Abstract
Malaria is a major public health problem in many developing countries, with the malignant tertian parasite Plasmodium falciparum causing the most malaria-associated mortality. Extensive research, especially with the advancement of genomics and transfection tools, has highlighted the fundamental importance of chromatin-mediated gene regulation in the developmental program of this early-branching eukaryote. The Plasmodium parasite genomes reveal the existence of both canonical and variant histones that make up the nucleosomes, as well as a full collection of conserved enzymes for chromatin remodeling and histone posttranslational modifications (PTMs). Recent studies have identified a wide array of both conserved and novel histone PTMs in P. falciparum, indicating the presence of a complex and divergent “histone code.” Genome-wide analysis has begun to decipher the nucleosome landscape and histone modifications associated with the dynamic organization of chromatin structures during the parasite's life cycle. Focused studies on malaria-specific phenomena such as antigenic variation and red cell invasion pathways shed further light on the involvement of epigenetic mechanisms in these processes. Here we review our current understanding of chromatin-mediated gene regulation in malaria parasites, with specific reference to exemplar studies on antigenic variation and host cell invasion.
Malaria continues to be a major cause of mortality and morbidity in tropical countries, bringing a death toll of ∼1 million each yqear. Four human parasites (Plasmodium falciparum, P. vivax, P. malariae, and P. ovale) and a monkey parasite (P. knowlesi) have been found to infect humans naturally. P. falciparum causes the malignant form of malaria and is responsible for most malaria-associated human deaths. Intensified research in the past decade has greatly improved our understanding of the parasites and the disease they cause, especially with the sequencing of multiple parasite genomes. A striking finding from global microarray analyses of parasite gene expression is the tightly regulated transcription program during the parasite's life cycle, with genes expressed in a “just-in-time” manner (12, 93). Bioinformatic analysis has recognized a general conservation of the basal transcription machinery (15), but the scarcity of recognizable specific transcription factors in the parasite genome has led to speculation about the existence of a divergent transcription mechanism used by the malaria parasites (2, 25). However, this speculation may be inaccurate, as recent studies revealed the presence of the AP2 family of transcription factors (4, 35), which have undergone lineage-specific expansion in apicomplexan parasites (75). Nevertheless, the full complement of chromatin-modifying proteins encoded in apicomplexan parasite genomes underlines the significance of epigenetic mechanisms in transcription regulation (2, 66, 75, 140). While epigenetic mechanisms are being intensively studied in model eukaryotes, only recently have they been explored in the malaria parasites. Two recent reviews have provided an excellent update on transcription control in malaria parasites (22, 68). Here we focus on recent advances in chromatin-mediated epigenetic mechanisms in the malaria parasites.
MALARIA PARASITE HISTONES AND “HISTONE CODE”
The word “epigenetics,” coined by Conrad Waddington in an attempt to unite the genetics and development fields, was used to describe biological events that could not be explained by genetic principles (152). The contemporary term “epigenetics” has taken on a much broader meaning, referring to changes in phenotype or gene expression that are inheritable but are caused by mechanisms other than changes in the underlying DNA sequence (6). With a tremendous number of publications each year, the epigenetics field has begun to have a significant impact on many biological disciplines (62). As the epigenetic landscape unfolds, studies on epigenetic mechanisms in eukaryotic organisms have revealed the fundamental importance of chromatin-mediated regulation of the developmental program. Chromatin is the physiological substrate for many cellular events, such as DNA replication, repair, and transcription. The chromatin-based epigenetic mechanism entails DNA methylation, covalent and noncovalent modifications of chromatin, and noncoding RNA (62). Thus far, there is no evidence of DNA methylation in P. falciparum despite the presence of a gene containing the DNA methyltransferase motif (19, 60, 145). Therefore, alteration of chromatin structure in Plasmodium is achieved mainly through chromatin remodeling, posttranslational modifications (PTMs) of histones, and replacement of core histones by histone variants.
Plasmodium nucleosome and histones.
The building block of chromatin is the nucleosome, consisting of ∼160 bp of DNA wrapped in about two superhelical turns around a histone octamer made of one H3/H4 tetramer and two H2A/H2B dimers. The malaria parasite chromosomes have a typical nucleosomal organization with a phasing of ∼155 bp (17, 88). Nucleosomal organization extends into the telomeric repeats, and nonnucleosomal chromatin occurs only at the extreme ends of the telomeric repeats (49). Nucleosomes are generally considered repressive for transcription. Earlier transfection assays to characterize Plasmodium promoter activities carried on episomal plasmids have demonstrated that the expected temporal pattern of gene expression requires the pass of S phase after the assembly of nucleosomes (32, 67).
The P. falciparum genome encodes four evolutionarily conserved, canonical core histones, H2A, H2B, H3, and H4 (101), and four variant histones, H2A.Z, H2Bv, H3.3, and CenH3, all of which have been confirmed by mass spectrometry (MS) (109, 147). The linker histone H1 has not been recognized, which partially explains the lack of higher-order compaction of nuclear DNA in P. falciparum. Consistent with required histone deposition during DNA replication, the core histones are highly expressed in late trophozoite and schizont stages (109), coinciding with DNA synthesis (64, 71). H2Bv is lineage-specific and is found in trypanosomes and apicomplexan parasites (141). The replacement of canonical histones with histone variants can influence the nucleosome stability and chromatin patterns (95, 123). Interestingly, in both Trypanosoma brucei and Toxoplasma gondii, H2A.Z interacts with H2Bv and is associated with active genes (31, 99). In T. brucei, variant histones are used to mark boundaries of polycistronic transcription units, and nucleosomes containing two variant histones (H2A.Z and H2Bv) are highly enriched at the putative polymerase II transcription start sites (TSS) (137). These studies revealed novel functions of the variant histones in these early-branching protozoans. The functions of histone variants in Plasmodium are yet to be investigated.
Covalent modifications of histones.
Canonical and variant histones contain a diverse array of PTMs, most of which are located on the N-terminal tails. These PTMs facilitate the establishment of a global chromatin environment and orchestrate DNA-related biological processes. The most common PTMs include acetylation, methylation, phosphorylation, ubiquitination, poly-ADP-ribosylation, and sumoylation (83). These modifications can alter the chromatin structure directly by modulating the interactions of proteins with DNA and affecting chromatin structures (cis effects) and serve as an epigenetic marking system to recruit specialized “effector” proteins (trans effects). Specific combinations of these different modifications of histone tails create a “histone code” (77). The “histone code” hypothesis posits that the totality of the histone modifications, in both kind and number, dictates a particular biological outcome (138).
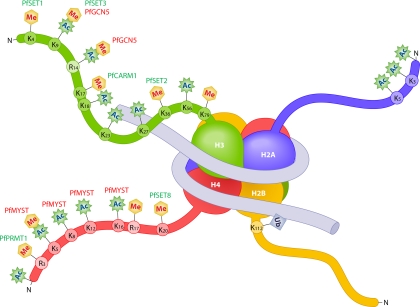
MS analyses of acid-extracted histones have identified an immense number of PTMs in P. falciparum (109, 147). Miao et al. (109) identified a total of 20 PTMs on P. falciparum histones by liquid chromatography-tandem MS (LC-MS/MS) using quadruple time-of-flight (Q-TOF), and Trelle et al. (147) later expanded this list to >44 using a more sensitive method through a combination of Q-TOF and linear trap quadrupole-Fourier transform (LTQ-FT) MS (Fig. 1; Table 1). Like those of other unicellular eukaryotes (58), P. falciparum histones contain more activation marks than silencing marks. The most abundant marks are histone lysine acetylation and methylation. Histone acetylation is linked largely to active genes, whereas histone lysine methylation is involved in both transcriptional activation and silencing. P. falciparum H3 (PfH3), PfH3.3, and PfH4 contain a number of marks for active genes, such as acetylated lysines and methylated lysine (H3K4) and arginines (H3R17 and H4R3). Marks normally associated with silent genes include trimethylated H3K9 and H4K20. The histone variant H2A.Z N-terminal tail is multiply acetylated on a repeated GGK motif (126), and this applies to PfH2A.Z as well (109, 147). Of the two divergent subtypes of H2B, PfH2Bv is abundantly acetylated, in contrast to the scarcity of similar modifications on PfH2B. It is noteworthy that many of the PTMs on Plasmodium histones are densely clustered, probably exerting synergy on the epigenetic regulatory circuits. There is no evidence of PfH3S10 phosphorylation yet, as is commonly observed in model eukaryotes. For other histone PTMs, H4 is found sumoylated, while H2B is ubiquitinated at K112 (74, 147). Taken together, the sheer number of conserved and novel PTMs on canonical and variant histones in P. falciparum demonstrates the potential complexity of the “histone code” in this parasite.
Fig. 1.
Schematic drawing of a nucleosome with the four canonical histones (H3, H4, H2A, and H2B) in P. falciparum. The covalent PTMs of the histone tails (methylation [Me], acetylation [Ac], and ubiquitination [Ub]) and enzymes catalyzing the addition of the PTMs (PfGCN5, PfSET1, PfSET2, PfCARM1, PfPRMT1, PfMYST, and PfSET8) are highlighted. PTMs on the tails of variant histones are listed in Table 1.
