Abstract
Methylenetetrahydrofolate reductases (MTHFRs) play a key role in biosynthesis of methionine and S-adenosyl-l-methionine (SAM) via the recharging methionine biosynthetic pathway. Analysis of 32 complete fungal genomes showed that fungi were unique among eukaryotes by having two MTHFRs, MET12 and MET13. The MET12 type contained an additional conserved sequence motif compared to the sequences of MET13 and MTHFRs from other eukaryotes and bacteria. Targeted gene replacement of either of the two MTHFR encoding genes in Fusarium graminearum showed that they were essential for survival but could be rescued by exogenous methionine. The F. graminearum strain with a mutation of MET12 (FgΔMET12) displayed a delay in the production of the mycelium pigment aurofusarin and instead accumulated nor-rubrofusarin and rubrofusarin. High methionine concentrations or prolonged incubation eventually led to production of aurofusarin in the MET12 mutant. This suggested that the chemotype was caused by a lack of SAM units for the methylation of nor-rubrofusarin to yield rubrofusarin, thereby imposing a rate-limiting step in aurofusarin biosynthesis. The FgΔMET13 mutant, however, remained aurofusarin deficient at all tested methionine concentrations and instead accumulated nor-rubrofusarin and rubrofusarin. Analysis of MET13 mutants in F. graminearum and Aspergillus nidulans showed that both lacked extracellular reduction potential and were unable to complete mycelium pigment biosynthesis. These results are the first to show that MET13, in addition to its function in methionine biosynthesis, is required for the generation of the extracellular reduction potential necessary for pigment production in filamentous fungi.
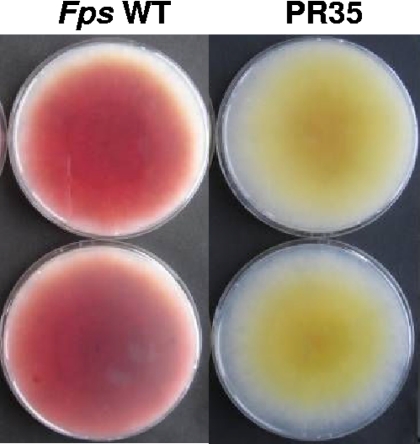
The biosynthetic pathway of aurofusarin has been shown to require the action of nine genes found within a 25-kb gene cluster (9). The enzymes include a polyketide synthase (PKS12p), an S-adenosyl-l-methionine (SAM)-dependent O-methyltransferase (AurJp), a flavin-dependent monooxygenase (AurFp), an oxidoreductase (AurOp), a laccase (GIP1p), a major facilitator pump (AurTp), a transcription factor (AurR1p), and two novel proteins that do not show homology to any previously characterized proteins (9, 20, 25). Aurofusarin is formed by a five-step pathway in which PKS12p catalyzes the initial condensation of one acetyl-coenzyme A (CoA) molecule and six malonyl-CoA molecules, resulting in the yellow compound nor-rubrofusarin. Nor-rubrofusarin is converted into rubrofusarin by an O-methylation reaction, catalyzed by AurJp with SAM as a carbon donor. The order of the remaining steps in the conversion of rubrofusarin to aurofusarin remains unclear as the Δgip1, ΔaurO, and ΔaurF mutants all accumulate rubrofusarin (9). In a study of mycelium pigment biosynthesis in Fusarium pseudograminearum, a mutant that accumulates a yellow pigment instead of aurofusarin was identified (Fig. 1) (25). The mutation responsible for this was genetically mapped by inverse PCR, and sequence analysis showed that the F. pseudograminearum MET13 (FpsMET13) gene had been disrupted. This is the first evidence that genes outside the cluster were directly involved in pigment biosynthesis.
Fig. 1.
Phenotype of the F. pseudograminearum MET13 disruption mutant (PR35.1). The cultures were grown for 10 days on DFM without (top) and with (bottom) 60 μg/ml l-methionine.
The recharging methionine biosynthetic pathway is dependent on methylenetetrahydrofolate reductases ([MTHFRs] EC 1.5.1.20), MET12 and MET13 in yeast, that irreversibly reduce 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate using flavin adenine dinucleotide (FAD) as the cofactor and NAD(P)H as the reducing agent (13). The formed 5-methyltetrahydrofolate is subsequently used as a single carbon donor for methylation of l-homocysteine, resulting in the formation of l-methionine, catalyzed by methionine synthase (EC 2.1.1.13) (14). In addition to the synthesis of l-methionine, this pathway also provides the basic building block for de novo synthesis of nucleotides (adenine, guanine, and thymidine), generates single-carbon donor units in the form of SAM, and is a central part of sulfur metabolism. SAM is predicted to be the carbon donor for the conversion of nor-rubrofusarin into rubrofusarin, and this could explain the altered pigmentation in the mutant.
Methylenetetrahydrofolate reductases have been extensively studied due to their suspected involvement in several human diseases, such as hyperhomocysteinemia (38), neural tube defects (5), and the development of cardiovascular diseases (17). Most of our current knowledge of eukaryotic MTHFRs originates from studies of the porcine enzyme, which has been shown to consist of an N-terminal catalytic and a C-terminal regulatory domain (8, 22, 26, 37). Bacterial MTHFR shares extensive similarity to eukaryotic MTHFRs within the N′-terminal catalytic domain but lacks the C′-terminal regulatory domain (32).
The evolutionary history of MTHFRs, including sequences from plants, animals, fungi, bacteria, and archaea, has recently been published (19). The analysis confirmed the central role of MTHFR in primary metabolism and revealed several previously unrecognized conserved sequence motifs. Fungal species appear to be unique in having two MTHFR encoding genes: MET12 and MET13 in Saccharomyces cerevisiae (ScMET12 and ScMET13, respectively) (31), met9 and met11 in Schizosaccharomyces pombe (Spmet9 and Spmet11, respectively) (28), and metA and metF in Aspergillus nidulans (AnmetA and AnmetF, respectively) (33). The MET13 homologs appear to provide approximately 80% of the MTHFR activity in the analyzed fungi, and the requirement for MET12 activity remains elusive.
In the present study, we carry out in-depth sequence and phylogenomic analyses of MTHFR from the publicly available fungal genomes. To gain an understanding of the function of the fungal MET13 MTHFRs in pigment biosynthesis, F. graminearum MET12 (FgMET12) and FgMET13 are replaced in F. graminearum. The aurofusarin biosynthesis pathway is studied as a model at the genetic and metabolite level. The effects on the plasma membrane redox system and its role for mycelium pigment biosynthesis are analyzed in F. graminearum and A. nidulans MTHFR mutants.
MATERIALS AND METHODS
Fungal strains and media.
F. graminearum PH-1 (NRRL 31084) was obtained from the Agriculture Research Service Culture Collection, National Center for Agricultural Utilization Research, Peoria, IL. The strain was stored as a conidia suspension in 10% glycerol at −70°C. For extraction of genomic DNA, the fungal strains were cultured in liquid yeast-peptone-galactose (YPG) medium for 3 days at room temperature at 150 rpm (6). For metabolic analysis and RNA extraction, the mutants and the wild-type (WT) strain of F. graminearum were cultivated on agar plates containing the medium described by Bell (4), with maltose as the C source and urea as the N source, at 25°C in darkness. Pure cultures of transformants were obtained using defined Fusarium medium (DFM) (43), in which the glucose content was reduced to 2% and urea was replaced with 10 μM NaNO3 and 40 μg/ml l-methionine; 150 μg/ml hygromycin B (Sigma-Aldrich) was added for selection.
