Abstract
The accumulation of bioenergy carriers was assessed in two starchless mutants of Chlamydomonas reinhardtii (the sta6 [ADP-glucose pyrophosphorylase] and sta7-10 [isoamylase] mutants), a control strain (CC124), and two complemented strains of the sta7-10 mutant. The results indicate that the genetic blockage of starch synthesis in the sta6 and sta7-10 mutants increases the accumulation of lipids on a cellular basis during nitrogen deprivation relative to that in the CC124 control as determined by conversion to fatty acid methyl esters. However, this increased level of lipid accumulation is energetically insufficient to completely offset the loss of cellular starch that is synthesized by CC124 during nitrogen deprivation. We therefore investigated acetate utilization and O2 evolution to obtain further insights into the physiological adjustments utilized by the two starchless mutants in the absence of starch synthesis. The results demonstrate that both starchless mutants metabolize less acetate and have more severely attenuated levels of photosynthetic O2 evolution than CC124, indicating that a decrease in overall anabolic processes is a significant physiological response in the starchless mutants during nitrogen deprivation. Interestingly, two independent sta7-10:STA7 complemented strains exhibited significantly greater quantities of cellular starch and lipid than CC124 during acclimation to nitrogen deprivation. Moreover, the complemented strains synthesized significant quantities of starch even when cultured in nutrient-replete medium.
Microalgae are able to efficiently convert sunlight, water, and CO2 into a variety of products suitable for renewable energy applications, including H2, carbohydrates, and lipids (11, 12, 16, 21, 38, 41, 44). The unicellular green alga Chlamydomonas reinhardtii has emerged as a model organism for studying algal physiology, photosynthesis, metabolism, nutrient stress, and the synthesis of bioenergy carriers (12, 15, 19, 24, 32). During acclimation to nitrogen deprivation, C. reinhardtii cells accumulate significant quantities of starch and form lipid bodies (4, 5, 8, 26, 28, 30, 34, 43, 46, 48). Despite the significance of these products in algal physiology and in biofuels applications, the metabolic, enzymatic, and regulatory mechanisms controlling the partitioning of metabolites into these distinct carbon stores in algae are poorly understood. Several C. reinhardtii starch mutants with various phenotypic changes in starch content and structure have been isolated (2–4). Two of these, the sta6 and sta7 mutants, contain single-gene disruptions that result in “starchless” phenotypes with severely attenuated levels of starch granule accumulation (2, 4, 34, 39, 40, 48).
The disrupted loci in the two isolated starchless mutants are distinct and each mutant has a unique phenotype (7, 40). In the sta6 mutant, the small, catalytic subunit of ADP-glucose pyrophosphorylase (AGPase-SS) is disrupted (2, 4, 48), and this mutant accumulates less than 1% of the starch observed in wild-type (WT) cells under conditions of nitrogen deprivation. The sta7 mutant contains a disrupted isoamylase gene (7, 8, 10, 39, 40) and also has severely attenuated levels of starch, but it accumulates a soluble glycogen-like product (4, 9). In this study, we conducted an examination of the unique physiological acclimations that are utilized by these mutants to adapt to the loss of starch synthesis. As the genetic lesions in these two mutants are distinct and block starch synthesis via two very different mechanisms, we investigated the physiological consequences of starch inhibition in both of these mutants from a holistic bioenergy perspective, which included photosynthetic parameters and the overall yields of lipids and carbohydrates, the two primary bioenergy carriers in C. reinhardtii. Specifically, we examined whether the inability to synthesize starch would result in the accumulation of additional lipid, alter cellular growth or cell size, affect acetate utilization, and/or influence photosynthetic O2 evolution. Our data indicate that both the sta6 (BAFJ5) and sta7 (sta7-10) mutants accumulate more lipid than the CC124 control during nitrogen deprivation. However, the additional lipid does not completely offset the loss of starch synthesis from a complete energetic perspective. Increased lipid accumulation during nitrogen stress has also been reported for a variety of starch mutants in recent papers (26, 27, 46). A significant feature in both of the starchless mutants studied here is that O2 evolution and acetate utilization are diminished during nitrogen stress, which is undesirable from an overall bioenergy perspective. Remarkably, complementation of sta7-10 with genomic DNA encoding the wild-type isoamylase gene resulted in cells that were larger than those of the sta6, sta7-10, and CC124 strains, exhibited the highest total lipid levels during nitrogen deprivation, and overaccumulated starch even in nutrient-replete medium.
MATERIALS AND METHODS
Strains and culturing conditions.
CC124 was obtained from the Chlamydomonas Center, the sta6 mutant (BAFJ5) was kindly provided by Steven Ball (48), and the sta7 (sta7-10) mutant in the CC425 background was isolated as described previously (40). The sta7-10 complemented strains were obtained after transformation of the sta7-10 mutant with a construct carrying the WT STA7 gene (BamHI/KpnI fragment), which was cloned along with a Bler resistance cassette (29) into pUC19. The sta7-10[c19] clone was isolated previously (40), and the sta7-10[c5] clone was isolated as a part of this study from a separate transformation.
Cultures were grown to late log phase in nitrogen-replete Tris-acetate-phosphate (TAP) liquid medium and resuspended at 2.0 × 106 to 2.5 × 106 cells/ml in parallel in nitrogen-replete TAP medium or nitrogen-depleted TAP (TAP-N) medium, in which NH4Cl was omitted (14, 17, 18). Cells were grown under 50 μmol m−2 s−1 photosynthetically active radiation (PAR) constant illumination on an orbital shaker. Samples for analysis were taken immediately after resuspension (0 h) and at the indicated times. Cell counts and cell sizes were assessed using a Z2 Coulter Counter cell and particle counter (Beckman-Coulter, Brea, CA). Cells were assumed to be spherical for diameter calculations, and background and cellular debris were excluded in all cellular count, volume, and diameter assessments. Coulter cell counts were verified for representative samples using microscopy.
Chlorophyll measurements.
Chlorophyll was determined using ethanol extraction. One milliliter of culture was centrifuged at 6,000 × g for 5 min at room temperature (RT), the supernatant was saved for acetate quantification (see below), and the cell pellets resuspended in 95% ethanol and vortexed to extract pigments. Cellular debris was pelleted by centrifugation (14,000 × g) for 3 min, and absorption was read at 665 nm and 649 nm using a Jenway 6505 UV-visible spectrophotometer (Barloworld Scientific Ltd., Essex, United Kingdom). Calculations of total chlorophyll (μg/ml) were performed as described previously (18).
Microscopy.
The effects of nitrogen deprivation on nonpolar lipid accumulation were visually assayed using laser scanning confocal microscopy. After 96 h in TAP-N medium, all strains were stained with the nonpolar lipid fluorophore Bodipy 493/503 (13) (Molecular Probes, Invitrogen Corporation). To prepare the cells for imaging, 3 ml of each culture was centrifuged at 4,000 × g at RT for 5 min. The supernatant was removed, and 100 μl of the supernatant was used to resuspend the cell pellet. The concentrated cells were stained with 10 μg/ml Bodipy 493/503 for 5 min. To immobilize cells, 1% low-melting-temperature (LMT) agarose was heated to 65°C for use as mounting medium, and 5 μl of stained cell suspension was rapidly mixed with 5 μl of molten 1% LMT agarose. Five microliters of this mixture was immediately transferred to a coverslip, which was then inverted on a microscope slide and allowed to solidify. Coverslips were sealed with a clear epoxy (nail polish) to prevent evaporation of the mounting medium during the imaging process.