Table 1.
Summary of PTMs identified on P. falciparum histones
| Histone | PTM(s)a |
|---|---|
| H2A | N-term-ac, K3ac, K5ac |
| H2B | K112ub |
| H3 | K4me, K4me2, K4me3, K9ac, K9me, K9me3, K14ac, K14me, R17me, R17me2, K18ac, K23ac, K27ac, K36me3, K56ac, K79me3 |
| H4 | N-term-ac, R3me, R3me2, K5me, K5ac, K8ac, K12ac, K12me, K16ac, R17me, K20me, K20me2, K20me3 |
| H2A.Z | N-term-ac, K11ac, K15ac, K19ac, K25ac, K28ac, K30ac, K35ac |
| H2Bv | N-term-ac, K3ac, K8ac, K13ac, K14ac, K18ac, T85ph |
| H3.3 | K4me, K4me2, K4me3, K9ac, K14ac, R17me, R17me2, K18ac, K23ac, K27ac |
Bold indicates PTMs identified by antibodies only.
CHROMATIN MODIFICATIONS: DEPOSITION, RECOGNITION, AND FUNCTIONS
Histone modifications are dynamic and are controlled by the opposing actions of various enzymes for their addition and removal (83). These enzymes have emerged as promising targets for designing novel chemotherapeutics (84). The malaria parasite has a large repertoire of ATP-dependent remodelers, enzymes catalyzing covalent histone modifications, and proteins with PTM-binding modules (68).
Histone acetylation.
Histone lysine acetylation is catalyzed by histone acetyltransferases (HATs). The first identification of a Tetrahymena HAT as the yeast GCN5 homologue directly links histone acetylation to transcription regulation. Five families of HATs are currently recognized: GNATs (GCN5 N-acetyltransferases), MYSTs (MOZ, Ybf1/Sas3, Sas2, and Tip60), p300/CBP (CREB-binding protein), general transcription factor HATs, and nuclear hormone-related HATs (16, 92). At least four HATs are found in the malaria parasite genomes: PF08_0034, PF11_0192, PFL1345c, and PFD0795w (68). PfGCN5 (PF08_0034) preferentially acetylates H3K9 and K14 in vitro, and the HAT domain can partially rescue the yeast GCN5 mutant (46). It is encoded by an essential gene and, like its orthologues in other organisms, is present in a protein complex(es) in vivo (45). The unusually long N-terminal extension is probably involved in mediating protein complex formation and, like the T. gondii GCN5, appears to harbor the nuclear localization signal (9, 87). Consistent with a role in gene activation (129), the H3K9ac mark is enriched in the promoters of active genes (30, 82). Consequently, disturbance of this histone mark by attenuation of PfGCN5 activity with curcumin and anacardic acid leads to growth inhibition (28, 29). Treatment with 20 μM anacardic acid for 12 h induced 2-fold or greater changes in the expression of ∼5% of genes in P. falciparum trophozoites, among which 76% were downregulated (29). Another HAT protein, PfMYST (PF11_0192), with homology to the yeast protein ESA1, is also an essential gene in the malaria parasite (J. Miao et al., unpublished data). The recombinant PfMYST prefers to acetylate histone H4 at K5, K8, K12, and, to a lesser extent, K16. PfMYST is involved in regulating the parasite cell cycle, and overexpression of this enzyme results in a defect in schizogony of the parasite. Two other HATs, PFL1345c and PFD0795w are probably the homologues of elp3 and Hat1 (a type B HAT), respectively.
In addition to HATs, three classes of histone deacetylases (HDACs) have been identified in P. falciparum. PfHDAC1 (PFI1260c) is a class I enzyme homologous to yeast Rpd3 and is a nuclear protein (79). PF14_069 and PF10_0078 are provisionally assigned to class II HDACs (68). HDACs have been evaluated as promising drug targets, and many HDAC inhibitors possess potent antimalarial activities (1). The HDAC inhibitor apicidin, which may affect both class I and II HDACs in the parasite, causes profound transcription changes in the parasite, and deregulation of transcription is evident at as early as 1 h posttreatment (18). Similar to the observation with the more specific HDAC inhibitor FR235222 in T. gondii (11), apicidin induces expression of stage-specific genes that are otherwise suppressed during that particular stage of the intraerythrocytic development cycle (IDC) in P. falciparum. Two class III enzymes, PfSir2A (PF13_0152) and PfSir2B (PF14_0489), also named sirtuins, have received considerable attention because of their roles in regulating the mutually exclusive expression of var genes. In vitro, PfSir2A catalyzes NAD+-dependent deacetylation of a number of acetyllysine peptides, including H3 and H4, and also shows ADP-ribosyltransferase activity on all histones (56, 107). The two PfSir2 paralogues are required for silencing the different var gene promoter subsets. PfSir2A plays a more significant role in silencing subtelomeric var genes transcribed toward the telomere and the intrachromosomal var genes (promoter types UpsA, UpsE, and UpsC), whereas PfSir2B silences the nonoverlapping group controlled by UpsB (146). As a result, genetic deletion of either PfSir2 leads to a general derepression of subsets of the var gene family (39, 108, 146). Loss of PfSir2 also affected the expression of other variant gene families, which is consistent with the notion that these clonally variant gene families in P. falciparum may share a transcriptional factor (70). PfSir2A plays a more important role in establishing heterochromatin in the subtelomeric regions and maintenance of telomere length (146).
Histone methylation.
The malaria parasites code for a large family of at least 10 members of histone lysine methyltransferases (HKMTs) containing the SET [Su(var), E(z), Trithorax] domain. Four of the HKMTs (PfSET1, -2, -3, and -8) are homologues of the well-characterized HKMTs that methylate H3K4, H3K36, H3K9, and H4K20, respectively (Fig. 1) (27). Only recombinant PfSET2 and PfSET8 are enzymatically active, and PfSET8 displays conserved activity to confer H4K20 mono-, di-, and trimethylation (27, 131). The H3K9 methylase PfSET3 (PF08_0012) is encoded by an essential gene and is localized to the heterochromatic nuclear periphery marked by CenH3, and H3K9me3-enriched genes also reside in this compartment (97, 149). Compared with the trypanosome parasite, which has two DOT1 (disruptor of telomeric silencing-1) homologues which methylate H3K79 (76), Plasmodium appears to lack these non-SET domain HKMTs. Two families of lysine demethylases employing two different molecular mechanisms of demethylation have been found to regulate histone methylation: the lysine-specific demethylases 1 (LSD1) and JmjC domain-containing histone demethylases (JHDMs) (135). P. falciparum encodes at least one LSD1 (PFL0575w) and two JHDMs (MAL8P1.111 and PFF0135w). In the case of arginine methylation, malaria parasites have three protein arginine methyltransferases (PRMTs): PfPRMT1 (PF14_0242), PfPRMT5 (PF13_0323), and PfCARM1 (PF08_0092) (47). Only PfPRMT1 has been characterized so far. This enzyme catalyzes monomethylation and asymmetric dimethylation of H4R3 and some nonhistone substrates, and its localization in both the nucleus and cytoplasm is consistent with its activities on different substrates (47). In addition, methylation of H3R17 is detected, which is presumably the substrate of PfCARM1. Since histone methylation marks are involved in gene regulation and maintenance of the subtelomeric heterochromatin, histone methylation is an important subject demanding further investigations.
Other histone modifications.
The malaria parasites encode ubiquitin and two ubiquitin-like proteins (Ubls), SUMO and Nedd8. Conjugation of these Ubls to substrates involves three separate enzyme activities: an ATP-dependent activating enzyme (E1), a conjugating enzyme (E2), and a protein isopeptide ligase (E3). The most commonly observed histone ubiquitinations are on H2A and H2B. Typically, H2A ubiquitination is considered a repressive mark, whereas H2B ubiquitination is involved in both transcription activation and silencing (153). In comparison, histone H4 sumoylation is associated with decreased gene expression (136). The malaria parasite genomes contain a number of protein homologues of E1, E2, and E3 and a number of proteases that might be involved in removing the Ubl modifications (118, 120). In an effort to identify the proteome of PfSUMO modified proteins, Issar et al. (74) identified sumoylated histone H4 from P. falciparum. Interestingly, PfSir2A is also a target for sumoylation, suggesting cross talk between these two epigenetic pathways. The reverse reaction is catalyzed by a diverse group of deubiquitinating proteases (DUBs) (3, 118). Using a specific probe design for deubiquitination enzymes, two enzymes, PfUCH54 (PF11_0177) and PfUCHL3 (PF14_0576), which show dual specificities for ubiquitin and Nedd8 (3, 57) have been identified in P. falciparum. Besides these studies, the significance of histone modifications by Ubls in parasite transcription regulation is largely unknown. Poly-ADP-ribosylation is probably absent in malaria parasites, since a poly-ADP-ribose polymerase homologue cannot be identified in the parasites.