The S. cerevisiae Met13 (YGL125w) replacement strain 10239a (Scmet13Δ) and the corresponding wild-type (10000F FY1679-01B) were purchased from the European Saccharomyces cerevisiae archive for functional analysis (EUROSCARF). For the methionine auxotrophy complementation analysis, strains were grown on salt and galactose (induction) medium (18) with and without l-methionine.
The A. nidulans W12 (pabaA1 pyrG89), A. nidulans ΔmetF ([AnΔmetF] (M115:pabaA1), and AnΔmetA (M124:pabaA2) strains were obtained from Marzena Sienko at the Institute of Biochemistry and Biophysics, Department of Genetics, Warsaw, Poland (33). The strains were maintained on minimal medium (MM) with Hutner's trace element solution (2) supplemented with 1 μg/ml p-aminobenzoate, 0.5 μg/ml pyridoxine-HCl, 250 μg/ml l-proline, 800 μg/ml Na-thiosulfate, and variable concentrations of l-methionine (0, 20, 40, and 60 μg/ml).
Bacterial strains.
High-efficiency chemically competent Escherichia coli JM109 cells (>108 CFU/μg) for Xi cloning were purchased from Promega. Chemically competent E. coli Top10 cells for Topo cloning were purchased from Invitrogen. For targeted gene replacement in F. graminearum, Agrobacterium tumefaciens LBA4404, carrying the virulence plasmid pTi4404, was used as host for the binary vector pAg1-H3 (44), obtained from J. S. Tkacz, Merck Research Laboratories, Rahway, NJ.
Bacteria strains were propagated in LB medium supplemented with 25 μg/ml kanamycin. E. coli cells were grown at 37°C and 250 rpm, at a pH of 7.2, while A. tumefaciens cells were grown at 28°C at a pH of 7.4 at 100 rpm to minimize filamentous growth.
Sequence analysis.
The F. graminearum database (http://mips.gsf.de/genre/proj/fusarium/) (15), PEDANT (http://mips.gsf.de/genre/proj/fungi/fungal_overview.html) at the Munich Information Center for Protein Sequences (MIPS), and the NCBI fungal genome service (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=fungi) were used for sequence retrieval. MTHFR homologs in complete Ascomycota genomes were identified in the listed databases using the blastp and tblastn algorithms with default settings (1). All Pezizomycotina tblastn hits were annotated using FGENEH-2 from Softberry (Mount Kisco, NJ) with FgMET13p as a template for the gene structure and supported by expressed sequence tag (EST) data when available. Saccharomycetes tblastn hits were annotated using GENSCAN with ScMet12p and ScMet13p as templates. The available annotated Basidiomycota, Microsporidia, Apicomplexa, Dictyosteliida, Diplomonadida, Arthropoda, Mammalia, and Nematoda genomes were analyzed by blastp using FgMET13p, and reannotations when deemed necessary were done using FGENESH. Amino acid sequences for identified MTHFRs were aligned using ClustalX (40) run on a local central processing unit (CPU), and the resulting alignments were manually checked for errors using BioEdit (16). The phylogenetic analysis was conducted using both the neighbor-joining (NJ) and parsimony (P) algorithms using the MEGA4 software package (39). The robustness of the constructed trees was tested by bootstrapping with 1,000 iterations, and for trees constructed with the parsimony algorithm, consensus trees were calculated. Newick files were displayed using TreeView (Win32), version 1.6.6 (30), and Phylodendon (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html). Hidden Markov models were built using Meta-MEME, version 3.4, allowing for a total of 50 different motifs ranging between 25 and 300 amino acids (aa), with zero to one occurrence per sequence (12).
Vector constructs were designed using Vector NTI Advance, version 10.0.1 (Invitrogen). Information on the S. cerevisiae deletion strains and protein-protein interaction information were retrieved from the Saccharomyces genome database ([SGD] (http://www.yeastgenome.org). Background literature for enzymes was retrieved using BRENDA—The Comprehensive Enzyme Information System (release 2006.2 [http://www.brenda.uni-koeln.de/]) (3).
Enzymes, kits, and apparatus.
For cloning procedures the proofreading Phusion DNA polymerase (Finnzyme, Expoo, Finland) was used, while Sigma Taq DNA polymerase was used for screening. Restriction enzymes and alkaline phosphatase (calf intestinal phosphatase [CIP]) were purchased from New England Biolabs (MA). Plasmid DNA was isolated from liquid 10-ml cultures using a Qiagen miniprep kit (QIAgen). Purification of PCR products was performed using an Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences). PCRs were performed using an Eppendorf Mastercycler ep; optimizations were performed in temperature gradient mode with a constant MgCl2 concentration. DNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer in nucleic acid mode using MilliQ water as a reference. The production of aurofusarin and rubrofusarin was analyzed using high-performance liquid chromatography–diode array detection (HPLC-DAD) as described by Malz (25).
Random mutagenesis and identification of the integration site.
From a library of random A. tumefaciens-generated F. pseudograminearum transformants (25), a mutant (PR35.1), which displayed a yellow phenotype, was isolated. The transfer DNA (T-DNA) integration site was determined by inverse PCR (25).
Construction of replacement vectors using Xi cloning (bacterial in vivo homologous recombination).
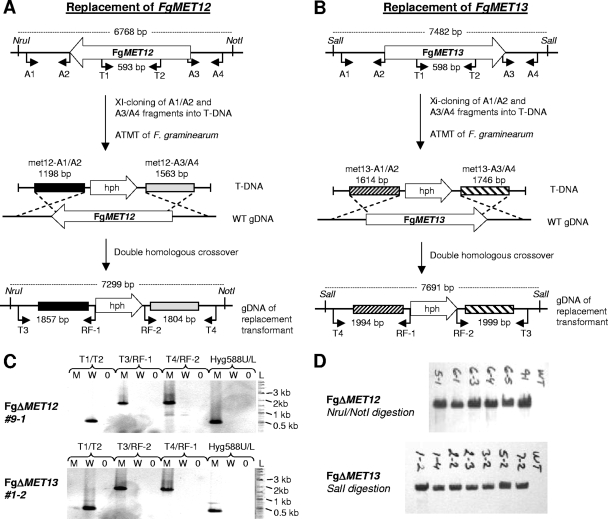
Vectors for targeted replacement of FgMET12 and FgMET13 in F. graminearum were constructed with pAg1-H3 as a backbone, using the two unique, blunt-end cutting enzymes SmaI and SwaI (Fig. 2 A and B). A total of 500 ng of vector was digested overnight with SmaI, dephosphorylated using CIP, and purified using the GFX purification system. The first recombination flank was amplified by PCR using the Phusion DNA polymerase with F. graminearum PH-1 as template and gene-specific primers (gene X-A1/A2, where gene X is the gene of interest) (Table 1). The amplicon was dephosphorylated and GFX purified. Fusion of the first insert and the vector was mediated by Xi cloning (23): 50 ng of digested vector (6.4 kb) and 150 ng of insert (1.5 to 2 kb) were transformed into chemically competent JM109 cells following the manufacturer's recommendations. Transformants were selected on LB plates supplemented with 25 μg/ml kanamycin. Resulting colonies were screened by colony PCR using the gene X-A1/A2 primers and Sigma Taq DNA polymerase. Positive colonies were grown overnight in 10 ml of liquid LB medium supplemented with 25 μg/ml kanamycin. The plasmids were purified, and the sizes of the vectors were confirmed by restriction enzyme digestion. The second recombination flank was introduced as described above but using SwaI for digestion of the vector and gene X-A3/A4 primers for the insert. The inserts in the final deletion vector constructs were sequenced using RF-1 and RF-2 primers to confirm correct insertions (GATC, Germany).