Images were acquired using a Nikon Eclipse E800 microscope equipped with a Nikon D-Eclipse C1 laser scanning confocal imaging system using a Melles Griot Kyma 488 series 85-BCD-010 solid-state laser for fluorescence excitation and light transmission as well as a SPOT RT KE color mosaic charge-coupled device (CCD) camera for bright-field imaging. The laser output power was 10 mW, with an emission wavelength of 488 ± 0.5 nM. Laser emission was controlled at 25% of maximum with an ND4 optical filter. Chlorophyll autofluorescence was detected using a 685/70 band-pass optical filter, and Bodipy 493/503 fluorescence was detected using a 515/30 band-pass optical filter. The small-pinhole confocal configuration was used to filter out-of-focus fluorescence emission light.
Starch assays.
Cellular glucose levels contained in starch were determined using amyloglucosidase digestion and the Sigma glucose (HK) assay kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Cells were concentrated by centrifugation of 10 ml of culture at 3,600 × g for 10 min. The supernatant was discarded, and cells were frozen at −80°C. Samples were then resuspended in 100 mM sodium acetate (pH 4.5), autoclaved to solubilize starch, and then digested with amyloglucosidase overnight at 60°C to liberate glucose. To visually assess starch content in colonies on agar plates, iodine vapor staining was performed by placing solid I2 pellets on the surfaces of agar plates to initiate sublimation.
FAME quantification.
Glycerolipids were converted to fatty acids for GC-FID analysis as described previously (36, 37). Lipids were extracted and derivatized from liquid culture at the indicated times. Briefly, 1.0 ml methanol saturated with NaOH was added to 0.5 ml culture and heated in tightly sealed vials at 100°C for 2 h, resulting in cell lysis and lipid saponification. Acid-catalyzed methylation was accomplished by adding 2 ml 1:1.2 6 N HCl/MeOH and incubating at 80°C for 2 h, followed by overnight incubation at 60°C. Fatty acid methyl esters (FAMEs) were extracted into 1 ml 1:1 hexane-methyl tertiary butyl ether (MTBE) via gentle inversion. Extracts were washed with distilled water and analyzed directly by gas chromatography-flame ionization detection (GC-FID) using an Agilent 7890A gas chromatograph with a DB5-ms column (Agilent Technologies, Santa Clara, CA). Tridecanoic acid (13:0 fatty acid) was also spiked into representative samples, and recovery of this internal standard converted to FAME was above 90%. Although the majority of FAME analyses for algae involve the use of dried biomass, the sta6 and sta7-10 cells after nitrogen deprivation were extremely fragile and difficult to centrifuge into cellular pellets due to their low density, resulting in significant sample losses. Therefore, we modified existing protocols (35–37) for lipid extraction and FAME analysis from wet biomass directly from culture, which also significantly improved sample throughput. FAME recovery from representative samples from nutrient-replete cultures, which are easily harvested by centrifugation, was analyzed using the protocol described here and compared to that with a more conventional lipid extraction and derivatization technique consisting of lyophilizing cells pellets, solubilizing the lipids in chloroform-methanol (2:1, vol/vol), and subsequently transesterifying the lipids with HCl-methanol (5%, wt/vol) for 1 h at 80°C in the presence of a tridecanoic acid (C13) methyl ester internal standard (25). The resulting FAMEs were extracted with hexane at room temperature for 1 h and analyzed by GC-FID (method adapted from that of Lepage and Roy [25] and optimized with regard to reaction conditions and extraction time and temperature). Although the wet biomass extraction and derivatization protocol resulted in diminished FAME recovery (by approximately 15 to 30% variation when expressed on a volume basis), the difference in FAME recovery was consistent for all strains used and was deemed to provide an acceptable proxy for relative changes in lipid accumulation.
Acetate utilization.
Acetate remaining in culture media was quantified using high-pressure liquid chromatography (HPLC) as described previously (6). Both nitrogen-replete and nitrogen-depleted TAP contained 17.5 mM acetate prior to culturing (0 h). One milliliter of medium supernatant was filtered through a 0.45-μm nylon membrane prior to HPLC analysis and injected into an Agilent 1200 HPLC (Agilent Technologies, Santa Clara, CA) equipped with an Aminex HPX-87H (Bio-Rad, Hercules, CA) column (45°C), using a 0.6-ml/min flow rate and 4 mM H2SO4 as an isocratic mobile phase. UV-visible and refractive index detectors were used to detect and quantify acetate levels by comparison with standards.
Photosynthetic oxygen evolution rates.
Oxygenic photosynthesis was assessed by measuring in vivo O2 production using a custom-built Clark-type apparatus equipped with YSI5331 platinum electrodes (YSI Incorporated, Yellow Springs, OH). Cells were grown to late log phase under nitrogen-replete conditions, whereupon cultures were resuspended at approximately 1 × 107 cells/ml in TAP-N medium. Cell suspensions (0.8 ml) were taken immediately after removal from the orbital shaker and added to a temperature-controlled, water-jacketed glass reaction cell (Allen Scientific Glass, Boulder, CO) based on an earlier design by Gilson Inc. Probe polarization (0.6 V) and digital signal amplification were accomplished using a custom-built digital picoammeter circuit and acquired using a digital data acquisition card (National Instruments, Austin, TX) and custom software (22). Calibration of the electrode signal was done at the initiation of each experiment using 50 mM air-saturated TAP medium and argon-purged (General Air, Denver, CO) reference buffer. Oxygen photoproduction of the stirred cell suspensions was measured during a 3-min illumination with approximately 80 μmol photons m−2 s−1 from an incandescent Fiber-Lite high-intensity illuminator (Dolan-Jenner Industries, Lawrence, MA). Oxygen photoproduction rates were calculated from the change in O2 concentration over the 3-min illumination phase. Normalizations were done based on the amount of chlorophyll (μg) or the cell density of each sample.
Chlorophyll fluorescence measurements.
The intensity of chlorophyll a fluorescence emission was measured during excitation at 618 ± 10 nm with 695-nm (high-pass) and 780-nm (low-pass) cutoff filters using the Photon Systems Instrument (PSI) FluorCam 800MF (Crno, Czech Republic). The light intensity at the sample surface (500 μmol photons m−2 s−1) was sufficient to saturate the variable yield of fluorescence emission, Fv, as evidenced by the lack of change in maximum fluorescence signal upon further increasing actinic light intensity. Fv fluorescence arises from photosystem II (PSII) and reaches a maximum yield (Fm = Fv + Fo) when PSII primary charge separation occurs and closes the reaction center by preventing further photochemical quenching (45). Fo reports on the nonvariable yield of chlorophyll a emission. The CC124, sta6, and sta7-10 strains (five replicates each) were plated on TAP and TAP-N agar and dark adapted initially for 15 min before any measurements and subsequently for a 1-min dark time following the initial flash sequence to reestablish a dark-adapted state (open reaction centers). Fo and Fm were measured directly from each colony and are reported here as Fv/Fm, where Fv = Fm − Fo.
RESULTS
Distinct growth rates and cell sizes in the starchless mutants and complemented strains.