ATP-dependent chromatin remodelers.
Chromatin-remodeling complexes such as the ATP-dependent SWI2/SNF2 protein complex can mobilize the nucleosomes along the DNA with the use of energy from ATP hydrolysis (103). The functions of SWI2/SNF2 family chromatin remodelers in protozoan parasites are not well understood. In trypanosomes, a protein related to SWI2/SNF2 is involved in the de novo synthesis of the modified thymine base J within the telomeric DNA, which correlates with the epigenetic silencing of variant surface glycoproteins (VSGs) (36). P. falciparum contains at least 11 SWI2/SNF2 ATPases (68, 145). Despite the fact that the global nucleosome positioning in P. falciparum is relatively stable throughout the IDC, var gene activation is associated with changes in local chromatin structure and reduced nucleosomal occupancy at the promoters (39, 151, 154). Therefore, it is important to determine whether P. falciparum SWI2/SNF2 ATPases also participate in antigenic switching.
Recognition of PTMs by histone code “readers.”
One function of the histone tail modifications is to act as recognition sites for effector modules, facilitating downstream events via the recruitment or stabilization of chromatin-related protein complexes. In the past decade, a number of conserved protein domains that specifically bind histone PTMs have been identified (144). These protein modules are classified into several subgroups, including the bromodomain, Royal superfamily, plant homeodomain (PHD) fingers, WD40 repeats, and 14-3-3 proteins (34, 144). Bromodomain is an evolutionarily conserved acetyllysine-binding module found in many chromatin-associated proteins (158). PHD fingers and the Royal superfamily protein fold, including chromodomain, double chromodomain, double or tandem tudor domain, and malignant brain tumor (MBT) repeats, are methyllysine-binding modules (105). The chromatin modification and remodeling pathways are also replete with WD40 repeat proteins. The WD40 repeat protein WDR5, a common component of the SET1 complex, binds unmodified H3R2 (128). The mammalian 14-3-3 proteins make up a family of phosphoserine-binding modules that regulate diverse cellular functions. A 14-3-3 isoform was found to bind phosphoserines in H3 with high affinity (100).
The malaria parasites possess an extensive catalogue of proteins with PTM-binding modules (Table 2). Proteins with a single bromodomain have been identified in P. falciparum, including PfGCN5 HAT and PfSET1 HKMT. For methyllysine recognition, Plasmodium contains a number of Royal superfamily and PHD finger proteins. Of the four chromodomain proteins, PfMYST and heterochromatin protein 1 (PfHP1) contain a single chromodomain, while the chromodomain-helicase-DNA-binding protein 1 homologue contains double chromodomains. Plasmodium has two genes with a single tudor domain (69); one is the Staphylococcus aureus nuclease homologue PfTSN (PF11_0374). This protein possesses nuclease activity toward single-stranded RNA (ssRNA), and the tudor domain binds RNA. Since PfTSN is localized mainly in the parasite nucleus, it would be interesting to determine whether it also binds modified histones. There are at least 10 PHD domain proteins in P. falciparum, including two HKMTs (PfSET1 and PfSET2), a putative SUMO ligase (MAL13P1.302), and a putative ISWI homologue (PFF1185w), which are potentially involved in chromatin physiology. There are more than 90 P. falciparum proteins containing the WD40 motif, including two putative chromatin assembly factors (PFA0520c and PFD0455w). In addition, there are three putative 14-3-3 proteins. Despite the great number of potential histone PTM-binding modules in Plasmodium, only PfHP1 has been characterized (52, 115). PfHP1 contains a chromodomain and a chromo-shadow domain, which are involved in H3K9me3 binding and dimerization, respectively. Chromatin immunoprecipitation (ChIP) analysis indicates that this protein is associated with the H3K9me3 mark in the subtelomeric regions and especially with both subtelomeric and intrachromosomal silent var genes (115). PfHP1 is essential for IDC, and overexpression of PfHP1 leads to enhancement of variegated gene expression (52). The ability of this protein to dimerize is probably responsible for aggregating nucleosomes in the subtelomeric regions and thus for the formation of the subtelomeric heterochromatin.
Table 2.
Proteins containing histone PTM-binding modules in P. falciparum
| Binding module/PTM mark | Gene | Annotation/PTM mark |
|---|---|---|
| Bromodomain/Kac | PF08_0034 | PfGCN5 |
| PFF1440w | PfSET1 | |
| PFA0510w | ||
| PFL0635c | ||
| PFL1645w | ||
| PF10_0328 | ||
| PF14_0724 | ||
| Royal superfamily/Kme | ||
| Chromodomain | PFL1005c | PfHP1/H3K9me3 |
| PF11_0192 | PfMYST | |
| PF11_0418 | ||
| Double chromodomain | PF10_0232 | PfCHD1 |
| Tudor domain | PF11_0374 | PfTSN |
| PFC1050w | PfSMN | |
| PHD fingers/Kme | PFF1440w | PFSET1 |
| MAL13P1.122 | PFSET2 | |
| MAL13P1.302 | ||
| PFC0425w | ||
| PF10_0079 | ||
| PF11_0429 | ||
| PFL1010c | ||
| PF14_0315 | ||
| PFL0575w | ||
| WD40 repeat (>90 genes) | PFA0520c | CAF-1 |
| PFD0455w | CAF-1 | |
| 14-3-3 proteins/Sph | MAL8P1.69 | |
| MAL13P1.309 | ||
| PF14_0220 |
As expected from the histone code hypothesis, distinct histone modifications can generate synergistic or antagonistic affinities for chromatin-associated proteins, which in turn dictate the dynamics of the chromatin states. In parallel to the coexisting marks on the same nucleosomes or the same histones, many chromatin-associated complexes, or even the same proteins, contain multiple histone-binding modules (127). This is also true for many chromatin-associated proteins in Plasmodium (27). Such multivalent chromatin binding may afford enhanced affinity, composite specificity, and cross talk among the chromatin-associated complexes.
GENOMIC AND NUCLEAR ORGANIZATION
Genome-wide epigenetic landscape.
Although the malaria parasite chromosomes do not show the typical condensation during mitosis, several lines of evidence indicate that the parasite chromosomes are also divided into euchromatin and heterochromatin (132). Genome sequencing has provided a blueprint for the definition of the epigenetic modifications across the genome: “the epigenome.” This is accomplished through the use of high-throughput technologies such as the combination of ChIP with DNA microarrays (ChIP-chip) and with massive parallel sequencing (ChIP-seq). Recently, the global nucleosomal occupancy in P. falciparum during the parasite's IDC has been mapped using a tiling array (154), and the formaldehyde-assisted isolation of regulatory elements to extract protein-free DNA (FAIRE) and the MNase-mediated purification of mononucleosomes to extract histone-bound DNA (MAINE) coupled to high-throughput sequencing (119). These studies have revealed that coding regions are generally densely packed with nucleosomes, while telomeres harbor a region with the highest nucleosomal density. This is consistent with the more condensed nature of subtelomeric regions determined by measuring the distance between fluorescent in situ hybridization sites within the same chromosome and the presence of heterochromatin mark H3K9me3 in this region (55, 97). tRNA and basal transcription factor genes showed low nucleosomal occupancy at all times, suggesting a perpetually permissive chromatin structure that might be required for constitutively high levels of expression. Unlike in model eukaryotes where nucleosomes are usually absent only at transcription start sites (TSS) and intergenic regions are condensed, P. falciparum intergenic regions are relatively devoid of nucleosomes. This is most likely attributable to the extreme AT richness in the intergenic regions, which may exclude nucleosomes (142). As a result, the small nucleosome-free regions often observed at TSS in model organisms (91, 156) cannot be conclusively identified. Yet, the prediction of core promoters in P. falciparum based solely on the DNA physiochemical properties certainly deserves more detailed mapping of nucleosome occupancy at the core promoters (13). The relaxed promoter regions are likely permissive for the preassembly of the transcription preinitiation complex on promoters of all IDC-expressed genes (63). The nucleosome mapping pattern did not change for most genes throughout the IDC, suggesting that individual nucleosome positions might be fixed for most coding regions, consistent with the presence of intrinsic signals for nucleosome positions encoded across the genome (72, 133). While the ChIP-chip analysis detected only large chromatin structure changes in the subtelomeric regions (154), the FAIRE- and MAINE-sequencing results indicate that chromatin architecture undergoes drastic upheavals throughout the IDC, with chromatin generally loosening in the ring stage and condensing in schizonts (119). This genome-wide nucleosome map provides a framework for further elaboration of the role of specific histone PTMs and the interaction of nucleosomes with DNA-binding proteins and transcription machinery. With the recent identification of the AP2 domain transcription factors in Plasmodium (35, 157), it would be interesting to determine whether functional transcription factor-binding sequences are nucleosome free. Recent analysis of nucleosome occupancy at putative AP2 domain-binding motifs suggests that these loci follow the general pattern of nucleosome occupancy changes for most genes (119), but these data require further scrutiny.