Fig. 2.
Strategy for targeted replacement of MET12 (A) and MET13 (B) in F. graminearum. (C) PCR-based analysis of transformants using four different primer pairs (listed above the lanes) for each of the two loci. Lanes L, 2 log ladder; lanes M, mutant; lanes W, wild type; lanes 0 water control. (D) Southern blot analysis of T-DNA copy number in the selected mutant indicated by laboratory designations using a 789-bp fragment of the hph gene as probe.
Table 1.
Primer sequencesa
| Primer function and name | Primer sequence (5′ to 3′) | Size of amplicon (bp) |
|---|---|---|
| Replacement of FgMET12 and FgMET13 | ||
| Fg-MET12-A1 | CCAGTGAATTCGAGCTCGGTACCAAGGCCCAGAGCTCCCTTCCAGTCCATCC | 1,198 |
| Fg-MET12-A2 | CTTGCGCGCCTAGGCGGCCGTGGCCAGCCCGAAGTTCTGTCAAGTTAAGCAAGGTGTCTA | |
| Fg-MET12-A3 | GGCCGGCCGGCGCGCCGTTTAAACGGATTTATGGGGTATTTATTCGTGGTATGTGAGTAG | 1,563 |
| Fg-MET12-A4 | GCATGCCTGCAGGTCGACATTAATTAATTTGCCAGTTGATTTTCCCAGCCAGTTCTTT | |
| Fg-MET13-A1 | CCAGTGAATTCGAGCTCGGTACCAAGGCCCAACCGCGCCATTTCCATTTTATCATTACC | 1,614 |
| Fg-MET13-A2 | CTTGCGCGCCTAGGCGGCCGTGGCCAGCCCGGGGGAGGGGAACTGCAACGGAAACTAT | |
| Fg-MET13-A3 | GGCCGGCCGGCGCGCCGTTTAAACGGATTTCGGCGGGCGCAGACGACTTTATGT | 1,746 |
| Fg-MET13-A4 | GCATGCCTGCAGGTCGACATTAATTAATTTCGGTGCGCGAGCTCCTTTTTCTGC | |
| Verification and RT-PCR | ||
| Fg-MET12-T1 (CDS)c | CCTCTAGGCCTGTCGCTTCCCACTCG | 593 (in wild type) |
| Fg-MET12-T2 (CDS) | CGGCGCTGACTTCATACAAACCCAACTC | No product in FgΔmet12 |
| Fg-MET12-T3 | CAGGTCGAGTGGTGAATACTTCG | 1,857 (with T3/RF-1) |
| Fg-MET12-T4 | GCCCTCAGAAATCGCTGCCT | 1,804 (with T4/RF-2) |
| Fg-MET13-T1 (CDS) | CCGACGACAACAAGAACGAGGACGAAC | 598 (in wild type) |
| Fg-MET13-T2 (CDS) | CCAAAAGCGGGAGAGCGAGAATCACC | No product in FgΔmet13 |
| Fg-MET13-T3 | CATCCAATCCCATGAATCCTTG | 1999 (with T3/RF-2) |
| Fg-MET13-T4 | GTTATACGGATCGCGGATCGC | 1994 (with T4/RF-1) |
| Hyg588U | AGCTGCGCCGATGGTTTCTACAA | 588 (with Hyg588U/Hyg588L) |
| Hyg588L | GCGCGTCTGCTGCTCCATACAA | |
| RF-1 | AAATTTTGTGCTCACCGCCTGGAC | NAb |
| RF-2 | TCTCCTTGCATGCACCATTCCTTG | NA |
| Probe for Southern analysis | ||
| Hyg789U | AATAGCTGCGCCGATGGTTTCT | 789 |
| Hyg789L | GCTTCTGCGGGCGATTTGTGTA | |
| Complementation of Scmet13D strain | ||
| Fg-MET12-H1 | AGTGGATCCAACATGGACAAGATCACCGACCGCATCG | 2,019 |
| Fg-MET12-H2 | AGTGCGGCCGCTTAACTTGACAGAACTTCCCATAGA | |
| Fg-MET13-H1 | AGTGGATCCAACATGCATATCAAGGATATGCTCAACG | 1,866 |
| Fg-MET13-H2 | AGTGCGGCCGCTTAGTTCGCAGCGGCAGTGACGGCA |
Primers for construction of replacement vectors include 30-bp overhangs, shown in boldface, for Xi cloning.
NA, not applicable (used for sequencing).
CDS, coding sequence.
A. tumefaciens-mediated transformation was carried out as described by Frandsen et al. (9), with the exception that the medium was yeast extract with supplements (YES) instead of DFM to compensate for the expected methionine auxotrophy of the mutants.
Verification of targeted F. graminearum deletion mutants.
The target locus was analyzed in the isolated hygromycin B-resistant F. graminearum transformants by PCR, using three primer pairs (Table 1): one pair to amplify the replaced region (gene X-T1/T2 CDS, where CDS is coding sequence) two other primer pairs, with one primer annealing internally in the hph gene and one in the genomic sequence surrounding the target locus (gene X-T3/RF-1 and gene X-T4/RF-2) (Fig. 2A and B). This primer combination allowed for the identification of ectopic, single-homologous crossover, and double-homologous crossover transformants. Five transformants for each of the two target genes, which were positive for double crossover, were selected for Southern blot analysis to determine the number of integration events. FgΔMET12 genomic DNA (gDNA) was digested with NruI/NotI, and FgΔMET13 gDNA was digested with SalI. The probe was prepared by PCR, using the primer pair Hyg789U/Hyg789L amplifying a 789-bp-large fragment of the hph coding sequence. Labeling and detection were carried out using an enhanced chemiluminescence (ECL) random primer system from Amersham Biosciences.
Chemical analysis.
The generated F. graminearum mutants were extracted with methanol-dichloromethane-ethyl acetate (1:2:3, vol/vol/vol, with 1% formic acid), evaporated to dryness, and redissolved in methanol (34). Metabolic profiles were the obtained by reversed-phase chromatography on an Agilent 1100 HPLC-DAD system (Waldbronn, Germany) scanning 200 to 500 nm, using a210-nm trace. The system was equipped with a GROM-SIL 120 ODS-5ST column (3-μm particle size; 60 by 4.6 mm; Grom Analytik, Rottenburg-Hailfingen, Germany), and metabolites were eluted with an acetonitrile-water gradient containing 0.1% O-phosphoric acid (25). Aurofusarin, rubrofusarin, and nor-rubrofusarin were identified based their retention time and full UV spectra reported in Frandsen et al. (9). Metabolite data were normalized to fungal biomass by measuring the ergosterol concentration (measured at 282 nm) in the extracts determined on the system described above but now using methanol containing 0.1% O-phosphoric acid at a rate of 1 ml/min for 12 min as eluent. The identities of detected metabolites were further verified by liquid chromatography coupled to an LCT orthogonal time-of-flight mass spectrometer (Waters-Micromass) operating in both positive- and negative-electrospray ionization mode (29, 42).
Growth rate and mycelium density.