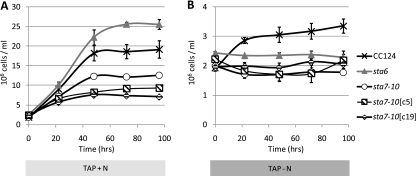
To examine the unique physiological acclimation mechanisms used in the absence of starch synthesis, we precultured the control strain (CC124), the starchless mutants (sta6 and sta7-10), and two sta7-10:STA7 complemented strains (denoted sta7-10[c5] and sta7-10[c19]) to late log phase in TAP medium. Cultures were then centrifuged and resuspended in either TAP or TAP-N medium at standardized cell numbers (∼ 2.0 × 106 to 2.5 × 106 cells/ml). CC124 was used as a control since the parental strains of the sta6 and sta7-10 mutants (330 and CC425, respectively) are auxotrophic for arginine, which could be used as a cellular nitrogen/carbon source. It should be noted that CC124 has an intact cell wall and that the mutants and complements lack a cell wall. The growth curves and cell diameter data in Fig. 1 and Table 1, respectively, show several distinguishing features. In nitrogen-replete medium, distinct final cell densities are achieved, with sta6 > CC124 > sta7-10 > sta7-10[c5] > sta7-10[c19]. Although the highest cell concentrations are attained in the sta6 mutant, this strain has the smallest average cell diameter (5.6 μm). However, it appears that this phenotype may be restricted to cells cultured in TAP medium, as sta6 cells cultured in HS medium do not exhibit smaller diameters (Ursula Goodenough, personal communication). In contrast, the sta7-10[c5] (6.2 μm) and sta7-10[c19] (6.8 μm) complemented strains have the largest average cell diameters but achieve the lowest cell densities. Intermediate cell diameters and final cell counts are observed for CC124 (5.9 μm) and sta7-10 (6.0 μm). The average cell diameters for each strain remained relatively stable for the entire 96-h culturing period in nitrogen-replete medium, with the exception of the sta7-10[c5] and sta7-10[c19] strains, which enter stationary phase prior to the other strains and become noticeably larger at the end of the 96-h culturing period (Table 1). Additionally, the results in TAP medium indicate that decreased rates of cell division correspond well with increased average cell diameters. In nitrogen-depleted medium, an increase in cell numbers was observed only for CC124 (∼1.7-fold increase), and none of the cell wall-less strains (sta6, sta7-10, sta7-10[c5], and sta7-10[c19]) showed any significant change in cell number. This indicates an arrest of cell division in the cell wall-less strains in nitrogen-depleted medium during the assay (Fig. 1B), which is consistent with a previous study (27). In contrast to the case for culturing in nutrient-replete medium, the average cell diameters increased for each strain during nitrogen stress. After 96 h of culturing in TAP-N, the largest cell diameters were observed for the sta7-10[c5] (7.5 μm) and sta7-10[c19] (7.5 μm) strains, followed by the CC124 (7.2 μm), sta7-10 (6.5 μm), and sta6 (6.0 μm) strains. Each strain exhibits an increase in average cell diameter during acclimation to nitrogen deprivation (Table 1), which is suggestive of an increase in cellular carbon product accumulation. It should be noted that all cultures contained a distribution of cell sizes for both culturing conditions, the majority of which were within 1.0 μm of the average cell diameter (Table 1). Additionally, cell diameters are calculated based on the assumption that the displaced fluid volume in the Coulter Counter corresponds to a sphere, which tends to underestimate the size of C. reinhardtii cells observed using transmission microscopy, as C. reinhardtii cells are not perfectly spherical.
Fig. 1.
Cell counts for CC124, the sta6 and sta7-10 mutants, and the sta7-10:STA7 complemented sta7-10[c5] and sta7-10[c19] strains. Growth curves from a representative experiment are shown and were constructed from cell counts at resuspension (0 h) until 96 h, as indicated, in TAP (A) or nitrogen-depleted TAP-N (B) medium. Each data point represents three replicates, except for sta7-10[c5], which represents two biological replicates. Error bars indicate standard deviations.
Table 1.
Average cell sizes for the CC124, sta6, sta7-10, sta7-10[c5], and sta7-10[c19] strains cultured in TAP (N+) or TAP-N (N−) medium
| Medium | Strain | 0 h |
96 h |
||||
|---|---|---|---|---|---|---|---|
| Cell diam (μm)a | Total cellular vol (μl/ml culture) | No. of cellsb | Cell diam (μm) | Total cellular vol (μl/ml culture) | No. of cells | ||
| N+ | CC124 | 5.9 ± 1 | 0.24 ± 0.05 | 6,524 | 5.7 ± 1.1 | 2.3 ± 0.2 | 106,695 |
| sta6 | 5.6 ± 0.7 | 0.15 ± 0.06 | 9,390 | 5.5 ± 0.7 | 2.4 ± 0.6 | 107,748 | |
| sta7-10 | 6 ± 0.9 | 0.18 ± 0.09 | 4,657 | 6.1 ± 0.9 | 1.7 ± 0.1 | 42,933 | |
| sta7-10 [c5] | 6.2 ± 0.9 | 0.21 ± 0.04 | 6,352 | 6.6 ± 0.9 | 1.9 ± 0.4 | 60,708 | |
| sta7-10 [c19] | 6.8 ± 1.1 | 0.27 ± 0.11 | 6,226 | 6.9 ± 1 | 1.7 ± 0.4 | 44,173 | |
| N− | CC124 | 5.2 ± 0.7 | 0.21 ± 0.05 | 9,172 | 7.2 ± 1.2 | 0.8 ± 0.2 | 17,157 |
| sta6 | 4.9 ± 0.7 | 0.11 ± 0.04 | 5,451 | 6 ± 0.8 | 0.4 ± 0.2 | 43,649 | |
| sta7-10 | 5.6 ± 0.9 | 0.15 ± 0.09 | 4,374 | 6.5 ± 0.8 | 0.5 ± 0.2 | 40,533 | |
| sta7-10 [c5] | 6.3 ± 0.9 | 0.18 ± 0.1 | 3,906 | 7.5 ± 1 | 0.4 ± 0.2 | 10,019 | |
| sta7-10 [c19] | 6.4 ± 1.1 | 0.16 ± 0.08 | 3,296 | 7.5 ± 1.1 | 0.3 ± 0.2 | 7,953 | |
Cell diameter data are reported as the mean value ± standard deviation of the normal distribution of cell diameters, which is representative of the distribution of cell sizes in each culture. Cell diameter data represent a minimum of at least four independent biological replicates, in which the standard deviation of replicate average diameters is less than 0.7 μm.
The number of cells was assessed with a Z2 Coulter Counter cell and particle counter at resuspension (0 h) and after 96 h in TAP and TAP–N media.
Excess starch accumulation in the sta7-10 complemented strains.