Genome-wide profiling of histone modifications has revealed some enlightening patterns. H3K9ac and H3K4me3 are found enriched in the 5′ ends of transcribed genes, while H3K36me3 is found at the middle and 3′ ends of transcribed genes. In contrast, H3K9me3 is observed mostly in broad domains over silenced regions (7, 125). Using low-density microarrays, Cui et al. (30) detected that H3K9ac is correlated with gene expression. However, this low-density array failed to properly identify enrichment of the H3K9me3 mark on restricted chromosome loci. Recently, several groups have reported mapping of histone PTMs in P. falciparum using either high-density or tiling microarrays (97, 129, 154). These studies have revealed an extremely euchromatic organization of the chromosomes with extensive distribution of H3K9ac (155) and H3K4me3, especially in the intergenic regions (129). Consistent with the paradigm found in most eukaryotes, their enrichment at the 5′ ends of active genes correlates with gene expression during the schizont stage (129). Intriguingly, these active marks are mostly lost and become evenly distributed across the genes in the ring stage. These studies are in general agreement with a study performed with T. gondii, which found an association of the activation marks (histone acetylation and H3K3me3) with active promoters (61). In stark contrast, the heterochromatin mark H3K9me3 is restricted to nucleosome-dense subtelomeric regions and some discrete intrachromosomal islands, where members of the multigene families are clustered (97, 129). Many of these gene families encode surface antigens that are involved in antigenic variation and immune evasion of the parasites, and there is evidence of differential expression of individual members of these gene families (132). This finding conforms to the early prediction of H3K9me3 as a memory mark for silent var genes (20, 96). Consistent with its conserved role in heterochromatin function, global ChIP-chip analysis of the H3K9me3-binding protein PfHP1 shows that its distribution is highly correlated with the two genome-wide H3K9me3 profiling studies (52, 97, 129), suggesting a potential role of PfHP1 in the maintenance and spread of the H3K9me3 mark. To investigate how the subtelomeric heterochromatin is established, Lopez-Rubio et al. (97) mapped H3K9me3 in the ΔPfSir2A mutant. Contrary to the expectation from a telomere position effect, PfSir2A deletion led to only a partial, discontinuous loss of H3K9me3 in the subtelomeric regions. In contrast to the distribution pattern of H3K9me3, another repressive histone mark, H4K20me3, has a broad distribution in P. falciparum, but its significance is yet to be determined (97). Taken together, these ChIP-chip studies demonstrate the feasibility of obtaining a comprehensive epigenetic map of histone modifications in Plasmodium.
Nuclear compartmentalization.
It has long been recognized that the eukaryotic nucleus is compartmentalized, and subnuclear localization of a gene influences its chromatin state and expression (143). The perinuclear compartment created by the telomere clusters in yeasts helps sequester silencing factors that promote and stabilize heterochromatin, whereas association of active genes with the nuclear pores creates another functional compartment. Recent studies have demonstrated that the malaria parasite nucleus is also structurally and functionally divided into compartments. Electron microscopy reveals that the parasite nuclear periphery consists of an electron-dense zone reminiscent of heterochromatin interspersed by electron-translucent space, presumably of noncondensed chromatin (124). Although the small size of the P. falciparum nucleus makes nuclear mapping difficult, the use of a set of nuclear markers still allows the recognition of several nuclear compartments. Scherf and colleagues first compared the localization of the PfSir2A protein to that of the nucleolar mark PfNop1 (50, 55). Further mapping was achieved through the use of nucleoporin Nup100 to define the outer limit of the nucleus, DAPI (4′,6′-diamidino-2-phenylindole) staining to label the most central nucleus, and CenH3 and other markers to define the nuclear peripheral space (97, 149). Localization studies show that the 28 telomeres form four to seven clusters at the nuclear periphery (55, 124). The clustered location of telomeres in P. falciparum may favor the maintenance of epigenetic “memory,” since the heterochromatin mark “writing” machineries such as PfSir2A and PfSET3 are also concentrated in such heterochromatic spaces (39, 55, 97). In addition, the H3K9me3-binding protein PfHP1 is clearly localized at an electron-dense, heterochromatin-like zone of the nuclear periphery and colocalized with the telomere clusters and subtelomeric PfSir2A protein (52, 115). The telomeric clusters are apparently tethered together by proteins (104), and a protein resembling the origin-of-recognition complex 1 (Orc1) is associated with telomere clusters at the nuclear periphery (102), which might play a role in nuclear positioning of the telomeres.
The nuclear localizations of various histone modifications in P. falciparum mapped using commercially available antibodies display tremendous differences (39, 73, 149). Acetylated H4 is restricted to the interior of the nucleus, overlapping extensively with DAPI staining, which is in sharp contrast to the perinuclear locations of telomere clusters (39). Similarly, H3K9ac is localized primarily in the central, DAPI-stained nucleus (presumably euchromatin), whereas the heterochromatin mark H3K9me3 is localized in the perinuclear space (97, 149). In mapping the locations of the methyllysine marks, Issar et al. (73) have found that the active chromatin mark H3K4me2 is distributed in the nucleus in punctate loci, whereas H3K4me3 labels the periphery of the DAPI-stained area. The repressive mark H4K20me3 has a similar but more peripheral labeling to H3K4me3, and in schizonts it is located in punctate spots (131). H3K9me3 is located in several discrete loci at the nuclear periphery outside the DAPI-stained area and does not overlap with the centromeres defined by CenH3 (73, 97, 149). With regard to two other methyllysine marks (H3K36me3 and H3K79me3), some discrepancies still exist. Though Plasmodium has a SET2 homologue that potentially methylates H3K36 (27), this mark has not been identified by MS (109, 147), and antibodies used to detect this mark yielded variable results (27, 73). The most intriguing result is the H3K79me3 labeling, which may define a new subnuclear compartment, since it does not overlap with the nucleolar protein PfNop1, the telomere clusters, or the active var locus (73). It is noteworthy that the typical enzyme Dot1, which catalyzes the methylation of H3K79, has not been identified in Plasmodium, and H3K79me3 was not detected by MS (147).
To date, the nuclear localizations of a number of chromatin-associated proteins have been determined. PfSir2A partially overlaps with the nucleolar marker PfNop1, but its localization is quite distinct from that of acetylated H4 (55). Volz et al. investigated the nuclear locations of 12 P. falciparum proteins, which are assigned to three compartments: a DAPI-stained central area with proteins potentially involved in chromatin remodeling (e.g., ISWI); an area surrounding and partially overlapping with DAPI staining where PfLSD1 is localized; and the nuclear periphery overlapped with CenH3, where PfSET2, PfSET3, and a nucleosome assembly protein are localized (97, 149). This nuclear space division will help further dissect the functional nuclear compartments involved in epigenetic regulation in P. falciparum.
Formation and inheritance of the chromatin structures.
One property of the epigenetic trait is the ability to perpetuate through generations, which includes propagation and maintenance of the states of active or repressive chromatin. As discussed above, positioning in different nuclear compartments may facilitate the establishment of the necessary environments for a chromatin state. Conceivably, the formation of a chromatin state may involve an “initiator,” which recruits protein complexes and coordinates establishment of the chromatin structure. It has been shown that the var promoter, introduced into the malaria parasite by transfection, is able to mediate the nucleation and spread of silenced chromatin (150). Studies with other eukaryotes have demonstrated the involvement of noncoding RNAs and RNA interference (RNAi) in the formation of specialized chromatin domains (8). Although RNAi is not functional in the malaria parasite due to a lack of the RNAi machinery components (5), the parasite does transcribe abundant antisense and noncoding RNAs (65, 112, 114). Also notably, noncoding RNAs produced by the bidirectional promoters of the centromeric regions and var introns are associated with chromatin, which might play a role in the formation of heterochromatin at these sites (44, 94). Once established, the chromatin structure can be maintained for generations. Using promoter titration, Dzikowski and Deitsch demonstrated that active transcription is required for maintaining the “memory” of an active var gene (40). Forcing transfected parasites to express increasing numbers of unregulated episomal var promoters leads to downregulation of the endogenous, active var gene, most likely due to titration of a factor(s) required for the maintenance of the active chromatin state. Interestingly, this also erases the epigenetic memory of individual var genes. “Readers” of histone marks also play essential roles in reinforcing and spreading the chromatin states through recruitment of chromatin-associated complexes. Furthermore, many histone PTMs are combined to specify a chromatin state, and considerable cross talk exists among different chromatin modification pathways. In P. falciparum, two proteins that establish the active chromatin marks H3K9ac (PfGCN5) and H3K4me3 (PfSET1) both contain a bromodomain that binds acetyllysines. These two proteins are potentially linked in a chromatin-associated network (87), suggesting that they may cooperate in the establishment of a euchromatin environment.
MODELS OF EPIGENETIC REGULATION DURING DEVELOPMENT
Antigenic variation.