The radial growth rate of the Fusarium WT and MTHFR mutants on DFM supplemented with different amounts of methionine was measured over a 7-day period by mapping the edge of the mycelium each day with a marker pen. The increase in area from day to day was determined by picture analysis using ImageJ, version 1.41 (NIH), and the growth rate (mm per hour) was calculated using Microsoft Excel. The experiments were performed with eight replicates, and a Student's t test was used to test for significance. The mycelium density (mycelia per volume of solid medium) of the mutants compared to the wild type was determined by cultivating the strains on DFM plates (fixed volume) for 10 days with different methionine concentrations. Half of the medium was cut out and weighed, and ergosterol was extracted and measured as described above.
Expression analysis.
The expression of genes in the aurofusarin cluster, FgMET12 and FgMET13, was determined in the F. graminearum and F. pseudograminearum mutants by reverse transcriptase PCR (RT-PCR), using the primers listed in Table 1 and the aurofusarin gene cluster-specific primers (9, 25).
Reduction potential.
F. graminearum PH-1 (WT), FgΔMET12, and FgΔMET13 strains were grown for 7 days in darkness at 25°C on DFM plates supplemented with different l-methionine concentrations. The A. nidulans W12, AnΔmetA, and AnΔmetF strains were grown for 14 days in darkness at 25°C on MM plates, supplemented with different concentrations of l-methionine. The reduction potential was determined based on the reduction of tetrazolium to formazan by visual inspection (35). One mg/ml Nitro Blue Tetrazolium (NBT) and 0.1 mg/ml phenazine methosulfate (PMS) were mixed in water just before use (27). Whatman 47-mm glass fiber GF/C filters were dipped in the NBT-PMS solution and placed on top of the fungal mycelium. Air bubbles, trapped under the filters, were removed using a Drigalski spatula. The reaction was allowed to proceed for 1 h at 25°C, at which time the results were recorded.
Yeast complementation.
To test if both FgMET12 and FgMET13 were functional methylenetetrahydrofolate reductases, their ability to complement the methionine auxotroph Scmet13Δ strain was determined. The coding sequences (CDS) of FgMET12 and FgMET13 were PCR amplified using the primer pairs Fg-MET12-H1/H2 and Fg-MET13-H1/H2 (Table 1). The PCR conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 66°C for 20 s, and 72°C for 2 min, with a final 2 min at 72°C. The PCR amplicons were Topo cloned (Invitrogen), and correct transformants were identified by colony PCR using the CDS primers. The subcloned fragments were recovered by digesting the vectors with BamHI/NotI and gel purification. The pYES2 vector was digested with BamHI/NotI and GFX purified. The inserts (50 μg) and vector (100 μg) were ligated using T4 DNA ligase (New England Biolabs) and transformed into E. coli DH5α cells. Correct transformants were identified by colony PCR using the CDS primers. The pYES2, pYES2::FgMET12, and pYES2::FgMET13 vectors were transformed into the Scmet13Δ yeast strain, and pYES2 was transformed into the yeast wild-type strain, using the protocol described in the pYES2 manual (18). The wild-type (+pYES2), Scmet13Δ (+pYES2), and complemented Scmet13Δ/FgMET12 and Scmet13Δ/FgMET13 strains were grown for 5 days at 30°C on induction medium with (20 μg/ml) and without l-methionine to test the ability of the introduced genes to complement the methionine deficient yeast mutant.
RESULTS
The yellow F. pseudograminearum mutant is disrupted in a MTHFR gene.
Mapping of the T-DNA integration site in F. pseudograminearum PR35.1 (25) was performed by inverse PCR and projection onto the F. graminearum genome sequence. The integration site was located between position 85290 and 85291 on contig 1.395 in the promoter of FG09572, a gene that shows homology to MET13 from S. cerevisiae. Following the standard nomenclature, FG09572 was therefore renamed FgMET13, and the ortholog in F. pseudograminearum was named FpsMET13. The F. graminearum gene consists of two exons of 1,699 bp and 167 bp, separated by a 56-bp-long intron, giving a total coding sequence of 1,922 bp (621 aa).
Chemical analyses by HPLC-DAD showed that the mutant produced nor-rubrofusarin, the precursor of the red pigment aurofusarin. Nor-rubrofusarin was identified by its [M+H]− ion at m/z 259.0598 (calculated mass, 259.0606; deviation, −3.1 ppm). Semiquantitative expression analysis by RT-PCR showed that all nine genes in the aurofusarin cluster were expressed at normal levels in the mutant. Based on these preliminary results, we speculated that the loss of aurofusarin biosynthesis was caused by a lack of S-adenosylmethionine units required for conversion of nor-rubrofusarin into rubrofusarin.
Fungi have two distinct classes of MTHFRs.
The derived amino acid sequence of FgMET13 was used in a blastp analysis of the F. graminearum genome, resulting in the identification of a second MTHFR, FgMET12 (FG07127), a gene located on contig 1.300. FgMET12 consists of a single 2,019-bp-long exon, equivalent to 672 amino acids. A global alignment of the two derived amino acid sequences showed that FgMET12 and FgMET13 share 242 (33%) amino acid positions and that 377 (46%) have similar properties.
The FgMET13 sequence was used for a tblastn search of all fully sequenced Pezizomycetes, Saccharomycetes, and Schizosaccharomycetes genomes available at NCBI and PEDANT, as of November 2006, plus selected complete genome sequences for other eukaryotic organisms. The majority of the analyzed fungal species had not yet been annotated, and identified sequences including 5 kb up- and downstream were annotated using FGENESH and GENSCAN. The annotated and reannotated fungal MET12p and MET13p proteins are given in File S1 in the supplemental material.
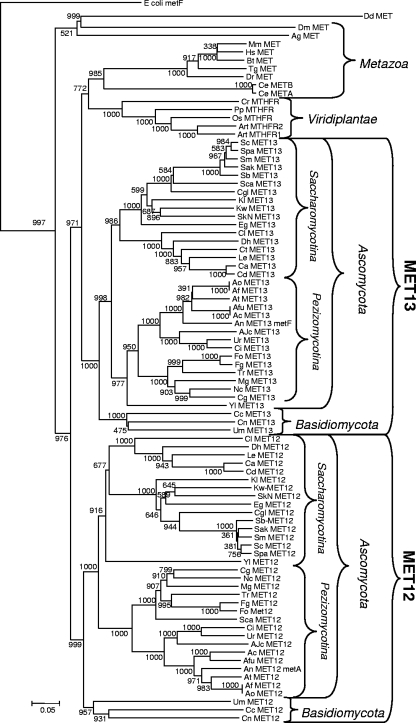
A global alignment (ClustalW) of the 86 identified MTHFR sequences confirmed that bacterial genes contain only the N-terminal part of the eukaryotic genes, as described by Sheppard et al. (32). Construction of phylogenies by neighbor-joining and maximum-parsimony methods showed that the fungal sequences clustered into two groups, MET12p and MET13p. The analyzed fungal species had one member in each group (Fig. 3), as also reported by Sienko and coworkers (33). The topology of the constructed tree was very robust as the major branching points yielded high bootstrap values (above 950 per 1,000 iterations) in the neighbor-joining tree. In addition, the topology of the tree is in good agreement with the generally accepted evolutionary history of the included organisms.
Fig. 3.