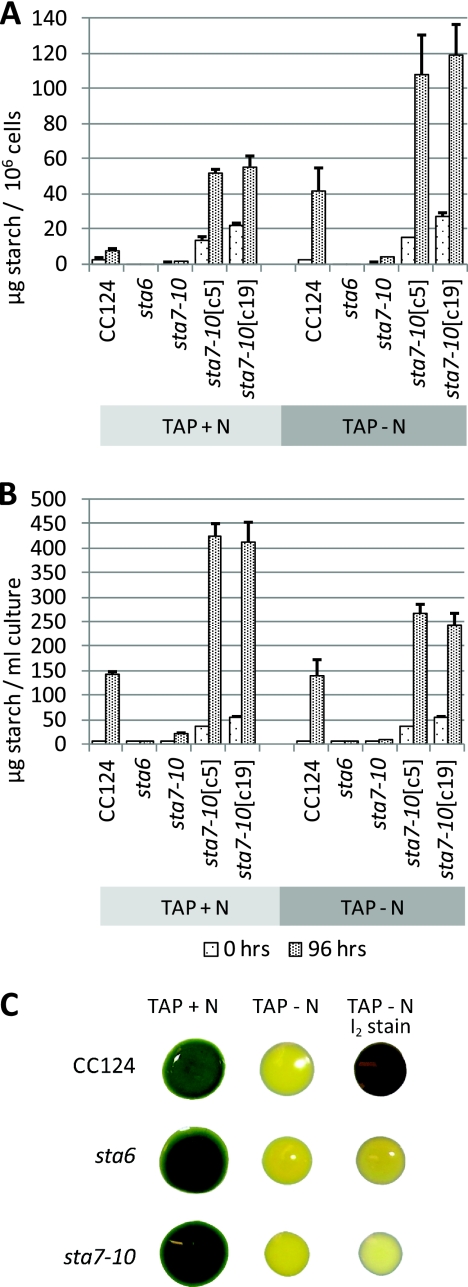
To assess whether the observed differences in growth rates and cell sizes could be correlated with the accumulation of starch and lipid, we first measured the levels of starch-derived glucose after treatment with amyloglucosidase (Fig. 2). In TAP medium, CC124 cells contained 7.8 ± 1.0 μg starch/106 cells, measured as glucose equivalents, whereas after 4 days in nitrogen-depleted medium, an approximately 6-fold cellular increase to 41.9 ± 13.0 μg starch/106 cells was observed, values that are consistent with those recently recorded by Chochois et al. (5). As reported previously (8, 34, 39, 40), both sta6 and sta7-10 mutant cells contained severely attenuated levels of starch and essentially no starch-derived glucose was detected in the sta6 mutant, while sta7-10 cells contained 1.7 ± 0.2 μg starch/106 cells in TAP medium and 4.0 ± 0.3 μg starch/106 cells after 4 days in TAP-N medium. Interestingly, after 96 h in nutrient-replete medium, the sta7-10[c5] and sta7-10[c19] complemented strains had 51.6 ± 2.1 μg starch/106 cells and 55.1 ± 6.3 μg starch/106 cells, respectively, cellular levels that are near those observed in CC124 only after transfer to nitrogen-depleted medium. In fact, as shown in Fig. 2B, the highest yields of starch on a culture volume basis were attained in nitrogen-replete sta7-10[c5] and sta7-10[c9] cultures, with yields exceeding 400 mg/liter after 4 days in cultures inoculated at 2.5 × 106 cells/ml. These data indicate that complementation of the sta7-10 mutant causes significantly altered starch accumulation and that despite transformation with a genomic copy of the STA7 gene containing native 5′ and 3′ untranslated regions (UTRs) and promoters, enzyme activity is occurring outside the native context, resulting in modulated starch accumulation.
Fig. 2.
Starch analyses. Cells were analyzed for glucose derived from starch after amyloglucosidase digestion at resuspension (0 h) and after 96 h in nitrogen-replete TAP or nitrogen-depleted TAP-N medium. (A) Starch-derived glucose per million cells. (B) Starch-derived glucose per ml of culture. Error bars indicate standard deviations. (C) Verification of starch phenotype in the CC124, sta6, and sta7-10 strains. Cells were spotted on TAP and TAP-N agar plates, as indicated. After 7 days the cells were imaged on TAP plates, and the TAP-N plate was stained with iodine vapors and imaged (far right) to indicate the presence of starch (deep purple color).
Increased cellular lipid levels in sta6, sta7-10, and complemented strains.
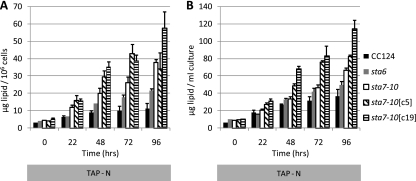
The other major carbon resource observed during nitrogen stress in C. reinhardtii is the formation of lipid bodies (5, 26, 34, 46, 48). Since the blockage of starch synthesis in the starchless mutants creates the potential for diverting metabolic precursors into lipid biosynthetic pathways, we investigated whether lipids were differentially accumulated in the starchless mutants by quantifying lipid-derived fatty acid methyl esters (FAMEs) using GC-FID. Lipids were extracted, derivatized, and quantified from liquid cultures of the CC124, sta6, sta7-10, sta7-10[c5], and sta7-10[c19] strains at the indicated times of acclimation to nitrogen deprivation (Fig. 3). The results indicate that the greatest levels of lipid amenable to conversion into FAMEs using the described transesterification protocol are observed in the sta7-10[c19], sta7-10[c5], and sta7-10 strains, followed by the sta6 and CC124 strains. The starchless mutants contained approximately 2- to 4-fold more lipids per cell than CC124, indicating that additional lipid accumulated in these strains relative to CC124. This is consistent with recent reports regarding increased lipid synthesis in the sta6 mutant relative to control strains (26, 27, 46). However, on a bioenergetic basis, it should be noted that CC124 cell numbers continued to increase in nitrogen-depleted medium. Overall, the increased lipid content in the starchless mutant cultures does not completely offset the loss of starch (140 μg starch/ml of culture at 96 h in TAP-N CC124 cultures) from an energetic standpoint, despite the increased energy density of lipids relative to starch. It should also be noted that our cultures were inoculated at low cell densities and that by standardizing to cell counts, the smaller sta6 cells are slightly underrepresented from an initial biomass perspective in these experiments.
Fig. 3.
GC-FID quantification of fatty acid methyl esters derived from CC124, sta6, sta7-10, sta7-10[c5], and sta7-10[c19] lipids at the indicated times of culturing in nitrogen-depleted TAP-N medium. Values are representative of triplicate biological samples. Lipid was quantified per million cells (A) or by ml of culture (B). Error bars indicate standard deviations.
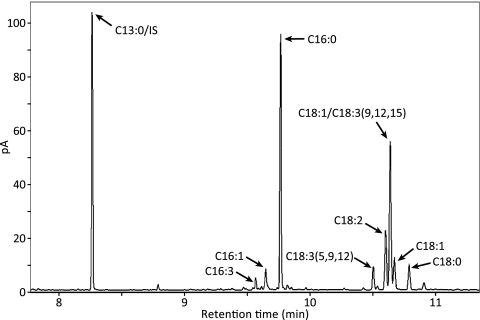
The major fatty acids observed in all strains cultured in either TAP or TAP-N medium were 16:0, 16:1, 18:0, 18:1, 18:2, and 18:3, which is consistent with previously reported results (26, 33, 46). A representative GC-FID chromatogram with a C13:0 internal standard is shown in Fig. 4. We observed only minor differences in the fatty acid profiles among the different strains under the culturing conditions used (Table 2).
Fig. 4.
Representative GC-FID chromatogram. Fatty acid methyl esters derived from the sta7-10 mutant after 96 h of nitrogen deprivation are shown. 18:1/C18:3(9,12,15) indicates a peak containing both 18:1 and 18:3(9,12,15). 13:0/IS indicates a tridecanoate internal standard.
Table 2.