Antigenic variation is a recurrent theme evolved in many pathogens of mammalian hosts as a strategy to evade the host defense (33, 106). In P. falciparum, PfEMP1 is the major variable antigen and adhesin expressed on the surface of infected erythrocytes and is encoded by a family of ∼60 hypervariable var genes. PfEMP1-mediated binding of infected red blood cells to host endothelium leads to sequestration of the infected cells in deep tissues, removing them from circulation and clearance by the spleen. This cytoadherence is responsible for several severe disease syndromes, most notably cerebral malaria and pregnancy-associated malaria (110). The var family members exhibit strictly mutually exclusive expression: only one member is expressed in a given cell, while the remainder of the family is silenced. The recent finding that duplicated var2csa gene copies in the HB3 strain are simultaneously expressed suggests that duplicated and highly similar var genes may be an exception to this dogma (14). Because of the significance of PfEMP1 in disease pathology, focused research has led to a staggering number of publications, including several recent reviews on the mechanisms of var gene expression (21, 41, 53, 85, 98, 106, 132). Here we provide an updated review of new findings on var gene regulation.
Different var genes appear to have intrinsically varied switching rates. During in vitro culture, var2csa is among the var genes with a higher “on” rate (111). Binding of trans regulatory factors to var promoter motifs may be responsible for establishing an active or repressive chromatin state (150) and for dictating the “on-off” states of individual var genes (54, 111). Voss and coworkers showed that an AP2 domain protein, PfSIP2 (PFF0200c), binds to the cis-acting SPE2 motif in the UpsB var promoters and the SPE2 arrays in subtelomeric domains and is involved in heterochromatin formation and genome integrity (51). Apart from var promoters, the var intron's bidirectional promoter activity produces the long sense and antisense noncoding RNAs, which are associated with chromatin and might play a role in the formation of heterochromatin (44). Genome-wide mapping of epigenetic marks of histone PTMs found significant enrichment of silent marks (histone hypoacetylation and H3K9me3) in a large subtelomeric domain and internal var genes, lending support to the hypothesis that var transcription “memory” is maintained by epigenetic mechanisms (132). These silent marks are likely formed and maintained by recruitment of special chromatin modification complexes such as PfSir2s and PfSET3 (97). Studies on individual var genes such as var2csa have highlighted that var gene activation is linked to reduced nucleosome occupancy at the var promoter, increased histone acetylation, and H3K4 methylation (96, 154). Interestingly, the H3K4me2 mark may serve to index the var gene that is poised for activation during the subsequent cycle. var gene expression is further influenced by the nuclear microenvironment. While silent var genes are localized mostly at the nuclear periphery (97, 150), activation of a var gene involves its dissociation from the telomere clusters and relocation to a transcriptionally competent compartment within the nuclear periphery (14, 42, 97, 111, 124, 150). Such a perinuclear var expression site is selective for promoters (70) and could accommodate at least two simultaneously active var promoters (14, 42). In summary, as evidence for a multilayer regulation of var expression accumulates, the molecular mechanism of mutually exclusive var expression appears to involve the interactions between var genetic elements, chromatin structure, noncoding RNA, and subnuclear localization.
Besides the var gene family, P. falciparum also contains additional multicopy gene families, including approximately 160 rif (repetitive interspersed family), 39 stevor (subtelomeric variable open reading frame), and 13 pfmc-2tm (Maurer's cleft 2-transmembrane domain proteins) genes (43, 89). Data gathered so far suggest that the three 2TM families are potentially involved in antigenic variation and pathogenesis: first, at least some members of these multigene families have been shown to be expressed on the surface of infected erythrocytes (10, 81, 89, 113, 116, 117, 130); second, they appear to be clonally variable (48, 86); and third, the stevor and pfmc-2tm families undergo expression switching (90, 113). These variant gene families also appear to follow an ordered pattern of expression: var transcription is active early during IDC (12 to 18 h after invasion), followed by that of rif (18 to 27 h), stevor (22 to 32 h), and pfmc-2tm (26 to 30 h) genes. It is unclear how such coordinated expression is achieved, but the unique patterns of H3K9me3 enrichment at these families suggests the involvement of epigenetics (97). The physical proximity of these variant genes on chromosomes and the effect of deletions of PfSir2 genes on both var and rif expression (146) suggest that these gene families are coregulated. Yet, the fact that switching of var expression by selection does not affect the repertoire of transcribed stevor genes argues against this speculation (37, 134). To further address this question, Howitt et al. (70) used the promoter titration technique and found that by forcing transfected parasites to express increasing numbers of episomal constructs containing either a var, rif, stevor, or pfmc-2tm promoter, not only genes of the corresponding families but also members of all four families are affected, suggesting that they all share a var-specific titratable factor(s). It remains to be determined whether these variant genes share similar mechanisms of expression switching.
Erythrocyte invasion.
The malaria parasite P. falciparum uses multiple redundant pathways to invade erythrocytes, which provide parasites clear advantages in fully exploring the repertoire of the erythrocyte receptors, in coping with their polymorphism and variability, and possibly in immune evasion (26). Two families of parasite protein, the Duffy binding-like (DBL) family and the reticulocyte homology (PfRh) family, play key roles in parasite invasion. DBL proteins include EBA-175, EBA-140/BAEBL, and EBA-181/JSEBL, which bind erythrocytes in a sialic acid-dependent manner. The Rh protein family includes PfRh1, PfRh2a, PfRh2b, and PfRh4, some of which mediate erythrocyte binding in a sialic acid-independent way. Members of these gene families undergo clonal variation in both laboratory-selected parasites and field isolates and are responsible for different invasion phenotypes (23). The first example of variant expression of proteins involved in erythrocyte invasion was described for the rodent malaria parasite P. yoelli. The Py235 gene family (PfRh orthologues) includes ∼14 genes, and each of the merozoites derived from a single schizont can express a different member (121). Moreover, different subsets of Py235 genes are expressed in sporozoites, hepatic schizonts, and erythrocyte schizonts, suggesting resetting of the expression program at each of these invasive stages (122). Similarly, PfRh proteins all display marked differences in expression between strains (38, 148), and strains selected to switch to the sialic acid-independent pathway are linked to PfRh4 expression (59, 139). It has been demonstrated recently that two isogenic lines of 3D7 parasites selected to exhibit drastically different invasion phenotypes differentially express EBA-140 and RhopH1/Clag (24). The five-member RhopH1/Clag gene family is a component of the RhopH complex with a potential function in erythrocyte invasion (80). Two members of this gene family, clag3.1 and clag3.2, display a pattern of mutually exclusive expression. Like for the var genes, silencing or activation of these invasion-associated genes does not show detectable DNA alterations, suggesting an epigenetic mechanism (24). To understand the mechanism underlying the marked upregulation of PfRh4 during switching of the parasite to a sialic-acid-independent invasion pathway, Jiang et al. (78) observed eviction of the +1 nucleosome at the PfRh4 transcription start site and enrichment of the euchromatin marks such as histone acetylation and H3K4me3 at the schizont stage. Consistent with H3K9me3 as a heterochromatin mark, silenced PfRh4 in the unselected Dd2 strain is associated with persistent H3K9me3 throughout the IDC. From this and the studies of var genes, it seems that the malaria parasites employ converging epigenetic mechanisms to regulate variant gene expression.
CONCLUDING REMARKS
The study of the epigenetic mechanism in malaria parasites is still in its infancy, and most knowledge gathered thus far is from focused studies of antigenic variation in P. falciparum. The available information clearly indicates that chromatin-mediated mechanisms underlie many cellular processes in the parasite's development. From an evolutionary point of view, the malaria parasites have a typical nucleosome organization, numerous histone PTMs, and a large catalogue of conserved chromatin modification and remodeling machineries and histone-binding modules, suggesting conserved epigenetic mechanisms in these early-branching protozoan parasites. Therefore, in-depth studies of the epigenome of the malaria parasites will contribute to a better comprehension of how chromatin-mediated mechanisms have evolved. In addition, the malaria parasite also possesses distinct histone PTMs and divergent histone variants, and their combination is expected to generate a complex yet different “histone code.” Besides, among the “writers” and “readers” of the “histone code” in this parasite, many are unique or contain unique domains, but only a countable few have been characterized so far. Thus, elucidation of the epigenetic pathways in malaria parasites not only will help us to understand gene regulation in this unicellular parasite but also will provide insights into many parasite-specific phenomena such as antigenic variation and alternative invasion pathways, which may ultimately lead to the development of novel control measures targeting host-parasite interactions. More importantly, the distinction of the parasite's chromatin modification machinery from those of its mammalian hosts makes it a promising target for antimalarial chemotherapy. For example, HDACs have been explored as potential candidates for antimalarials, and many HDAC inhibitors have potent antimalarial activity (1). With the identification of most of the PTMs, modifiers, and “readers,” future studies will need to characterize and integrate these components and to advance toward a mechanistic understanding of chromatin-mediated gene regulation in the malaria parasite.