Phylogeny made with the neighbor-joining algorithm, based on a global alignment (ClustalW) of the identified MTHFR amino acid sequences from the following fungi, bacteria (E. coli), metazoans, Viridiplantae, and Dictyosteliida (species abbreviations): Ajellomyces capsulatus NAm1 (Ajc), Ascosphaera apis USDA-ARSEF 7405 (Aa), Aspergillus clavatus NRRL 1 (Ac), Aspergillus flavus NRRL3357 (Af), Aspergillus fumigatus Af293 (Afu), Aspergillus nidulans FGSC A4 (An), Aspergillus oryzae RIB40 (Ao), Aspergillus terreus ATCC 20542 (At), Botryotinia fuckeliana B05.10 (Bf), Candida albicans SC5314 (Ca), Candida dubliniensis CD36 (Cd), Candida glabrata CBS138 (Cgl), Candida tropicalis MYA-3404 cont1.60 (Ct), Chaetomium globosum CBS 148.51 (Cg), Clavispora lusitaniae ATCC 42720 cont1.29 (Cl), Coccidioides immitis H538.4 (Ci), Debaryomyces hansenii CBS767 (Dh), Eremothecium gossypii (Eg), Gibberella moniliformis 7600 (GM), Gibberella zeae PH-1 (Fg), Kluyveromyces lactis NRRL Y-1140 (Kl), Kluyveromyces waltii NCYC 2644 cont306 (Kw), Lodderomyces elongisporus NRRL YB-4239 cont1.24 (Le), Magnaporthe grisea 70-15 (MG), Neosartorya fischeri NRRL 181 (Nf), Neurospora crassa (Nc), Phaeosphaeria nodorum SN15 (Pn), Saccharomyces bayanus MCYC 623 (Sb), Saccharomyces castellii NRRL Y-12630 (Sca), Saccharomyces cerevisiae (Sc), Saccharomyces kluyveri NRRL Y-12651 (SkN), Saccharomyces kudriavzevii IFO1802 (Sak), Saccharomyces mikatae IFO1815 (Sm), Saccharomyces paradoxus NRRL Y-17217 (Spa), Schizosaccharomyces pombe 972 h− (Sp), Sclerotinia sclerotiorum 1980 (Ss), Stagonospora nodorum SN15 (Sn), Trichoderma reesei QM9414 (Tr), Uncinocarpus reesii 1704 (Ur), Yarrowia lipolytica CLIB122 (Yl), Ustilago maydis 521 (Us), Coprinopsis cinerea okayama 7#130 (Cc), Cryptococcus neoformans var. neoformans B-3501A (Cn), Dictyostelium discoideum AX4 (Dd), Malassezia globosa CBS 7966 (Mag), Homo sapiens (Hs), Mus musculus (Mm), Bos taurus (Bt), Taeniopygia guttata (Tg), Danio rerio (Dr), Caenorhabditis elegans (Ce), Gallus gallus (Gg), Drosophila melanogaster (Dm), Anopheles gambiae strain PEST (Ag), Arabidopsis thaliana (Art), Physcomitrella patens subsp. patens (Pp), Oryza sativa (Os), and Chlamydomonas reinhardtii (Cr).
The clustering was based on conserved differences between members of the two groups. Members of the MET12 group shared 111 fully conserved positions (FCP), and the MET13p group shared 110 FCP, whereas the two groups only had 76 FCP in common. The identified fungal sequences were used to build hidden Markov models, using META-MEME, to identify conserved sequence motifs in the unaligned sequences. A total of 14 motifs with high information content were identified, and their occurrence in fungi, E. coli, and Homo sapiens MTHFRs was determined (Fig. 4; see also File S2 in the supplemental material). The sequence alignment and distribution of conserved sequence motifs showed that the MET12p and MET13p MTHFR types shared motifs 1 to 12, situated along their entire lengths, and that the main difference was the presence of an additional motif (number 13) in the MET12p group. The core of this motif consists of the highly conserved arginines (R9 and R10), serines (S13 and S16), and isoleucine (I23) which were found in all analyzed MET12p sequences (Fig. 4).
Fig. 4.
(A) Distribution of conserved sequence motifs in MET12, MET13, H. sapiens MTHFR, and E. coli MTHFR. Motifs 2 and 14 share many conserved amino acid positions but are not completely identical (see File S2 in the supplemental material). Analyses of the sequence between motifs 7 and 9 in H. sapiens MTHFR showed that it did not display a significant level of similarity to motif 8, found in MET12 and MET13 sequences. (B) Sequence logo describing motif number 13, which is unique for the MTHFR belonging to the MET12 group.
FgMET12 and FgMET13 both complement the function of ScMET13.
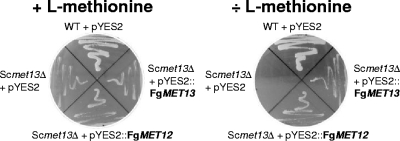
To confirm that FgMET12 and FgMET13 are functional MTHFRs, they were constitutively expressed in an l-methionine auxotroph Scmet13Δ strain. The two heterologous expression strains were both able to grow on minimal medium without l-methionine supplementation (Fig. 5), confirming that the FgMET12 and FgMET13 genes are functional MTHFRs capable of catalyzing the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate.
Fig. 5.
Phenotype of S. cerevisiae wild-type, Scmet13Δ, and the two complementation strains Scmet13Δ/FgMET12 and Scmet13Δ/FgMET13, grown with and without l-methionine in the medium.
Replacement of FgMET12 and FgMET13 leads to methionine auxotrophy and affects pigment biosynthesis.
To confirm the aurofusarin-deficient phenotype of the F. pseudograminearum PR35 mutant, targeted replacement of FgMET12 and FgMET13 was conducted in F. graminearum using A. tumefaciens-mediated transformation. Thirty-seven FgΔMET12 and 26 FgΔMET13 mutants were obtained, with a double-homologous recombination frequency of approximately 70%. Five FgΔMET12 and five FgΔMET13 mutants that tested positive for double-homologous crossover by PCR (Fig. 2C) were selected for Southern analysis (Fig. 2D), which showed that the selected transformants contained only a single copy of the hph gene.
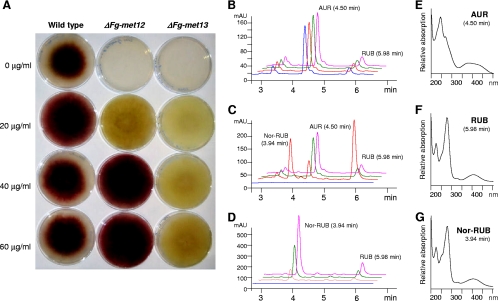
The FgΔMET12 and FgΔMET13 strains were both methionine auxotrophs and affected in aurofusarin biosynthesis (Fig. 6A). The growth rate of the wild type was positively affected by concentrations of 20 μg/ml l-methionine while higher concentrations reduced the growth rate. The addition of increasing concentrations of l-methionine to the minimal medium showed that concentrations above 20 μg/ml were able to restore aurofusarin production in the FgΔMET12 strain but not in the FgΔMET13 strain (Fig. 6A). The production of aurofusarin in FgΔMET12 was positively correlated with incubation time and temperature. The two mutants displayed growth rates similar to the growth rate of the wild type at l-methionine levels above 20 mg/ml. However, the mycelium density, measured as the amount of ergosterol per medium volume, was reduced to between 9 and 16% in both mutant strains under the tested methionine conditions. The FgΔMET12 strain produced nearly wild-type levels of aurofusarin when supplemented with methionine levels higher than 40 μg/ml, but at the low concentrations the strain accumulated increased levels of nor-rubrofusarin and rubrofusarin and lower levels of aurofusarin (Fig. 6C). The FgΔMET13 strain remained aurofusarin deficient throughout the tested l-methionine range and instead accumulated high concentrations of nor-rubrofusarin and rubrofusarin (Fig. 6D). The levels of nor-rubrofusarin and rubrofusarin were positively correlated with increasing methionine concentration and incubation time (Fig. 6D). Analysis of expression of the aurofusarin gene cluster in the two F. graminearum mutants and wild type, by reverse transcriptase PCR, showed that the genes had wild-type levels of expression in both mutants (see File S4 in the supplemental material).