Fatty acid composition during early (22 h) and late (96 h) nitrogen deprivation
| Strain | ha | Total lipid (μg)b | Fatty acid (% of total lipids)c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C16:2 | C16:3 | C18:0 | C18:1–3d | Other | |||
| CC124 | 22 | 18 ± 2.2 | 1.7 ± 1 | 37.2 ± 0.9 | 3.7 ± 0.7 | 1.2 ± 0.1 | 2.5 ± 0.4 | 5.4 ± 0.8 | 43.7 ± 0.7 | 4.7 ± 0.6 |
| 96 | 36.8 ± 8 | 0.6 ± 0.1 | 44.1 ± 1.7 | 2.9 ± 0.2 | 1.3 ± 0.2 | 0.6 ± 0.1 | 3.5 ± 0.1 | 44.7 ± 1.4 | 2.4 ± 0.3 | |
| sta6 | 22 | 15.4 ± 0.6 | 0.6 ± 0.1 | 33.2 ± 0.5 | 6.4 ± 0 | 1.1 ± 0 | 1 ± 0.1 | 4.3 ± 0.2 | 48 ± 0.4 | 5.4 ± 0 |
| 96 | 49.7 ± 3.7 | 0.3 ± 0 | 31.7 ± 1.2 | 5.3 ± 0.2 | 1.4 ± 0.1 | 0.3 ± 0.1 | 3.6 ± 0.2 | 55.6 ± 0.8 | 1.8 ± 0.2 | |
| sta7-10 | 22 | 20.3 ± 1.5 | 1.1 ± 0.3 | 32.3 ± 0.6 | 5.3 ± 0.2 | 0.6 ± 0.1 | 3 ± 0.2 | 4.5 ± 0.7 | 48.7 ± 0.2 | 4.4 ± 0.5 |
| 96 | 66.8 ± 2.2 | 0.4 ± 0.1 | 37 ± 0.7 | 4.6 ± 0.3 | 0.5 ± 0 | 1 ± 0.1 | 3.8 ± 0.1 | 48.5 ± 0.3 | 2.2 ± 0.3 | |
| sta7-10[c5] | 22 | 27.3 ± 1.7 | 1.5 ± 0.2 | 40.1 ± 0.7 | 5.2 ± 0 | 0.7 ± 0.1 | 0.7 ± 0.1 | 4.7 ± 0.3 | 43.8 ± 1.8 | 3.2 ± 1.9 |
| 96 | 83 ± 1 | 0.3 ± 0 | 33.5 ± 0.7 | 5.6 ± 0 | 0.8 ± 0 | 2.6 ± 0.1 | 3.6 ± 0.1 | 51 ± 0.7 | 2.6 ± 1.4 | |
| sta7-10[c19] | 22 | 31.3 ± 2.3 | 0.5 ± 0.1 | 37.6 ± 0.5 | 6.6 ± 0.6 | 0.7 ± 0.1 | 0.7 ± 0 | 3.6 ± 0.1 | 46.9 ± 0.8 | 3.3 ± 0.8 |
| 96 | 118.3 ± 0.7 | 0.3 ± 0 | 31.5 ± 1.1 | 7.3 ± 0.1 | 0.8 ± 0 | 1.8 ± 0.1 | 3.1 ± 0 | 53.6 ± 0.9 | 1.7 ± 0.2 | |
Hours of culture in nitrogen-depleted medium.
Total lipid-derived FAME per ml culture.
Values are averages of triplicate measurements, with standard deviations, obtained using GC-FID for quantification.
C18:1, C18:2, and C18:3 together.
Lipid body formation during nitrogen deprivation visualized by fluorescence imaging.
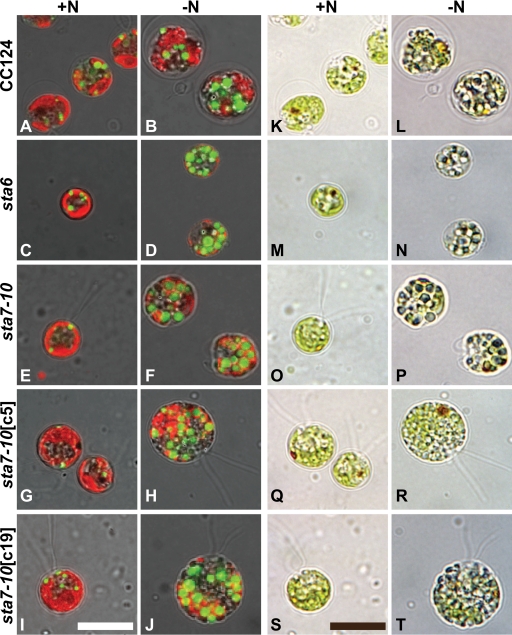
To investigate lipid droplet formation after acclimation to nitrogen deprivation, all strains were visually assayed for nonpolar lipid accumulation using laser scanning confocal microscopy after incubation with the nonpolar lipid fluorophore Bodipy 493/503. As shown in Fig. 5, nonpolar lipid body formation, depicted by green Bodipy 493/503 fluorescence, increases in nitrogen-stressed cells relative to cells in nutrient-replete medium, which is consistent with previous reports on the induction of lipid droplet formation in C. reinhardtii as a consequence of nitrogen limitation (26, 33, 46). Although the greatest density of lipid droplet formation appears in sta6 cells, substantial accumulation of nonpolar lipid bodies is observed in all strains (Fig. 5).
Fig. 5.
Laser scanning confocal fluorescence microscopy images merged with transmitted light images. CC124, sta6, sta7-10, sta7-10[c5], and sta7-10[c5] cells are shown. The two leftmost columns show cells stained with the nonpolar lipid fluorophore Bodipy 493/503. Nonpolar lipid bodies are visualized by green Bodipy fluorescence after 96 h (left, TAP; right, TAP-N). The two rightmost columns show differential interference contrast images after 96 h (left, TAP; right, TAP-N). Oil bodies are visible inside the cells as blue-green-tinted vesicles. Chlorophyll autofluorescence is red. All scale bars represent 10 μm.
Attenuation of photosynthetic oxygen evolution and acetate uptake in the starchless mutants during nitrogen deprivation.
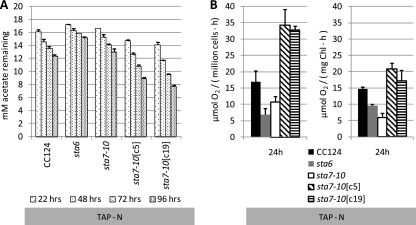
To further probe the underlying mechanisms of differential carbohydrate and lipid accumulation during nitrogen deprivation and to determine whether these parameters are correlated to lipid and starch synthesis, we assessed the utilization of acetate and quantified the levels of photosynthetic O2 evolution of each strain during acclimation to nitrogen deprivation (Fig. 6). Interestingly, significant quantities of acetate remained in the medium for all strains after 96 h of nitrogen deprivation when cultures were inoculated at 2.0 × 106 to 2.5 × 106 cells/ml (Fig. 6A). More complete acetate utilization would be anticipated if cultures were inoculated at higher cell densities. Under nutrient-replete conditions, acetate is completely consumed within 48 h (data not shown) when cells are inoculated at 2.0 × 106 to 2.5 × 106 cells/ml, and acetate is presumably used to synthesize proteins, membrane lipids, and nucleic acids or to support respiration. These results indicate an overall attenuation of anabolic processes in all strains during nitrogen deprivation. However, in accordance with the starch and lipid accumulation data, acetate utilization was the greatest in the sta7-10[c5] and sta7-10[c19] strains, followed by the CC124, sta7-10, and sta6 strains. The increased use of acetate in the complemented sta7-10[c5] and sta7-10[c19] strains relative to the other strains at the cell concentrations used indicates that anabolic processes in the sta7-10[c5] and sta7-10[c19] strains are less severely affected during nitrogen stress.
Fig. 6.
(A) Concentrations of acetate remaining in CC124, the sta6 and sta7-10 mutants, and the sta7-10:STA7 complemented sta7-10[c5] and sta7-10[c19] strains in TAP-N medium at the indicated culturing times. Cultures were inoculated at approximately 2.0 × 106 to 2.5 × 106 cells/ml. The initial acetate concentration in TAP and TAP-N media was 17.5 mM. Values are representative of triplicate biological samples. (B) Oxygen evolution from 0.8-ml aliquots of the indicated strains after 24 h in nitrogen-depleted TAP-N medium. Oxygen evolution is shown on the basis of cell number and chlorophyll (Chl). Cultures were inoculated at approximately 1.0 × 107 cells/ml to produce O2 at levels sufficient for detection. Error bars indicate standard deviations.
