300 ± 160 s−1 (not shown) or based on the slope in Figure 9 and an estimate of papb is ~500 s−1. In either case, the time scale is faster than the overall catalytic rate constant indicating that these motions would not present a bottleneck to enzyme turnover. Dispersion experiments at a second static magnetic field will resolve this ambiguity.
Despite this progress, the allosteric mechanism in IGPS remains to be determined. The observation that binding of HisH to HisF results in quenching of motions in HisF is consistent with the KNF model of allosterism that requires consideration of a subunit-subunit interaction constant, K (Koshland et al. 1966). However, the CPMG relaxation dispersion data that suggest a propensity of HisF to exchange between ligand bound and ligand free conformations is consistent with the MWC model (Monod et al. 1965), though not exclusive of the KNF model of allosterism (Yu et al. 2001). Lastly as noted, several mutations disrupt the 1:1 stoichiometry between the two active sites. All of these mutations are at or near sites that exhibit flexibility, either evidenced by CPMG dispersion profiles or by exchange broadening. To address these questions additional experiments to examine the effects of PRFAR and to investigate how ligand binding alters the physical properties of HisH are required.
Acknowledgements
JPL acknowledges support from the NIH (R01-GM070823), JML was supported by an NIH biophysical training grant (5T32GM008283). We thank Joanna Dunn (Yale University) for help in constructing His-tagged HisH and Simon Lipchock for delaying his birth until after completion of this manuscript. This manuscript is published in Journal of Biomolecular NMR (2009) volume 45, pages 73–84. The original publication is available at www.springerlink.com.
Abbreviations
- AICAR
5-aminoimidazole-4-carboxamide ribotide
- CPMG
Carr-Purcell-Meiboom-Gill
- HisF-IGPS
isotopically labeled HisF in complex with HisH
- IGP
Imidazole glycerol phosphate
- IGPS
Imidiazole glycerol phosphate synthase (a heterodimer of HisF and HisH)
- PRFAR
N’-[(5’-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide-ribonucleotide
References
- Amaro RE, Myers RS, Davisson VJ, Luthey-Schulten ZA. Structural elements in IGP synthase exclude water to optimize ammonia transfer. Biophys J. 2005;89:475–487. doi: 10.1529/biophysj.104.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro RE, Sethi A, Myers RS, Davisson VJ, Luthey-Schulten ZA. A network of conserved interactions regulates the allosteric signal in a glutamine amidotransferase. Biochemistry. 2007;46:2156–2173. doi: 10.1021/bi061708e. [DOI] [PubMed] [Google Scholar]
- Beach H, Cole R, Gill M, Loria JP. Conservation of µs - ms enzyme motions in the apo- and substrate-mimicked state. J. Am. Chem. Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Kurochkin AV, Yip GN, Zhang Y, Bertelsen EB, Zuiderweg ER. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JA, Lee AL. Monitoring aromatic picosecond to nanosecond dynamics in proteins via 13C relaxation: expanding perturbation mapping of the rigidifying core mutation, V54A, in eglin c. Biochemistry. 2008;47:4876–4886. doi: 10.1021/bi702330t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschweiler S, Schanda P, Kloiber K, Brutscher B, Kontaxis G, Konrat R, Tollinger M. Direct Observation of the Dynamic Process Underlying Allosteric Signal Transmission. J Am Chem Soc. 2009 doi: 10.1021/ja809947w. [DOI] [PubMed] [Google Scholar]
- Chaudhuri BN, Lange SC, Myers RS, Chittur SV, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites. Structure. 2001;9:987–997. [PubMed] [Google Scholar]
- Chittur SV, Chen Y, Davisson VJ. Expression and purification of imidazole glycerol phosphate synthase from Saccharomyces cerevisiae. Protein Expr Purif. 2000;18:366–377. doi: 10.1006/prep.2000.1207. [DOI] [PubMed] [Google Scholar]
- Chittur SV, Klem TJ, Shafer CM, Davisson VJ. Mechanism for acivicin inactivation of triad glutamine amidotransferases. Biochemistry. 2001;40:876–887. doi: 10.1021/bi0014047. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano WL. MacPyMOL: a PyMOL-based molecular graphics application for MacOS X. South San Francisco, CA, USA: DeLano Scientific LLC; 2005. [Google Scholar]