Fig. 6.
Phenotypes and chemotypes of F. graminearum wild-type, FgΔmet12, and FgΔmet13 strains. (A) Picture of wild-type and mutants grown for 5 days on DFM with variable l-methionine concentrations. (B to D) HPLC-DAD analysis at 280 nm of wild-type (B), FgΔmet12 (C), and FgΔmet13 (D) strains grown for 10 days on DFM with variable l-methionine concentrations (blue, 0 μg/ml; red, 20 μg/ml; green, 40 μg/ml; and pink, 60 μg/ml). (E to G) Recorded UV/visible light spectra for aurofusarin (E), rubrofusarin (F), and nor-rubrofusarin (G).
Extracellular reduction potential.
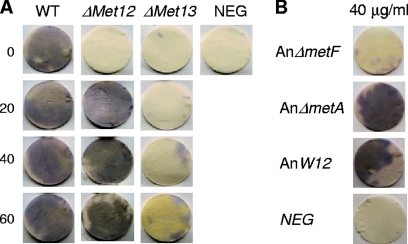
The accumulation of rubrofusarin in FgΔMET13 at high methionine concentrations suggested that the mutant had lost the AurFp (P450 monooxygenase) activity required for conversion of rubrofusarin to aurofusarin (9). Monooxygenase activity is highly dependent on the reduction power normally provided by a P450 reductase. AurF is predicted to be situated at the exterior surface of the plasma or endoplasmic reticulum membrane. The mutant's ability to catalyze extracellular reduction reactions via the plasma membrane redox system was tested using Nitro Blue Tetrazolium (NBT). The FgΔMET13 strain had lost its ability to perform extracellular reduction reactions at all methionine concentrations tested, while the FgΔMET12 strain performed as the wild type at methionine levels above 20 μg/ml (Fig. 7A). To eliminate the possibility that aurofusarin was involved in the generation of the measured reduction potential, three different aurofusarin-deficient mutants (ΔGIP1, ΔaurS, and ΔaurF) were also tested, showing that the loss of reduction potential was not correlated with the lack of aurofusarin (data not shown). To determine whether the loss of extracellular reduction potential in the ΔMET13 strain was specific to F. graminearum or a general feature in filamentous fungi, the corresponding A. nidulans mutants (AnΔmetA and AnΔmetF) and wild-type strain were also analyzed. The analysis showed that the AnmetF (MET13) mutant had also lost its ability to perform extracellular reduction reactions on all tested l-methionine concentrations, while the AnmetA (MET12) strain and wild type were both able to perform the reaction (Fig. 7B). Similarly to the FgΔMET13 mutant, the corresponding Aspergillus mutant (AnΔmetF) had also lost its ability to complete mycelium pigment production, resulting in a white mycelium at low methionine concentrations and the accumulation of a yellow water-soluble pigment at high methionine concentrations, compared to a purplish-red pigmentation of the wild type (see File S3 in the supplemental material).
Fig. 7.
Measurements of the ability of the F. graminearum and A. nidulans mutants to perform extracellular reduction reactions. (A) F. graminearum mutants grown on DFM with different l-methionine levels. (B) A. nidulans mutants grown on MM with 40 mg/ml l-methionine.
DISCUSSION
The identification of two MTHFR types in all analyzed fungal genomes, combined with the conserved domain architecture, suggests that MET12p and MET13p serve distinct and vital functions during the fungal life cycle. Otherwise, one or the other would have been expected to be lost during evolution. The MET13p group is most likely the ancestral form, based to its similarity to MTHFRs from other nonfungal species (Fig. 4). And the MET12 type probably originates from a duplication of the MET13 gene in a common fungal ancestor, followed by mutation and adaptation to serve new functions. The extra motif identified in the MET12p sequences is located between the predicted catalytic and regulatory domains previously described for mammalian MTHFRs (26). To resolve whether it affects the regulatory or catalytic properties of the enzyme or both, direct kinetics studies of purified enzymes are required. The identification of many highly conserved residues in the MET12-specific motif supports the theory that the motif does have a general biological function in fungi.
MET13 has the highest MTHFR activity in fungi.
The targeted replacement of the FgMET12 and FgMET13 genes resulted in methionine auxotroph strains, showing that both enzymes are required for the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate in F. graminearum (Fig. 6). This indicates that the two enzymes have overlapping catalytic properties, which is further supported by the ability of both genes to complement the methionine auxotroph Scmet13Δ strain (Fig. 5). The function of the two MTHFR types in fungi has been analyzed in relation to their involvement in the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate. Raymond and coworkers (31) showed that replacement of ScMET13 resulted in methionine auxotrophy, which could be rescued by l-methionine, while deletion of ScMET12 had no phenotypic effect. Complementation analysis of the Scmet13Δ mutant showed that even multiple copies of ScMET12 were unable to complement the mutant. Also ScMet13p appeared to be responsible for approximately 85% of the observed MTHFR in vivo activity. However, heterologous expression of ScMET12 in an E. coli ΔmetF strain confirmed that it encodes a functional MTHFR. The situation in S. pombe was analyzed by Naula and coworkers (28), who found that replacement of either the MTHFR-encoding gene SpMET11 (equivalent to ScMET12) or SpMET9 (ScMET13) resulted in methionine auxotrophy but that the Spmet11Δ mutant was leaky. Cross-complementation showed that SpMET11 was unable to complement the Spmet9Δ strain and vice versa. Analysis of the MTHFR activity showed that SpMET9p was responsible for approximately 80 to 85% of the recorded MTHFR activity. Similar results were obtained for A. nidulans by Sienko and coworkers, as targeted replacement of AnmetA (approximately equivalent to ScMET12) and AnmetF (ScMET13) resulted in methionine auxotrophs, and cross-complementation analysis using the natural promoters showed that neither of the genes could replace the function of the other. Measurement of MTHFR activity showed that AnMetFp was responsible for the majority of activity. Supplementation with exogenous l-homocysteine (1 mM) allowed the AnΔmetA mutant to grow without l-methionine due to a transcriptional upregulation of the AnmetF gene, resulting in higher total MTHFR activity. Mutations in the main cysteine synthesis pathway (cysA and cysB) or the negative regulatory complex for this pathway (SCON) were also found to be suppressive of the AnmetA mutation but not the AnmetF, most likely due to a transcriptional upregulation (33). In F. graminearum l-homocysteine was not able to rescue either of the mutants (data not shown), suggesting that regulation differs between A. nidulans and F. graminearum.
It appears that the MET13 type provides the majority of the MTHFR activity in fungi but that MET12p also is involved in l-methionine biosynthesis. None of the previous studies determined MET12p and MET13p concentrations, making it impossible to conclude whether the difference in activity is due to kinetics or the in vivo concentration. The results for A. nidulans and S. cerevisiae show that a single functional MTHFR (AnmetF [approximately equivalent to MET13] and ScMET13) in some organisms is sufficient for survival.