Photosynthetic O2 evolution was also monitored, and all strains exhibited attenuated levels of O2 evolution (50 to 70% reduction [data not shown]) after 24 h of nitrogen deprivation (Fig. 6B) relative to those during nutrient-replete culturing. This is consistent with previous observations that demonstrated attenuated levels of O2 evolution in C. reinhardtii and other algae as a consequence of a variety of nutrient (N, P, and S) stresses (20, 30, 31, 47). As shown in Fig. 6B, O2 evolution in the sta6 and sta7-10 strains was more severely attenuated than in the sta7-10[c5], sta7-10[c19], and CC124 strains during the first 24 h of nitrogen deprivation. These O2 evolution data, in combination with the acetate utilization results, indicate diminished anabolic activity in the starchless mutants relative to CC124, activity that is reestablished and possibly even augmented in the complemented strains. Although sta6 cultures showed the lowest levels of acetate utilization and photosynthetic activity, it should be noted again that because the average diameter of these cells is less than those of the other strains and because we standardized cultures according to cell number, less cellular volume and chlorophyll were initially present in the sta6 cultures. However, even when the acetate utilization and photosynthetic activity data are adjusted to the same starting chlorophyll, O2 evolution and acetate uptake are still attenuated in the sta6 strain relative to the CC124, sta7-10[c5], and sta7-10[c19] strains.
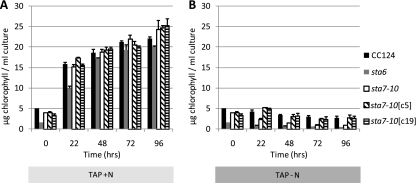
As shown in Fig. 7, chlorophyll levels in the sta6 and sta7-10 strains decrease faster than in the CC124, sta7-10[c5], and sta7-10[c19] strains during culturing in TAP-N medium. The accelerated loss of chlorophyll in the starchless mutants is consistent with the more severely attenuated O2 evolution activities observed in these strains. Under nutrient-replete conditions, similar amounts of chlorophyll are attained in all strains.
Fig. 7.
Chlorophyll content per ml of culture in a representative experiment at the specified time points for the CC124, sta6, sta7-10, sta7-10[c5], and sta7-10[c19] strains in nitrogen-replete (TAP) (A) or nitrogen-depleted (TAP-N) (B) medium. Values are representative of triplicate biological samples. Cells were precultured to late log phase and then resuspended at 2.0 × 106 to 2.5 × 106 cells/ml in either TAP or TAP-N medium. Error bars indicate standard deviations.
Chlorophyll fluorescence is able to distinguish the starchless mutants from CC124.
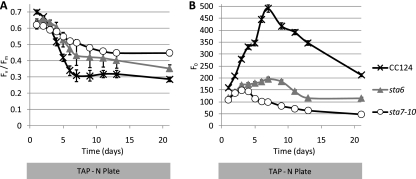
It is clear that photosynthetic and anabolic processes are significantly modulated in the starchless mutants under stress conditions. Nevertheless, the blockage of starch synthesis and the ability to channel metabolic precursors into other biosynthetic pathways, including lipid synthesis, will make these mutants very useful platforms for further metabolic engineering. The ability to assess photosynthetic performance in a high-throughput fashion should allow the screening of secondary mutant populations in the starchless backgrounds with the aim of isolating strains that have water oxidation capacity restored as the result of increased CO2 fixation. Of particular interest is generating strains that divert starch precursors to the production and secretion of targeted hydrocarbons (1, 41). To ascertain whether the starchless phenotype could be discriminated from that of wild-type cells using chlorophyll fluorescence techniques, we probed the variable fluorescence ratio Fv/Fm (23) in the sta6, sta7-10, and CC124 strains. Measurements were recorded periodically after plating cells using the PSI FluorCam imager, which is amenable to high-sample-throughput applications. As shown for a representative experiment in Fig. 8, when strains are plated on TAP-N agar, Fv/Fm ratios are consistently higher for the starchless mutants (sta7-10 slightly higher than sta6), whereas Fv/Fm ratios are similar on nutrient-replete TAP plates (data not shown). This increase indicates differential acclimation of the photosynthetic apparatus in the starchless mutants relative to the wild type after several days of nitrogen stress.
Fig. 8.
(A) Variable chlorophyll a fluorescence (Fv/Fm) of the CC124, sta6, and sta7-10 strains plated on TAP-N agar (five replicate colonies for each strain) as measured by the PSI FluorCam imager indicated. (B) Nonvariable chlorophyll a emission (Fo) of the CC124, sta6, and sta7-10 strains plated on TAP-N agar (five replicate colonies for each strain) as measured by the PSI FluorCam imager. Error bars indicate standard deviations.
DISCUSSION
Significant remodeling of metabolic processes occurs both in the starchless mutants and, surprisingly, in the sta7-10:STA7 complemented strains. From a holistic bioenergy perspective, it is critical that high levels of photosynthetic activity are maintained during illumination and that culturing conditions are optimized for water oxidation and CO2 reduction. Metabolic pathway engineering strategies must focus on improving the overall yields of all algal bioenergy carriers, and efforts that optimize the accumulation of a single bioenergy feedstock should not sacrifice overall fitness and energy carrier yields.
The underlying hypothesis of this study was that by genetically blocking the synthesis of starch, the dominant carbon storage product in nutrient-deprived C. reinhardtii cells, major compensatory mechanisms would result, providing new insights into the acclimation mechanisms used in starchless mutants relative to control strains during nitrogen deprivation. These insights could then be leveraged to further engineer this alga for improved bioenergy phenotypes using rational or random approaches. Moreover, because sta6 and sta7-10 represent the most severe carbohydrate mutations but block starch synthesis at different levels, it was anticipated that unique acclimation mechanisms that could be uniquely leveraged in bioenergy applications would emerge in these strains.
To investigate the acclimation mechanisms used in the starchless mutants, we focused primarily on two potential metabolic outcomes: (i) the partitioning of carbon precursors normally used for starch synthesis to lipid biosynthetic pathways and (ii) the attenuation of cellular anabolic processes (e.g., photosynthesis and carbon storage product synthesis/accumulation). A third possible outcome, the secretion of soluble sugars, was briefly assessed, but under our experimental conditions we were unable to detect evidence for the secretion of significant quantities of soluble sugars (data not shown). However, both of the first two potential acclimation mechanisms were observed during nitrogen deprivation, when the greatest levels of starch and nonpolar lipid accumulation occur. First, increased lipid accumulation on a cellular basis was seen in both the sta6 and sta7-10 mutants relative to CC124 during nitrogen deprivation. Lipid-derived FAMEs under our experimental conditions were approximately 2- and 4-fold greater in the sta6 and sta7-10 mutants, respectively, than in CC124 on a per-cell basis. This increase was more modest on a culture volume basis (∼ 1.4- to 1.8-fold, respectively), as CC124 cells continued to undergo limited cell division during the first 24 h of acclimation to nitrogen stress. It is currently unclear why CC124 cell numbers continued to increase relative to those of the cell wall-less strains used in this study, but is likely a consequence of CC124 having a fully assembled cell wall and not due to an aspect of carbohydrate metabolism, as the sta7-10[c5] and sta7-10[c19] complemented strains did not continue to divide in nitrogen-deprived medium. Attempts to cross sta7-10 into a cell wall background have been unsuccessful to date in our hands.
The second significant acclimation mechanism observed during nitrogen stress in the starchless mutants was a decrease in overall anabolic processes, reflected by decreased levels of O2 evolution activity and acetate utilization. Additional research is required to understand the regulatory mechanisms controlling these processes, which must be reversed to fully realize the potential of shifting metabolic flux to alternative pathways in the absence of starch synthesis.