ACKNOWLEDGMENT
We acknowledge grant support (R01 AI064553) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Andrews K. T., Tran T. N., Wheatley N. C., Fairlie D. P. 2009. Targeting histone deacetylase inhibitors for anti-malarial therapy. Curr. Top. Med. Chem. 9:292–308 [DOI] [PubMed] [Google Scholar]
- 2.Aravind L., Iyer L. M., Wellems T. E., Miller L. H. 2003. Plasmodium biology: genomic gleanings. Cell 115:771–785 [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas K., Misaghi S., Comeaux C. A., Catic A., Spooner E., Duraisingh M. T., Ploegh H. L. 2006. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol. Microbiol. 61:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaji S., Babu M. M., Iyer L. M., Aravind L. 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33:3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum J., Papenfuss A. T., Mair G. R., Janse C. J., Vlachou D., Waters A. P., Cowman A. F., Crabb B. S., de Koning-Ward T. F. 2009. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. 2009. An operational definition of epigenetics. Genes Dev. 23:781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein B. E., Meissner A., Lander E. S. 2007. The mammalian epigenome. Cell 128:669–681 [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E., Allis C. D. 2005. RNA meets chromatin. Genes Dev. 19:1635–1655 [DOI] [PubMed] [Google Scholar]
- 9.Bhatti M. M., Sullivan W. J., Jr 2005. Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J. Biol. Chem. 280:5902–5908 [DOI] [PubMed] [Google Scholar]
- 10.Blythe J. E., Yam X. Y., Kuss C., Bozdech Z., Holder A. A., Marsh K., Langhorne J., Preiser P. R. 2008. Plasmodium falciparum STEVOR proteins are highly expressed in patient isolates and located in the surface membranes of infected red blood cells and the apical tips of merozoites. Infect. Immun. 76:3329–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bougdour A., Maubon D., Baldacci P., Ortet P., Bastien O., Bouillon A., Barale J. C., Pelloux H., Menard R., Hakimi M. A. 2009. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206:953–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozdech Z., Llinas M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brick K., Watanabe J., Pizzi E. 2008. Core promoters are predicted by their distinct physicochemical properties in the genome of Plasmodium falciparum. Genome Biol. 9:R178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brolin K. J., Ribacke U., Nilsson S., Ankarklev J., Moll K., Wahlgren M., Chen Q. 2009. Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biol. 10:R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callebaut I., Prat K., Meurice E., Mornon J. P., Tomavo S. 2005. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics 6:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrozza M. J., Utley R. T., Workman J. L., Cote J. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321–329 [DOI] [PubMed] [Google Scholar]
- 17.Cary C., Lamont D., Dalton J. P., Doerig C. 1994. Plasmodium falciparum chromatin: nucleosomal organisation and histone-like proteins. Parasitol. Res. 80:255–258 [DOI] [PubMed] [Google Scholar]
- 18.Chaal B. K., Gupta A. P., Wastuwidyaningtyas B. D., Luah Y. H., Bozdech Z. 2010. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 6:e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S. W., Keyes M. K., Horrocks P. 2006. LC/ESI-MS demonstrates the absence of 5-methyl-2′-deoxycytosine in Plasmodium falciparum genomic DNA. Mol. Biochem. Parasitol. 150:350–352 [DOI] [PubMed] [Google Scholar]
- 20.Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A. Z., Hartl D. L., Deitsch K. W. 2007. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U. S. A. 104:899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chookajorn T., Ponsuwanna P., Cui L. 2008. Mutually exclusive var gene expression in the malaria parasite: multiple layers of regulation. Trends Parasitol. 24:455–461 [DOI] [PubMed] [Google Scholar]
- 22.Coleman B. I., Duraisingh M. T. 2008. Transcriptional control and gene silencing in Plasmodium falciparum. Cell. Microbiol. 10:1935–1946 [DOI] [PubMed] [Google Scholar]
- 23.Cortes A. 2008. Switching Plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol. 24:517–524 [DOI] [PubMed] [Google Scholar]
- 24.Cortes A., Carret C., Kaneko O., Yim Lim B. Y., Ivens A., Holder A. A. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 3:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulson R. M., Hall N., Ouzounis C. A. 2004. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 14:1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowman A. F., Crabb B. S. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755–766 [DOI] [PubMed] [Google Scholar]
- 27.Cui L., Fan Q., Cui L., Miao J. 2008. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol. 38:1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L., Miao J., Cui L. 2007. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob. Agents Chemother. 51:488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui L., Miao J., Furuya T., Fan Q., Li X., Rathod P. K., Su X. Z., Cui L. 2008. Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot. Cell 7:1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui L., Miao J., Furuya T., Li X., Su X. Z., Cui L. 2007. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell 6:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalmasso M. C., Onyango D. O., Naguleswaran A., Sullivan W. J., Jr., Angel S. O. 2009. Toxoplasma H2A variants reveal novel insights into nucleosome composition and functions for this histone family. J. Mol. Biol. 392:33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deitsch K. W., Calderwood M. S., Wellems T. E. 2001. Malaria. Cooperative silencing elements in var genes. Nature 412:875–876 [DOI] [PubMed] [Google Scholar]
- 33.Deitsch K. W., Lukehart S. A., Stringer J. R. 2009. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Microbiol. 7:493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Cruz X., Lois S., Sanchez-Molina S., Martinez-Balbas M. A. 2005. Do protein motifs read the histone code? Bioessays 27:164–175 [DOI] [PubMed] [Google Scholar]
- 35.De Silva E. K., Gehrke A. R., Olszewski K., Leon I., Chahal J. S., Bulyk M. L., Llinas M. 2008. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl. Acad. Sci. U. S. A. 105:8393–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiPaolo C., Kieft R., Cross M., Sabatini R. 2005. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol. Cell 17:441–451 [DOI] [PubMed] [Google Scholar]
- 37.Duffy M. F., Tham W. H. 2007. Transcription and coregulation of multigene families in Plasmodium falciparum. Trends Parasitol. 23:183–186 [DOI] [PubMed] [Google Scholar]
- 38.Duraisingh M. T., Triglia T., Ralph S. A., Rayner J. C., Barnwell J. W., McFadden G. I., Cowman A. F. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22:1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duraisingh M. T., Voss T. S., Marty A. J., Duffy M. F., Good R. T., Thompson J. K., Freitas-Junior L. H., Scherf A., Crabb B. S., Cowman A. F. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13–24 [DOI] [PubMed] [Google Scholar]
- 40.Dzikowski R., Deitsch K. W. 2008. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 382:288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dzikowski R., Deitsch K. W. 2009. Genetics of antigenic variation in Plasmodium falciparum. Curr. Genet. 55:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S., Wellems T. E., Deitsch K. W. 2007. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 8:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dzikowski R., Templeton T. J., Deitsch K. 2006. Variant antigen gene expression in malaria. Cell. Microbiol. 8:1371–1381 [DOI] [PubMed] [Google Scholar]
- 44.Epp C., Li F., Howitt C. A., Chookajorn T., Deitsch K. W. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Q., An L., Cui L. 2004. PfADA2, a Plasmodium falciparum homologue of the transcriptional coactivator ADA2 and its in vivo association with the histone acetyltransferase PfGCN5. Gene 336:251–261 [DOI] [PubMed] [Google Scholar]
- 46.Fan Q., An L., Cui L. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3:264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan Q., Miao J., Cui L., Cui L. 2009. Characterization of PRMT1 from Plasmodium falciparum. Biochem. J. 421:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez V., Hommel M., Chen Q., Hagblom P., Wahlgren M. 1999. Small, clonally variant antigens expressed on the surface of the Plasmodium falciparum-infected erythrocyte are encoded by the rif gene family and are the target of human immune responses. J. Exp. Med. 190:1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Figueiredo L. M., Pirrit L. A., Scherf A. 2000. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 106:169–174 [DOI] [PubMed] [Google Scholar]
- 50.Figueiredo L. M., Rocha E. P., Mancio-Silva L., Prevost C., Hernandez-Verdun D., Scherf A. 2005. The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus. Nucleic Acids Res. 33:1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B. T., Moes S., Bozdech Z., Jenoe P., Stunnenberg H. G., Voss T. S. 2010. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 6:e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A. M., Alako B. T., Ehlgen F., Ralph S. A., Cowman A. F., Bozdech Z., Stunnenberg H. G., Voss T. S. 2009. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 5:e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank M., Deitsch K. 2006. Activation, silencing and mutually exclusive expression within the var gene family of Plasmodium falciparum. Int. J. Parasitol. 36:975–985 [DOI] [PubMed] [Google Scholar]