A possible explanation for the coepistatic nature of the two genes could be that the two enzyme types interact to form a common heterodimer complex. This is supported by reports for S. cerevisiae, where two independent large-scale protein-protein interaction projects have showed that ScMET12p and ScMET13p copurify (10, 21), suggesting the existence of a heterodimer complex. This is in agreement with reports for other eukaryotic MTHFRs, such as the porcine MTHFR that has been proven to function as a dimer (8). The findings that the MET13p type, when expressed at sufficient levels by the addition of l-homocysteine in A. nidulans, is adequate for survival suggest that the ancestral MET13p MTHFR type is able to form functional homodimers for the reduction of 5,10-methylenetetrahydrofolate. This does not appear to be the case for the MET12 type, which may be due to the presence of the extra motif.
Lack of SAM may prevent the synthesis of aurofusarin.
The initial visual inspections of the FgΔMET12 strain, grown on minimal medium supplemented with low concentrations of l-methionine, suggest that the strain was unable to synthesize aurofusarin. However, the HPLC-DAD analysis revealed that aurofusarin was produced but at low concentrations (Fig. 6A to C). The production of aurofusarin was positively correlated with an increasing methionine concentration, suggesting that the reduced production rate was caused by a lack of substrate for the biosynthetic pathway. The intermediate nor-rubrofusarin is not observed in the wild type, showing that its conversion to rubrofusarin, catalyzed by AurJp with SAM as a methyl donor, is not the rate-limiting step in the biosynthetic pathway under normal conditions. The FgΔMET12 strain accumulates nor-rubrofusarin to high concentrations. This can be explained by a lower availability of SAM due to the effect the mutation has on the recharging the methionine biosynthetic pathway, making conversion of nor-rubrofusarin to rubrofusarin the rate-limiting step in the biosynthetic pathway. Supplementation with higher concentrations of l-methionine restores wild-type SAM conditions in the mutant, resulting in a chemotype similar to that of the wild type.
MET13p is required for the extracellular reduction potential and pigment biosynthesis.
The FgΔMET13 strain remained aurofusarin deficient throughout the tested methionine range (Fig. 6D) and accumulated high concentrations of nor-rubrofusarin and rubrofusarin. At low methionine concentrations the observed phenotype is similar to that of the FgΔMET12 strain, accounting for the accumulation of nor-rubrofusarin. At high methionine concentrations, equivalent to more SAM in the cell, rubrofusarin would be expected to be the dominant metabolite; however, this is not the case. The accumulation of multiple upstream intermediates in mutants has been reported for other biosynthetic systems but not for the aurofusarin pathway, where all mutants to date have accumulated a single intermediate (9, 20). The complete lack of aurofusarin in the FgΔMET13 strain demonstrates that the function of MET13p differs from that of MET12p.
That the pathway stalls at rubrofusarin suggests that the FgΔMET13 is unable to catalyze the conversion of rubrofusarin to aurofusarin, a multistep process that has been proposed to be catalyzed by the protein complex AurFp-GIP1p-AurOp located at the extracellular part of the plasma membrane (9). Laccases (GIP1p) act independently of the cell by utilizing molecular oxygen as a terminal electron acceptor, while the action of P450s, such as AurFp, depends on cellular reduction power, supplied by P450 reductases in eukaryotic systems (41). The mechanism that powers the AurFp enzyme at the plasma remains elusive but could be provided by the generic trans-plasma membrane redox system (11). The observed absence of extracellular reduction power in the FgΔMET13 strain presents a likely explanation for the lack of aurofusarin biosynthesis. Attempts to replicate the FgΔMET13 phenotype in the wild type, by offering an easily reducible substrate in the form of ferricyanide as used by Stahl et al. (36), proved unsuccessful as toxic levels were reached prior to any observable effect on aurofusarin biosynthesis (data not shown).
The involvement of MET13p activity in extracellular reduction reactions and thereby pigment biosynthesis was confirmed by the analysis of the A. nidulans mutants, suggesting that this is a general feature in fungal systems. In addition, the data suggest that mycelium pigment biosynthesis in A. nidulans and F. graminearum is dependent on similar mechanisms. This is the first demonstration that the MET13 type may have additional functions in filamentous fungi not covered by the MET12p class of MTHFRs.
Recent studies on compartmentalization of aflatoxin biosynthesis in Aspergillus parasiticus (7, 24) has highlighted the need for further in-depth studies of the subcellular localization of different PKS pathways. This is also true for pathways responsible for the formation of fungal pigments including aurofusarin, where experimental evidence on subcellular localization would aid in the development of a biosynthesis model.
Conclusion.
Analyses of 32 complete fungal genome sequences confirmed that fungi are unique in having two MTHFR types, MET12 and MET13. The two fungal MTHFR types can be distinguished by the presence of an additional highly conserved fungus-specific motif in the MET12 type. The observed lack of aurofusarin production in F. graminearum MTHFR mutants cultivated at low methionine concentrations is best explained by a lack of SAM required for the conversion of nor-rubrofusarin into rubrofusarin catalyzed by the methyltransferase AurJp. The inability of FgΔMET13 strains to form aurofusarin even at high methionine concentrations appears to be due to the loss of trans-plasma membrane reduction power, required to drive the extracellular P450 monooxygenase AurFp. Future work should be directed at understanding the link between MET13 activity and the extracellular reduction potential.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks are due to laboratory technician Kirsten Henriksen and the 2006 spring genetics class for their involvement in making the yeast complementation analysis. Ulrich Güldener is thanked for his continued work with the FGDB. Softberry is acknowledged for granting us extended access to their gene prediction programs. We are grateful to Marzena Sienko from the Institute of Biochemistry and Biophysics, Department of Genetics, Warsaw, Poland, for supplying us with the MTHFR A. nidulans mutant strains.
This research was supported by the Danish Ministry for Food, Agriculture and Fishery and The Danish Research Council, Technology and Production, grant 274-06-0371.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 11 June 2010.