- Douangamath A, Walker M, Beismann-Driemeyer S, Vega-Fernandez MC, Sterner R, Wilmanns M. Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure. 2002;10:185–193. doi: 10.1016/s0969-2126(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Eaton WA, Henry ER, Hofrichter J, Mozzarelli A. Is cooperative oxygen binding by hemoglobin really understood? Nat Struct Biol. 1999;6:351–358. doi: 10.1038/7586. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 2000;97:3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem TJ, Chen Y, Davisson VJ. Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase. J Bacteriol. 2001;183:989–996. doi: 10.1128/JB.182.3.989-996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem TJ, Davisson VJ. Imidazole Glycerol Phosphate Synthase - the Glutamine Amidotransferase in Histidine Biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- Kneller DG, Kuntz ID. Ucsf Sparky - an Nmr Display, Annotation and Assignment Tool. Journal of Cellular Biochemistry. 1993:254–254. [Google Scholar]
- Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Krahn JM, Kim JH, Burns MR, Parry RJ, Zalkin H, Smith JL. Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry. 1997;36:11061–11068. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- Kuenzler M, Balmelli T, Egli CM, Paravicini G, Braus GH. Cloning, primary structure, and regulation of the HIS7 gene encoding a bifunctional glutamine amidotransferase: cyclase from Saccharomyces cerevisiae. J Bacteriol. 1993;175:5548–5558. doi: 10.1128/jb.175.17.5548-5558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- Lipchock J, Loria JP. 1H,15N, and 13C resonance assignment of imidazole glycerol phosphate (IGP) synthase protein HisF from Thermotoga maritima. Biomol. NMR Assign. 2008;2:219–221. doi: 10.1007/s12104-008-9125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG. A Relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- Loria JP, Rance M, Palmer AG. A TROSY CPMG Sequence for Characterizing Chemical Exchange in Large Proteins. J. Biomol. NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- Luz Z, Meiboom S. Nuclear magnetic resonance study of the protolysis of trimethylammonium ion in aqueous solution—order of the reaction with respect to solvent. J. Chem. Phys. 1963;39:366–370. [Google Scholar]
- Massi F, Wang C, Palmer AG., 3rd Solution NMR and computer simulation studies of active site loop motion in triosephosphate isomerase. Biochemistry. 2006;45:10787–10794. doi: 10.1021/bi060764c. [DOI] [PubMed] [Google Scholar]
- Massiere F, Badet-Denisot MA. The mechanism of glutamine-dependent amidotransferases. Cell Mol Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- Myers RS, Jensen JR, Deras IL, Smith JL, Davisson VJ. Substrate-induced changes in the ammonia channel for imidazole glycerol phosphate synthase. Biochemistry. 2003;42:7013–7022. doi: 10.1021/bi034314l. [DOI] [PubMed] [Google Scholar]
- Omi R, Mizuguchi H, Goto M, Miyahara I, Hayashi H, Kagamiyama H, Hirotsu K. Structure of imidazole glycerol phosphate synthase from Thermus thermophilus HB8: open-closed conformational change and ammonia tunneling. J Biochem. 2002;132:759–765. doi: 10.1093/oxfordjournals.jbchem.a003284. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Stubbe J. Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry. 1995;34:2241–2250. doi: 10.1021/bi00007a019. [DOI] [PubMed] [Google Scholar]
- Schachman HK. Can a simple model account for the allosteric transition of aspartate transcarbamoylase? J Biol Chem. 1988;263:18583–18586. [PubMed] [Google Scholar]
- Sinha SC, Chaudhuri BN, Burgner JW, Yakovleva G, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold. J Biol Chem. 2004;279:15491–15498. doi: 10.1074/jbc.M312733200. [DOI] [PubMed] [Google Scholar]
- Stevens SY, Sanker S, Kent C, Zuiderweg ER. Delineation of the allosteric mechanism of a cytidylyltransferase exhibiting negative cooperativity. Nat Struct Biol. 2001;8:947–952. doi: 10.1038/nsb1101-947. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of carbamoyl phosphate synthetase: a journey of 96 A from substrate to product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- Velyvis A, Yang YR, Schachman HK, Kay LE. A solution NMR study showing that active site ligands and nucleotides directly perturb the allosteric equilibrium in aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 2007;104:8815–8820. doi: 10.1073/pnas.0703347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- Yu EW, Koshland DE., Jr Propagating conformational changes over long (and short) distances in proteins. Proc Natl Acad Sci U S A. 2001;98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H, Smith JL. Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol. 1998;72:87–144. doi: 10.1002/9780470123188.ch4. [DOI] [PubMed] [Google Scholar]
- Zhu G, Xia Y, Nicholson LK, Sze KH. Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]