Fv/Fm parameters are higher for the starchless mutants than for CC124 after several days on TAP-N agar plates. In liquid media during nitrogen stress, chlorophyll is degraded faster in the starchless mutants. The decrease in Fv/Fm under nitrogen-deplete conditions in CC124 during the first several days after plating (Fig. 8A) is primarily a consequence of an increase in Fo, the baseline-level fluorescence not involving PSII charge separation (Fig. 8B): Fv/Fm reaches its minimum on day 7, when Fo is at its maximum. Fo first increases, presumably due to the dissociation of PSII antenna complexes from the reaction center, then subsequently decreases as the cells lose pigmentation under nitrogen-depleted conditions. This effect is most pronounced in CC124, while the sta6 and sta7-10 mutants show a more gradual decline in Fo. These results indicate that the starchless mutants have distinct fluorescent signatures that will be useful in future studies both examining perturbations in the photosynthetic electron transport chain that occur as a consequence of the blockage in starch synthesis and identifying mutants in the starchless backgrounds that have improved photosynthetic properties.
During the preparation of this paper, studies by Wang et al. (46) and Li et al. (26, 27) characterized the effects of nitrogen depletion on the accumulation of nonpolar lipid bodies in the sta6 mutant. Our results regarding the sta6 mutant are consistent with the results reported in those studies and further extend the characterization of lipid overaccumulation to the sta7-10 mutant, which under our culturing conditions accumulates even more cellular lipid than the sta6 mutant. Increased levels of lipid accumulation have also been observed in a starchless mutant of the alga Chlorella pyrenoidosa (42). The study by Li et al. used light intensities 4-fold higher than those used here during acclimation to nitrogen deprivation and reports a 10-fold increase in triacylglycerol (TAG) content relative to that in the control strain, CC1690 (26), as a function of dry weight, which is a consequence of both increased TAG synthesis and loss of carbohydrate mass. The study by Wang et al. reported an approximately 2-fold increase in lipid bodies relative to their control (a potential revertant of 330 that is no longer an arginine auxotroph [46]), which is more reflective of the increase of overall lipid in the sta6 mutant that we observed.
Complementation of sta7-10 resulted in an unanticipated phenotype displaying significant differences in cell morphology, starch synthesis, and lipid accumulation relative to those for CC124 and the sta7-10 mutant. The sta7-10[c5] and sta7-10[c19] strains accumulate levels of cellular starch in nutrient-replete medium that are similar to those achieved in nitrogen-deprived CC124 cells. The mechanistic reasons for this phenotype are outside the scope of the present study but will be pursued in future research. It is possible that the isoamylase enzyme, which is typically part of a larger protein complex (8), is no longer regulated in the proper context or that enzyme levels have been perturbed, resulting in increased starch synthesis. The complemented strains also had the greatest quantities of lipid on a cellular basis, which is likely the result of larger cells with more volume for accumulation and/or additional quantities of membrane that can be converted into FAMEs. The increased accumulation of both starch and lipid in the complemented strains is consistent with increased acetate utilization and oxygenic photosynthesis in these strains during nitrogen deprivation. It should be noted that although sta7-10:STA7 complemented strains were analyzed for H2 production in a previous study (40), starch levels were assessed only visually using a qualitative iodine assay to determine whether starch accumulation was restored relative to the that in sta7-10 mutant. Starch levels in the complemented strains were not quantified as a part of the previous study, and the starch-excess phenotype was not readily observed until incorporation of the culturing and analytical methods used in this study.
The increases in starch content in the complemented strains during nutrient-replete culturing correlate with decreased growth rates but larger cell sizes. This phenotype is of particular interest as it represents a mechanism to increase the synthesis of a primary bioenergy carrier (starch) at the expense of other cellular constituents used for cell division (proteins and nucleic acids) without transferring cells to a nutrient-depleted medium.
It is important to reinforce that our cultures were standardized at relatively dilute cells numbers (2.5 × 106 cells/ml). As a consequence of the complete arrest of cell division in the cell wall-less strains cultured in nitrogen-deprived medium, the total amount of bioenergy carriers is smaller than those reported in other studies on a culture volume basis, but those levels could be achieved if we resuspended our cultures at higher cell numbers.
Our initial experiments have been conducted in continuous light using acetate as a heterotrophic boost. Although biased toward the accumulation of bioenergy carriers, these conditions provide insights into the capacity of these strains to synthesize starch and lipids and will serve as a useful reference point for further dissecting the metabolic pathways used for carbon product synthesis and examining metabolic adjustments as a consequence of genetic manipulation. Under the current growth conditions, significant quantities of acetate (320 μg/ml in CC124 [Fig. 6A]) are metabolized, which could account for a substantial quantity of the products observed during nitrogen deprivation.
In summary, our results indicate that from a bioenergy perspective (i) nitrogen deprivation is advantageous as it represents an effective means to organize cellular metabolites into the two principal biofuels feedstocks, starch and TAG, but a significant drawback of nitrogen deprivation is that it results in attenuated rates of photosynthesis and acetate uptake, as overall anabolic processes are diminished; (ii) the disruption of starch synthesis is an effective means to overaccumulate lipid in the sta6 and sta7-10 starchless mutants during nitrogen deprivation, but this is tempered by decreases in overall anabolic processes (primarily starch accumulation) in comparison to the other strains; and (iii) the isoamylase complemented strains accumulate significantly higher levels of lipids and starch on a cellular basis during nitrogen deprivation, and starch overaccumulation can be achieved in the sta7-10[c5] and sta7-10[c19] strains in nutrient-replete medium while these cells are actively dividing, albeit at a lower rate than the other strains in nitrogen-replete medium.
The single-gene alterations reported here result in dramatic carbon product accumulation phenotypes. The metabolic engineering of algae for improved biofuel productivity is just beginning, and it is noteworthy that the relatively simple genetic manipulations reported here resulted in metabolic alterations that significantly affected the accumulation of starch or lipid, the two most relevant algal bioenergy carriers. As engineering approaches in C. reinhardtii and other algae become more refined, additional advances are likely.
ACKNOWLEDGMENTS
We thank Steven Ball for generously providing his sta6 mutant previously.