REFERENCES
- 1.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2.Barratt R. W., Johnson G. B., Ogata W. N. 1965. Wild-type and mutant stocks of Aspergillus nidulans. Genetics 52:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthelmes J., Ebeling C., Chang A., Schomburg I., Schomburg D. 2007. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 35:D511–D514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell A. A., Wheeler M. H., Liu J. G., Stipanovic R. D. 2003. United States Department of Agriculture-Agricultural Research Service studies on polyketide toxins of Fusarium oxysporum f sp vasinfectum: potential targets for disease control. Pest Manag. Sci. 59:736–747 [DOI] [PubMed] [Google Scholar]
- 5.Blom H. J., Shaw G. M., den Heijer M., Finnell R. H. 2006. Neural tube defects and folate: case far from closed. Nat. Rev. Neurosci. 7:724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown D. W., McCormick S. P., Alexander N. J., Proctor R. H., Desjardins A. E. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 36:224–233 [DOI] [PubMed] [Google Scholar]
- 7.Chanda A., Roze L. V., Kang S., Artymovich K. A., Hicks G. R., Raikhel N. V., Calvo A. M., Linz J. E. 2009. A key role for vesicles in fungal secondary metabolism. Proc. Nat. Acad. Sci. U. S. A. 106:19533–19538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubner S. C., Matthews R. G. 1982. Purification and properties of methylenetetrahydrofolate reductases from pig livers, p. 165–172 InMassey V., Williams C. H. (ed.), Flavins and flavoproteins. Elsevier, New York, NY [Google Scholar]
- 9.Frandsen R. J. N., Nielsen N. J., Maolanon N., Sorensen J. C., Olsson S., Nielsen J., Giese H. 2006. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol. Microbiol. 61:1069–1080 [DOI] [PubMed] [Google Scholar]
- 10.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dumpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636 [DOI] [PubMed] [Google Scholar]
- 11.Gómez-Toribio V., Garciá-Matín A. B., Martínez M. J., Martiínez A. T., Guillén F. 2009. Induction of extracellular hydroxyl radical production by white-rot fungi through quinone redox cycling. Appl. Environ. Microbiol. 75:3944–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy W. N., Bailey T. L., Elkan C. P., Baker M. E. 1997. Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 13:397–406 [DOI] [PubMed] [Google Scholar]
- 13.Guenther B. D., Sheppard C. A., Tran P., Rozen R., Matthews R. G., Ludwig M. L. 1999. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat. Struct. Mol. Biol. 6:359–365 [DOI] [PubMed] [Google Scholar]
- 14.Guest J. R., Friedman S., Foster M. A., Tejerina G., Woods D. D. 1964. Transfer of methyl group from N5-methyltetrahydrofolates to homocysteine in Escherichia Coli. Biochem. J. 92:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guldener U., Mannhaupt G., Munsterkotter M., Haase D., Oesterheld M., Stumpflen V., Mewes H. W., Adam G. 2006. FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 34:D456–D458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 17.Hong Y. S., Lee M. J., Kim K. H., Lee S. H., Lee Y. H., Kim B. G., Jeong B., Yoon H. R., Nishio H., Kim J. Y. 2004. The C677 mutation in methylene tetrahydrofolate reductase gene: correlation with uric acid and cardiovascular risk factors in elderly Korean men. J. Korean Med. Sci. 19:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Invitrogen 2004. pYES2 manual. Invitrogen, Carlsbad, CA [Google Scholar]
- 19.Kasap M., Sazci A., Ergul E., Akpinar G. 2007. Molecular phylogenetic analysis of methylenetetrahydrofolate reductase family of proteins. Mol. Phylogenet. Evol. 42:838–846 [DOI] [PubMed] [Google Scholar]
- 20.Kim J. E., Kim J. C., Jin J. M., Yun S. H., Lee Y. W. 2008. Functional characterization of genes located at the aurofusarin biosynthesis gene cluster in Gibberella zeae. Plant Pathol. J. 24:8–16 [Google Scholar]
- 21.Krogan N. J., Cagney G., Yu H. Y., Zhong G. Q., Guo X. H., Ignatchenko A., Li J., Pu S. Y., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St. Onge P., Ghanny S., Lam M. H. Y., Butland G., M. taf-Ui A., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643 [DOI] [PubMed] [Google Scholar]
- 22.Kutzbach C., Stokstad E. L. 1971. Mammalian methylenetetrahydrofolate reductase:partial purification, properties, and inhibition by S-adenosylmethionine. Biochim. Biophys. Acta 250:459–477 [DOI] [PubMed] [Google Scholar]
- 23.Liang X., Teng A., Chen S., Xia D., Felgner P. L. August2005. Rapid and enzymeless cloning of nucleic acid fragments. U.S. patent 6936470. [Google Scholar]
- 24.Maggio-Hall L. A., Wilson R. A., Keller N. P. 2005. Fundamental contribution of beta-oxidation to polyketide mycotoxin production in planta. Mol. Plant Microbe Interact. 18:783–793 [DOI] [PubMed] [Google Scholar]
- 25.Malz S., Grell M. N., Thrane C., Maier F. J., Rosager P., Felk A., Albertsen K. S., Salomon S., Bohn L., Schafer W., Giese H. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42:420–433 [DOI] [PubMed] [Google Scholar]
- 26.Matthews R. G., Vanoni M. A., Hainfeld J. F., Wall J. 1984. Methylenetetrahydrofolate reductase: evidence for spatially distinct subunits domains obtained by scanning transmission electron microscopy and limited proteolysis. J. Biol. Chem. 259:11647–11650 [PubMed] [Google Scholar]
- 27.Mayer K. M. 2003. Colorimetric dehydrogenase screen based on NAD(P)H generation. Methods Mol. Biol. 230:183–191 [DOI] [PubMed] [Google Scholar]
- 28.Naula N., Walther C., Baumann D., Schweingruber M. E. 2002. Two non-complementing genes encoding enzymatically active methylenetetrahydrofolate reductases control methionine requirement in fission yeast Schizosaccharomyces pombe. Yeast 19:841–848 [DOI] [PubMed] [Google Scholar]
- 29.Nielsen K. F., Smedsgaard J. 2003. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography-UV-mass spectrometry methodology. J. Chromatogr. A 1002:111–136 [DOI] [PubMed] [Google Scholar]
- 30.Page R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 31.Raymond R. K., Kastanos E. K., Appling D. R. 1999. Saccharomyces cerevisiae expresses two genes encoding isozymes of methylenetetrahydrofolate reductase. Arch. Biochem. Biophys. 372:300–308 [DOI] [PubMed] [Google Scholar]
- 32.Sheppard C. A., Trimmer E. E., Matthews R. G. 1999. Purification and properties of NADH-dependent 5,10-methylenetetrahydrofolate reductase (MetF) from Escherichia coli. J. Bacteriol. 181:718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sienko M., Natorff R., Zielinski Z., Hejduk A., Paszewski A. 2007. Two Aspergillus nidulans genes encoding methylenetetrahydrofolate reductases are up-regulated by homocysteine. Fungal Genet. Biol. 44:691–700 [DOI] [PubMed] [Google Scholar]
- 34.Smedsgaard J. 1997. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A 760:264–270 [DOI] [PubMed] [Google Scholar]
- 35.Stahl J. D., Aust S. D. 1995. Properties of a transplasma membrane redox system of Phanerochaete chrysosporium. Arch. Biochem. Biophys. 320:367–374 [DOI] [PubMed] [Google Scholar]
- 36.Stahl J. D., Rasmussen S. J., Aust S. D. 1995. Reduction of quinones and radicals by a plasma membrane redox system of Phanerochaete chrysosporium. Arch. Biochem. Biophys. 322:221–227 [DOI] [PubMed] [Google Scholar]
- 37.Sumner J., Jencks D. A., Khani S., Matthews R. G. 1986. Photoaffinity-labeling of methylenetetrahydrofolate reductase with 8-azido-S-adenosylmethionine. J. Biol. Chem. 261:7697–7700 [PubMed] [Google Scholar]
- 38.Sunder-Plassmann G., Winkelmayer W. C., Fodinger M. 2006. Genetic aspects of hyperhomocysteinemia in chronic kidney disease. Semin. Nephrol. 26:8–13 [DOI] [PubMed] [Google Scholar]
- 39.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 40.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Brink H. M., Van Gorcom R. F., A. van den Hondel C., Punt P. J. 1998. Cytochrome P450 enzyme systems in fungi. Fungal Genet. Biol. 23:1–17 [DOI] [PubMed] [Google Scholar]
- 42.Wulff E., Sørensen J. L., Lübeck M., Nielsen K. F., Thrane U., Torp J. 2010. Fusarium spp. associated with rice Bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 12:649–657 [DOI] [PubMed] [Google Scholar]
- 43.Yoder W. T., Christianson L. M. 1998. Species-specific primers resolve members of Fusarium section Fusarium: taxonomic status of the edible “Quorn” fungus reevaluated. Fungal Genet. Biol. 23:68–80 [DOI] [PubMed] [Google Scholar]
- 44.Zhang A., Lu P., Dahl-Roshak A. M., Paress P. S., Kennedy S., Tkacz J. S., An Z. 2003. Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyeasis. Mol. Genet. Genomics 268:645–655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.