- 54.Frank M., Dzikowski R., Amulic B., Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 64:1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freitas-Junior L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., Mancio-Silva L., Leal-Silvestre R. J., Gontijo A. M., Shorte S., Scherf A. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36 [DOI] [PubMed] [Google Scholar]
- 56.French J. B., Cen Y., Sauve A. A. 2008. Plasmodium falciparum Sir2 is an NAD+-dependent deacetylase and an acetyllysine-dependent and acetyllysine-independent NAD+ glycohydrolase. Biochemistry 47:10227–10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frickel E. M., Quesada V., Muething L., Gubbels M. J., Spooner E., Ploegh H., Artavanis-Tsakonas K. 2007. Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution. Cell. Microbiol. 9:1601–1610 [DOI] [PubMed] [Google Scholar]
- 58.Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. 2007. Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282:7641–7655 [DOI] [PubMed] [Google Scholar]
- 59.Gaur D., Furuya T., Mu J., Jiang L. B., Su X. Z., Miller L. H. 2006. Upregulation of expression of the reticulocyte homology gene 4 in the Plasmodium falciparum clone Dd2 is associated with a switch in the erythrocyte invasion pathway. Mol. Biochem. Parasitol. 145:205–215 [DOI] [PubMed] [Google Scholar]
- 60.Gissot M., Choi S. W., Thompson R. F., Greally J. M., Kim K. 2008. Toxoplasma gondii and Cryptosporidium parvum lack detectable DNA cytosine methylation. Eukaryot. Cell 7:537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gissot M., Kelly K. A., Ajioka J. W., Greally J. M., Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldberg A. D., Allis C. D., Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128:635–638 [DOI] [PubMed] [Google Scholar]
- 63.Gopalakrishnan A. M., Nyindodo L. A., Ross Fergus M., Lopez-Estrano C. 2009. Plasmodium falciparum: preinitiation complex occupancy of active and inactive promoters during erythrocytic stage. Exp. Parasitol. 121:46–54 [DOI] [PubMed] [Google Scholar]
- 64.Gritzmacher C. A., Reese R. T. 1984. Protein and nucleic acid synthesis during synchronized growth of Plasmodium falciparum. J. Bacteriol. 160:1165–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gunasekera A. M., Patankar S., Schug J., Eisen G., Kissinger J., Roos D., Wirth D. F. 2004. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 136:35–42 [DOI] [PubMed] [Google Scholar]
- 66.Hakimi M. A., Deitsch K. W. 2007. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr. Opin. Microbiol. 10:357–362 [DOI] [PubMed] [Google Scholar]
- 67.Horrocks P., Pinches R., Kriek N., Newbold C. 2002. Stage-specific promoter activity from stably maintained episomes in Plasmodium falciparum. Int. J. Parasitol. 32:1203–1206 [DOI] [PubMed] [Google Scholar]
- 68.Horrocks P., Wong E., Russell K., Emes R. D. 2009. Control of gene expression in Plasmodium falciparum—ten years on. Mol. Biochem. Parasitol. 164:9–25 [DOI] [PubMed] [Google Scholar]
- 69.Hossain M. J., Korde R., Singh S., Mohmmed A., Dasaradhi P. V., Chauhan V. S., Malhotra P. 2008. Tudor domain proteins in protozoan parasites and characterization of Plasmodium falciparum tudor staphylococcal nuclease. Int. J. Parasitol. 38:513–526 [DOI] [PubMed] [Google Scholar]
- 70.Howitt C. A., Wilinski D., Llinas M., Templeton T. J., Dzikowski R., Deitsch K. W. 2009. Clonally variant gene families in Plasmodium falciparum share a common activation factor. Mol. Microbiol. 73:1171–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inselburg J., Banyal H. S. 1984. Synthesis of DNA during the asexual cycle of Plasmodium falciparum in culture. Mol. Biochem. Parasitol. 10:79–87 [DOI] [PubMed] [Google Scholar]
- 72.Ioshikhes I. P., Albert I., Zanton S. J., Pugh B. F. 2006. Nucleosome positions predicted through comparative genomics. Nat. Genet. 38:1210–1215 [DOI] [PubMed] [Google Scholar]
- 73.Issar N., Ralph S. A., Mancio-Silva L., Keeling C., Scherf A. 2009. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microb. Infect. 11:403–407 [DOI] [PubMed] [Google Scholar]
- 74.Issar N., Roux E., Mattei D., Scherf A. 2008. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell. Microbiol. 10:1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iyer L. M., Anantharaman V., Wolf M. Y., Aravind L. 2008. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 38:1–31 [DOI] [PubMed] [Google Scholar]
- 76.Janzen C. J., Hake S. B., Lowell J. E., Cross G. A. 2006. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell 23:497–507 [DOI] [PubMed] [Google Scholar]
- 77.Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 78.Jiang L., Lopez-Barragan M. J., Jiang H., Mu J., Gaur D., Zhao K., Felsenfeld G., Miller L. H. 2010. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc. Natl. Acad. Sci. U. S. A. 107:2224–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joshi M. B., Lin D. T., Chiang P. H., Goldman N. D., Fujioka H., Aikawa M., Syin C. 1999. Molecular cloning and nuclear localization of a histone deacetylase homologue in Plasmodium falciparum. Mol. Biochem. Parasitol. 99:11–19 [DOI] [PubMed] [Google Scholar]
- 80.Kaneko O. 2007. Erythrocyte invasion: vocabulary and grammar of the Plasmodium rhoptry. Parasitol. Int. 56:255–262 [DOI] [PubMed] [Google Scholar]
- 81.Kaviratne M., Khan S. M., Jarra W., Preiser P. R. 2002. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell 1:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Komaki-Yasuda K., Okuwaki M., Kano S., Nagata K., Kawazu S. 2008. 5′ sequence- and chromatin modification-dependent gene expression in Plasmodium falciparum erythrocytic stage. Mol. Biochem. Parasitol. 162:40–51 [DOI] [PubMed] [Google Scholar]
- 83.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 84.Kristeleit R., Stimson L., Workman P., Aherne W. 2004. Histone modification enzymes: novel targets for cancer drugs. Exp. Opin. Emerg. Drugs. 9:135–154 [DOI] [PubMed] [Google Scholar]
- 85.Kyes S. A., Kraemer S. M., Smith J. D. 2007. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot. Cell 6:1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kyes S. A., Rowe J. A., Kriek N., Newbold C. I. 1999. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 96:9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438:103–107 [DOI] [PubMed] [Google Scholar]
- 88.Lanzer M., de Bruin D., Wertheimer S. P., Ravetch J. V. 1994. Transcriptional and nucleosomal characterization of a subtelomeric gene cluster flanking a site of chromosomal rearrangements in Plasmodium falciparum. Nucleic Acids Res. 22:4176–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lavazec C., Sanyal S., Templeton T. J. 2006. Hypervariability within the Rifin, Stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Nucleic Acids Res. 34:6696–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lavazec C., Sanyal S., Templeton T. J. 2007. Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Mol. Microbiol. 64:1621–1634 [DOI] [PubMed] [Google Scholar]
- 91.Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900–905 [DOI] [PubMed] [Google Scholar]
- 92.Lee K. K., Workman J. L. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8:284–295 [DOI] [PubMed] [Google Scholar]
- 93.Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503–1508 [DOI] [PubMed] [Google Scholar]
- 94.Li F., Sonbuchner L., Kyes S. A., Epp C., Deitsch K. W. 2008. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J. Biol. Chem. 283:5692–5698 [DOI] [PubMed] [Google Scholar]
- 95.Lieb J. D., Clarke N. D. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 123:1187–1190 [DOI] [PubMed] [Google Scholar]
- 96.Lopez-Rubio J. J., Gontijo A. M., Nunes M. C., Issar N., Hernandez Rivas R., Scherf A. 2007. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lopez-Rubio J. J., Mancio-Silva L., Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microb. 5:179–190 [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Rubio J. J., Riviere L., Scherf A. 2007. Shared epigenetic mechanisms control virulence factors in protozoan parasites. Curr. Opin. Microbiol. 10:560–568 [DOI] [PubMed] [Google Scholar]
- 99.Lowell J. E., Kaiser F., Janzen C. J., Cross G. A. 2005. Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J. Cell Sci. 118:5721–5730 [DOI] [PubMed] [Google Scholar]
- 100.Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L. C. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol. Cell 20:199–211 [DOI] [PubMed] [Google Scholar]
- 101.Malik H. S., Henikoff S. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882–891 [DOI] [PubMed] [Google Scholar]
- 102.Mancio-Silva L., Rojas-Meza A. P., Vargas M., Scherf A., Hernandez-Rivas R. 2008. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 121:2046–2053 [DOI] [PubMed] [Google Scholar]
- 103.Martens J. A., Winston F. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136–142 [DOI] [PubMed] [Google Scholar]
- 104.Marty A. J., Thompson J. K., Duffy M. F., Voss T. S., Cowman A. F., Crabb B. S. 2006. Evidence that Plasmodium falciparum chromosome end clusters are cross-linked by protein and are the sites of both virulence gene silencing and activation. Mol. Microbiol. 62:72–83 [DOI] [PubMed] [Google Scholar]
- 105.Maurer-Stroh S., Dickens N. J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C. P. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28:69–74 [DOI] [PubMed] [Google Scholar]
- 106.Merrick C. J., Duraisingh M. T. 2006. Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 62:612–620 [DOI] [PubMed] [Google Scholar]
- 107.Merrick C. J., Duraisingh M. T. 2007. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot. Cell 6:2081–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Merrick C. J., Dzikowski R., Imamura H., Chuang J., Deitsch K., Duraisingh M. T. 2010. The effect of Plasmodium falciparum Sir2a histone deacetylase on clonal and longitudinal variation in expression of the var family of virulence genes. Int. J. Parasitol. 40:35–43 [DOI] [PubMed] [Google Scholar]