We acknowledge support from the Air Force Office of Scientific Research (grant FA9550-05-1-0365) (M.C.P.) and the U.S. Department of Energy BES and BER programs.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Atsumi S., Higashide W., Liao J. C. 2009. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27:1177–1180 [DOI] [PubMed] [Google Scholar]
- 2.Ball S. G. 1998. Regulation of starch biosynthesis, p. 549–567 InRochaix J.-D., Goldschmidt-Clermont M., Merchant S. (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 3.Ball S. G. 2002. The intricate pathway of starch biosynthesis and degradation in the monocellular alga Chlamydomonas reinhardtii. Aust. J. Chem. 55:49–59 [Google Scholar]
- 4.Ball S. G., Deschamps P. 2009. Starch metabolism, p. 1–40 InStern D. (ed.), The Chlamydomonas sourcebook, 2nd ed. Organellar and metabolic processes, vol. 2.Academic Press, Oxford, United Kingdom [Google Scholar]
- 5.Chochois V., Dauvillee D., Beyly A., Tolleter D., Cuine S., Timpano H., Ball S., Cournac L., Peltier G. 2009. Hydrogen production in Chlamydomonas: photosystem II-dependent and -independent pathways differ in their requirement for starch metabolism. Plant Physiol. 151:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datar R., Huang J., Maness P.-C., Mohagheghi A., Czernik S., Chornet E. 2007. Hydrogen production from the fermentation of corn stover biomass pretreated with a steam-explosion process. Int. J. Hydrogen Energy 32:932–939 [Google Scholar]
- 7.Dauvillee D., Colleoni C., Mouille G., Buleon A., Gallant D. J., Bouchet B., Morell M. K., d'Hulst C., Myers A. M., Ball S. G. 2001. Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 125:1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dauvillee D., Colleoni C., Mouille G., Morell M. K., d'Hulst C., Wattebled F., Lienard L., Delvalle D., Ral J. P., Myers A. M., Ball S. G. 2001. Biochemical characterization of wild-type and mutant isoamylases of Chlamydomonas reinhardtii supports a function of the multimeric enzyme organization in amylopectin maturation. Plant Physiol. 125:1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauvillee D., Colleoni C., Shaw E., Mouille G., D'Hulst C., Morell M., Samuel M. S., Bouchet B., Gallant D. J., Sinskey A., Ball S. 1999. Novel, starch-like polysaccharides are synthesized by an unbound form of granule-bound starch synthase in glycogen-accumulating mutants of Chlamydomonas reinhardtii. Plant Physiol. 119:321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauvillee D., Mestre V. V., Colleoni C., Slomianny M., Mouille G., Delrue B., d'Hulst C., Bliard C., Nuzillard J., Ball S. 2000. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Sci. 157:145–156 [DOI] [PubMed] [Google Scholar]
- 11.Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M., Posewitz M. C. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19:235–240 [DOI] [PubMed] [Google Scholar]
- 12.Ghirardi M. L., Posewitz M. C., Maness P. C., Dubini A., Yu J., Seibert M. 2007. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58:71–91 [DOI] [PubMed] [Google Scholar]
- 13.Gocze P. M., Freeman D. A. 1994. Factors underlying the variability of lipid droplet fluorescence in Ma-10 Leydig tumor-cells. Cytometry 17:151–158 [DOI] [PubMed] [Google Scholar]
- 14.Gorman D. S., Levine R. P. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Nat. Acad. Sci. U. S. A. 54:1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman A. R., Croft M., Gladyshev V. N., Merchant S. S., Posewitz M. C., Prochnik S., Spalding M. H. 2007. Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10:190–198 [DOI] [PubMed] [Google Scholar]
- 16.Hankamer B., Schenk P., Marx U., Posten C., Kruse O. 2007. The solar bio-fuels consortium: developing advanced bio-fuel production systems. Photosynth. Res. 91:136 [Google Scholar]
- 17.Harris E. 2009. Chlamydomonas in the laboratory, p. 241–262 InHarris E. (ed.), The Chlamydomonas sourcebook, 2nd ed. Organellar and metabolic processes, vol. 1.Academic Press, Oxford, United Kingdom [Google Scholar]
- 18.Harris E. (ed.). 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 19.Hemschemeier A., Melis A., Happe T. 2009. Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth. Res. 102:523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzig R., Falkowski P. G. 1989. Nitrogen limitation in Isochrysis-Galbana (Haptophyceae). 1. Photosynthetic energy-conversion and growth efficiencies. J. Phycol. 25:462–471 [Google Scholar]
- 21.Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54:621–639 [DOI] [PubMed] [Google Scholar]
- 22.Karns D. 2009. M.S. thesis. Colorado School of Mines, Golden, CO [Google Scholar]
- 23.Kolber Z. S., Prasil O., Falkowski P. G. 1998. Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: defining methodology and experimental protocols. Biochim. Biophys. Acta 1367:88–106 [DOI] [PubMed] [Google Scholar]
- 24.Kruse O., Rupprecht J., Mussgnug J. H., Dismukes G. C., Hankamer B. 2005. Photosynthesis: a blueprint for solar energy capture and biohydrogen production technologies. Photochem. Photobiol. Sci. 4:957–970 [DOI] [PubMed] [Google Scholar]
- 25.Lepage G., Roy C. C. 1986. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 27:114–120 [PubMed] [Google Scholar]
- 26.Li Y., Han D., Hu G., Dauvillee D., Sommerfeld M., Ball S., Hu Q. 2010. Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12:387–391 [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Han D., Hu G., Sommerfeld M., Hu Q. 2010. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. [Epub ahead of print.] doi:10.1002/bit.22807 [DOI] [PubMed] [Google Scholar]
- 28.Libessart N., Maddelein M. L., Koornhuyse N., Decq A., Delrue B., Mouille G., D'Hulst C., Ball S. 1995. Storage, photosynthesis, and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas. Plant Cell 7:1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lumbreras V., Stevens D. R., Purton S. 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14:441–447 [Google Scholar]
- 30.Martin N. C., Goodenough U. W. 1975. Gametic differentiation in Chlamydomonas reinhardtii. I. Production of gametes and their fine structure. J. Cell Biol. 67:587–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. 2000. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal- Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernandez E., Fukuzawa H., Gonzalez-Ballester D., Gonzalez-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riano-Pachon D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martinez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moellering E. R., Benning C. 2010. RNAi silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell. 9:97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouille G., Maddelein M. L., Libessart N., Talaga P., Decq A., Delrue B., Ball S. 1996. Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Fallon J. V., Busboom J. R., Nelson M. L., Gaskins C. T. 2007. A direct method for fatty acid methyl ester synthesis: application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 85:1511–1521 [DOI] [PubMed] [Google Scholar]
- 36.Pendergrass S., Jensen P. 1997. Application of the gas chromatography-fatty acid methyl ester system for the identification of environmental and clinical isolates of the family Micrococcaceae. Appl. Occup. Environ. Hyg. 12:543–546 [Google Scholar]
- 37.Pendergrass S. M. 1998. Aerobic bacteria by GC-FAME method 0801. NIOSH manual of analytical methods, 4th ed.NIOSH, Atlanta, GA [Google Scholar]
- 38.Posewitz M. C., Dubini A., Meuser J. E., Seibert M., Ghirardi M. L. 2009. Hydrogenases, hydrogen production and anoxia, p. 217–246 InStern D. (ed.), The Chlamydomonas sourcebook, 2nd ed. Organellar and metabolic processes, vol. 2.Academic Press, Oxford, United Kingdom [Google Scholar]
- 39.Posewitz M. C., King P. W., Smolinski S. L., Smith R. D., Ginley A. R., Ghirardi M. L., Seibert M. 2005. Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33:102–104 [DOI] [PubMed] [Google Scholar]
- 40.Posewitz M. C., Smolinski S. L., Kanakagiri S., Melis A., Seibert M., Ghirardi M. L. 2004. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell 16:2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radakovits R., Jinkerson R. E., Darzins A., Posewitz M. C. 2010. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramazanov A., Ramazanov Z. 2006. Isolation and characterization of a starchless mutant of Chlorella pyrenoidosa STL-PI with a high growth rate, and high protein and polyunsaturated fatty acid content. Phycol. Res. 54:255–259 [Google Scholar]
- 43.Sager R., Granick S. 1954. Nutritional control of sexuality in Chlamydomonas reinhardtii. J. Gen. Physiol. 37:729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schenk P., Thomas-Hall S., Stephens E., Marx U., Mussgnug J., Posten C., Kruse O., Hankamer B. 2008. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenerg. Res. 1:20–43 [Google Scholar]
- 45.Shinkarev V. P. 2004. Oxygen evolution and chlorophyll-a fluorescence induced by multiple flashes, p. 197–229 InPapageorgiou G., Govindjee (ed.), Chlorophyll-a fluorescence: a signature of photosynthesis. Springer, Norwell, MA [Google Scholar]
- 46.Wang Z. T., Ullrich N., Joo S., Waffenschmidt S., Goodenough U. 2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8:1856–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wykoff D. D., Davies J. P., Melis A., Grossman A. R. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zabawinski C., Van Den Koornhuyse N., D'Hulst C., Schlichting R., Giersch C., Delrue B., Lacroix J. M., Preiss J., Ball S. 2001. Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183:1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]