- 109.Miao J., Fan Q., Cui L., Li J., Li J., Cui L. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369:53–65 [DOI] [PubMed] [Google Scholar]
- 110.Miller L. H., Baruch D. I., Marsh K., Doumbo O. K. 2002. The pathogenic basis of malaria. Nature 415:673–679 [DOI] [PubMed] [Google Scholar]
- 111.Mok B. W., Ribacke U., Rasti N., Kironde F., Chen Q., Nilsson P., Wahlgren M. 2008. Default pathway of var2csa switching and translational repression in Plasmodium falciparum. PloS One 3:e1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mourier T., Carret C., Kyes S., Christodoulou Z., Gardner P. P., Jeffares D. C., Pinches R., Barrell B., Berriman M., Griffiths-Jones S., Ivens A., Newbold C., Pain A. 2008. Genome-wide discovery and verification of novel structured RNAs in Plasmodium falciparum. Genome Res. 18:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Niang M., Yan Yam X., Preiser P. R. 2009. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog. 5:e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Patankar S., Munasinghe A., Shoaibi A., Cummings L. M., Wirth D. F. 2001. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell 12:3114–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perez-Toledo K., Rojas-Meza A. P., Mancio-Silva L., Hernandez-Cuevas N. A., Delgadillo D. M., Vargas M., Martinez-Calvillo S., Scherf A., Hernandez-Rivas R. 2009. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 37:2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Petter M., Bonow I., Klinkert M. Q. 2008. Diverse expression patterns of subgroups of the rif multigene family during Plasmodium falciparum gametocytogenesis. PloS One 3:e3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Petter M., Haeggstrom M., Khattab A., Fernandez V., Klinkert M. Q., Wahlgren M. 2007. Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol. Biochem. Parasitol. 156:51–61 [DOI] [PubMed] [Google Scholar]
- 118.Ponder E. L., Bogyo M. 2007. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryot. Cell 6:1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ponts N., Harris E. Y., Prudhomme J., Wick I., Eckhardt-Ludka C., Hicks G. R., Hardiman G., Lonardi S., Le Roch K. G. 2010. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20:228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ponts N., Yang J., Chung D. W., Prudhomme J., Girke T., Horrocks P., Le Roch K. G. 2008. Deciphering the ubiquitin-mediated pathway in apicomplexan parasites: a potential strategy to interfere with parasite virulence. PloS One 3:e2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Preiser P. R., Jarra W., Capiod T., Snounou G. 1999. A rhoptry-protein-associated mechanism of clonal phenotypic variation in rodent malaria. Nature 398:618–622 [DOI] [PubMed] [Google Scholar]
- 122.Preiser P. R., Khan S., Costa F. T., Jarra W., Belnoue E., Ogun S., Holder A. A., Voza T., Landau I., Snounou G., Renia L. 2002. Stage-specific transcription of distinct repertoires of a multigene family during Plasmodium life cycle. Science 295:342–345 [DOI] [PubMed] [Google Scholar]
- 123.Raisner R. M., Mahdhani H. D. 2006. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 16:119–124 [DOI] [PubMed] [Google Scholar]
- 124.Ralph S. A., Scheidig-Benatar C., Scherf A. 2005. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. U. S. A. 102:5414–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rando O. J. 2007. Global patterns of histone modifications. Curr. Opin. Genet. Dev. 17:94–99 [DOI] [PubMed] [Google Scholar]
- 126.Ren Q., Gorovsky M. A. 2001. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell 7:1329–1335 [DOI] [PubMed] [Google Scholar]
- 127.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8:983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J., Verdine G. L. 2006. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salcedo-Amaya A. M., van Driel M. A., Alako B. T., Trelle M. B., van den Elzen A. M., Cohen A. M., Janssen-Megens E. M., van de Vegte-Bolmer M., Selzer R. R., Iniguez A. L., Green R. D., Sauerwein R. W., Jensen O. N., Stunnenberg H. G. 2009. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 106:9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sam-Yellowe T. Y., Florens L., Johnson J. R., Wang T., Drazba J. A., Le Roch K. G., Zhou Y., Batalov S., Carucci D. J., Winzeler E. A., Yates J. R., III 2004. A Plasmodium gene family encoding Maurer's cleft membrane proteins: structural properties and expression profiling. Genome Res. 14:1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sautel C. F., Cannella D., Bastien O., Kieffer S., Aldebert D., Garin J., Tardieux I., Belrhali H., Hakimi M. A. 2007. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol. Cell. Biol. 27:5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scherf A., Lopez-Rubio J. J., Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 62:445–470 [DOI] [PubMed] [Google Scholar]
- 133.Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I. K., Wang J. P., Widom J. 2006. A genomic code for nucleosome positioning. Nature 442:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sharp S., Lavstsen T., Fivelman Q. L., Saeed M., McRobert L., Templeton T. J., Jensen A. T., Baker D. A., Theander T. G., Sutherland C. J. 2006. Programmed transcription of the var gene family, but not of stevor, in Plasmodium falciparum gametocytes. Eukaryot. Cell 5:1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi Y., Whetstine J. R. 2007. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25:1–14 [DOI] [PubMed] [Google Scholar]
- 136.Shiio Y., Eisenman R. N. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 100:13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Siegel T. N., Hekstra D. R., Kemp L. E., Figueiredo L. M., Lowell J. E., Fenyo D., Wang X., Dewell S., Cross G. A. 2009. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 23:1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Strahl B. D., Allis C. D. 2000. The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- 139.Stubbs J., Simpson K. M., Triglia T., Plouffe D., Tonkin C. J., Duraisingh M. T., Maier A. G., Winzeler E. A., Cowman A. F. 2005. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309:1384–1387 [DOI] [PubMed] [Google Scholar]
- 140.Sullivan W. J., Jr., Hakimi M. A. 2006. Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 148:109–116 [DOI] [PubMed] [Google Scholar]
- 141.Sullivan W. J., Jr., Naguleswaran A., Angel S. O. 2006. Histones and histone modifications in protozoan parasites. Cell. Microbiol. 8:1850–1861 [DOI] [PubMed] [Google Scholar]
- 142.Suter B., Schnappauf G., Thoma F. 2000. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 28:4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Taddei A., Hediger F., Neumann F. R., Gasser S. M. 2004. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38:305–345 [DOI] [PubMed] [Google Scholar]
- 144.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Templeton T. J., Iyer L. M., Anantharaman V., Enomoto S., Abrahante J. E., Subramanian G. M., Hoffman S. L., Abrahamsen M. S., Aravind L. 2004. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 14:1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tonkin C. J., Carret C. K., Duraisingh M. T., Voss T. S., Ralph S. A., Hommel M., Duffy M. F., Silva L. M., Scherf A., Ivens A., Speed T. P., Beeson J. G., Cowman A. F. 2009. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 7:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Trelle M. B., Salcedo-Amaya A. M., Cohen A. M., Stunnenberg H. G., Jensen O. N. 2009. Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome Res. 8:3439–3450 [DOI] [PubMed] [Google Scholar]
- 148.Triglia T., Duraisingh M. T., Good R. T., Cowman A. F. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol. Microbiol. 55:162–174 [DOI] [PubMed] [Google Scholar]
- 149.Volz J., Carvalho T. G., Ralph S. A., Gilson P., Thompson J., Tonkin C. J., Langer C., Crabb B. S., Cowman A. F. 2010. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int. J. Parasitol. 40:109–121 [DOI] [PubMed] [Google Scholar]
- 150.Voss T. S., Healer J., Marty A. J., Duffy M. F., Thompson J. K., Beeson J. G., Reeder J. C., Crabb B. S., Cowman A. F. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439:1004–1008 [DOI] [PubMed] [Google Scholar]
- 151.Voss T. S., Tonkin C. J., Marty A. J., Thompson J. K., Healer J., Crabb B. S., Cowman A. F. 2007. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol. Microbiol. 66:139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Waddington C. H. 1942. The epigenotype. Endeavour 1:18–20 [Google Scholar]
- 153.Weake V. M., Workman J. L. 2008. Histone ubiquitination: triggering gene activity. Mol. Cell 29:653–663 [DOI] [PubMed] [Google Scholar]
- 154.Westenberger S. J., Cui L., Dharia N., Winzeler E., Cui L. 2009. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics 10:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Winter G., Kawai S., Haeggstrom M., Kaneko O., von Euler A., Kawazu S., Palm D., Fernandez V., Wahlgren M. 2005. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 201:1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yaragatti M., Basilico C., Dailey L. 2008. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Res. 18:930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yuda M., Iwanaga S., Shigenobu S., Mair G. R., Janse C. J., Waters A. P., Kato T., Kaneko I. 2009. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 71:1402–1414 [DOI] [PubMed] [Google Scholar]
- 158.Zeng L., Zhou M. M. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124–128 [DOI] [PubMed] [Google Scholar]